Abstract
Homocysteine thiolactone-induced protein modification (HTPM) is a unique post-translational protein modification that is recognized as an emergent biomarker for cardiovascular disease. HTPM involves the site-specific acylation of proteins at lysine residues by homocysteine thiolactone (HTL) to produce protein homocystamide, which has been found at elevated levels in patients with coronary heart disease. Herein, we report the development of a novel gold nanoparticle (GNP) biochemical sensor for detection of protein homocystamide in an in vitro serum protein-based model system. Human serum albumin (HSA) and human sera were subjected to HTPM in vitro to produce HSA-homocystamide or serum protein homocystamide, respectively, which was subsequently treated with citrate-capped GNPs. This GNP sensor typically provided instantaneous visual confirmation of HTPM in the protein model systems. Transmission electron microscopy images of the GNPs in the presence of HSA-homocystamide suggest that modification-directed nanoparticle assembly is the mechanism by which the biochemical sensor produces a colorimetric signal. The resultant nanoparticle-protein assembly exhibited excellent thermal and dilutional stability, which is expected for a system stabilized by chemisorption and intermolecular disulfide bonding. The sensor typically provided a linear response for modified human sera concentrations greater than ~5 mg/mL. The calculated limit of detection and calibration sensitivity for the method in human sera were 5.2 mg/mL and 13.6 AU · (μg/mL)−1, respectively.
Introduction
An elevated plasma level of the potentially toxic amino acid homocysteine (Hcy) is a known risk factor for cardiovascular1,2 and neurodegenerative3 diseases such as coronary heart disease and Alzheimer’s disease, respectively. Although the function of Hcy in the pathogenesis of these diseases is not yet fully understood, there is considerable evidence that suggests that Hcy thiolactone (HTL) may contribute to cardiovascular disease.4–7 HTL is a highly reactive thioester analogue of Hcy synthesized in vivo via enzymatic aminoacyl-tRNA synthase pathways at a rate proportional to extracellular Hcy concentration.8 Hence, it is likely that HTL production is elevated in individuals with elevated plasma levels of Hcy.9 Increased in vivo production of HTL is believed to initiate various detrimental physiological effects, including vascular tissue damage and formation of early atherosclerotic lesions.4,10 It has been shown that HTL readily and selectively reacts with proteins at the ε-amino groups of lysine residues under physiological conditions to yield highly labile protein homocystamide species.11,12 It is known that HTL-induced protein modification (HTPM) can be detrimental to protein function because it may lead to spontaneous intermolecular disulfide bonding to yield protein oligomers.11,13 HTPM may also result in decreased biological activity, decreased solubility, and increased immunogenicity for affected proteins.11,14 Moreover, a recent clinical study by Yang et al.6 reported the discovery of elevated levels of protein homocystamide in individuals with coronary heart disease. They found a positive correlation between plasma protein homocystamide levels and the number of major diseased coronary vessels in 254 subjects, which further supports the validity of protein homocystamide as a biomarker for cardiovascular disease.
Considering the status of HTPM as an emerging biomarker for cardiovascular disease, the development of novel detection methods for protein homocystamide and related cardiovascular disease biomarkers is of great importance. Our laboratory has developed several novel organic dye-based colorimetric and fluorimetric sensors for the detection of various amino thiol disease biomarkers, including cysteine and homocysteine.15–18 Several of these novel sensors show promise for amino thiol detection in human blood plasma. We also reported the use of redox indicators for electrochemical detection of glutathione, another important amino thiol biomarker.19 Our recent work explores new detection schemes and strategies for investigating the physiochemical properties of protein homocystamide. For example, we reported the development a rapid capillary electrophoresis (CE) method for separating mixtures containing oligomeric protein homocystamide derived from in vitro modified cytochrome c. This CE method was successfully applied to monitoring the inhibition of HTL-induced protein oligomerization via in situ pyridoxal tetrahydrothazine formation.20
Separation techniques such as TLC21 and HPLC22 have also been employed to quantify protein homocystamide, but few specific detection schemes have been reported. Enzyme-linked immunosorbent assays (ELISAs) are perhaps the most commonly used affinity technique for protein homocystamide determination.6,23 ELISAs are a highly sensitive and versatile format that can be configured to generate either a colorimetric or a fluorimetric response, provided that appropriate antibodies are available. Another advantage of ELISA is the wide commercial availability of 96-well plate kits, which provide a user-friendly interface that is amenable to high-throughput applications. However, one potential disadvantage of the use of ELISAs for the detection of HTPM is the time and expense required to produce and purify sufficient quantities of antibodies specific for protein homocystamide.24
Metallic nanoparticles may offer an alternative to conventional immunoaffinity methods for some biochemical sensing applications. One reason is because citrate-capped colloidal gold nanoparticles can be quickly synthesized using a facile one-step redox reaction and at a small fraction of the cost of a comparable quantity of monoclonal antibodies.25 In addition, gold nanoparticles (GNPs) are well suited for use in colorimetric detection applications because they exhibit intense surface plasmon bands in the visible region of the electromagnetic spectrum. For instance, dispersed colloidal gold particles having dimensions on the order of ~10 nm typically have an intense surface plasmon band at approximately 520 nm, resulting in a red-colored solution. Upon aggregation of the GNPs, red-shifted extended plasmon bands typically appear at ~620 nm, causing a characteristic transition in solution color from red to purple-violet.26
The concept of gold probe biochemical sensors originated more than 40 years ago when Bloomfield devised a colloidal gold test for plasma protein.27 Since then, a multitude of gold colloid-based biochemical sensors have been reported. This topic has been comprehensively reviewed in the literature.28–31 GNPs are capable of sensing a wide range of physiologically relevant small molecules including carbohydrates,32 amino thiol biomarkers,33,34 and DNA.35,36 GNP-based sensors have also proven useful for a variety of protein applications such as assessing protein structure37–41 and the detection of various protein-based analytes, including platelet-derived growth factors,42 steptavidin,43 and cholera toxin.44
In general, the utility of GNPs as biochemical colorimetric sensors is based on exploiting the appropriate biochemical mechanisms in order to stimulate nanoparticle assembly. Accordingly, it is often necessary to functionalize the nanoparticles with affinity ligands in order to achieve analyte-specific nanoparticle self-assembly.35,44,45 However, applications involving the detection of thiol-containing species do not necessarily require the use of functionalized nanoparticles since gold is intrinsically thiol reactive. For example, unfunctionalized citrate-capped GNPs have been successfully employed as colorimetric sensors for various amino thiols including Hcy,33 cysteine,34 and glutathione.34 Amino thiols tend to readily adsorb onto the surface of colloidal gold particles via chemisorption-type interactions. Nanoparticle assembly has been shown to occur via noncovalent electrostatic-type interactions between the amino and carboxylic acid groups.33,34
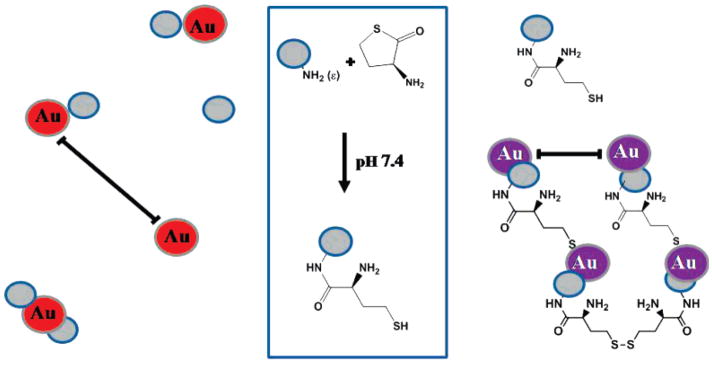
Herein, we report the use of citrate-capped colloidal GNPs as a sensor for HTPM. Scheme 1 illustrates the proposed mechanism for the colorimetric GNP sensor. In the presence of unmodified protein, the nanoparticles interact in a nonspecific manner with individual protein molecules but remain randomly dispersed throughout the protein solution. As a result, the sensor solution retains the characteristic red color of dispersed colloidal GNPs. In contrast, the nanoparticles bind with the HTL-modified proteins via chemisorption-type binding (Au-S-R), which can facilitate nanoparticle assembly via cross-linking. The nanoparticle assembly is further stabilized by intermolecular disulfide bonding (R-S-S-R) between protein homocystamide species. The scale bars are shown to emphasize the relative difference in interparticle distances for dispersed and assembled GNPs, respectively. UV-vis absorption spectroscopy and transmission electron microscopy (TEM) were employed to investigate the assembly behavior of colloidal gold nanoparticles in response to protein homocystamide. To the best of our knowledge, direct colorimetric detection of serum protein homocystamide using GNPs has not been previously reported.
Scheme 1.
Proposed Colorimetric Gold Nanoparticle Sensor for Homocysteine Thiolactone-Induced Protein Modification
Materials and Methods
Reagents
L-Homocysteine thiolactone hydrogen chloride, hydrogen tetrachloroaurate trihydrate, trisodium citrate, EDTA, and all other reagents used for the preparation of buffer and serum protein solutions were obtained from Sigma-Aldrich (St. Louis, MO) at the highest purity available and used as received. Ultrapure water (18.2 MΩ) was obtained from an Elga model PURELAB Ultra water filtration system.
Synthesis and Characterization of Gold Nanoparticles
Citrate-capped GNPs, ca. 13 nm diameter, were synthesized using the well-known Turkevich–Frens method.25,46 Briefly, a solution of hydrogen tetrachloroaurate trihydrate was prepared in water and heated to reflux while stirring, followed immediately by addition of a 40 molar excess of freshly prepared trisodium citrate–water solution, which initiated the reduction of the hydrogen tetrachloroaurate trihydrate. The aurate–citrate solution was allowed to reflux for approximately 25 min or until completion of the redox reaction as indicated by a change in solution color from faint yellow to dark red. The excess citrate in solution creates a negatively charged surface on the GNPs, thereby minimizing nonspecific GNP aggregation postsythesis.25 The resultant GNP solution was allowed to cool to room temperature prior to storage in an amber glass bottle. Aurate (1.0 mM) was reduced in 40 mM citrate, resulting in a GNP stock solution of 1.0 mM, assuming complete reduction of the gold. GNP solutions were purified using a 0.1 μm polyetrafluoroethylene syringe filter devices. Nanoparticle size was confirmed using dynamic light scattering (DLS) and transmission electron microscopy (TEM).
Preparation of Homocysteine Thiolactone-Modified Protein
The protein homocystamide stock solutions used in this study were synthesized using procedures similar to those previously reported by Jakubowski.11 Briefly, essentially fatty acid free human serum albumin purchased from Sigma-Aldrich (St. Louis, MO) was dissolved in pH 7.4, 100 mM, sodium phosphate buffer containing 0.2 mM EDTA and incubated at 25 °C in the presence of 4.6 mM HTL for 4 h at room temperature. The post-translational modification reaction was quenched by filtering the protein reaction solution through a 10 kDa molecular weight cutoff centrifugal membrane device (Millipore, Billerica, MA) to remove any unreacted HTL as well as other low molecular weight byproducts such as Hcy. The protein retentate was rinsed twice with 1 mL buffer aliquots to remove nonspecifically bound HTL or Hcy. The rinsed modified protein was promptly reconstituted with buffer to obtain the protein homocystamide stock solution.
Electrospray ionization mass (ESI-MS) was used to verify the post-translational modification of HSA by HTL. The mass spectrometry data indicates that the in vitro modification reaction protocol used in this study is efficient and typically yields an extensively modified HSA solution. The majority of the resultant HSA species typically contained ~1–4 Hcy per protein molecule postmodification. A substantial signal corresponding to dimeric HSA was also detected due to modification-induced oligomerization. On the basis of these findings, the concentrations of the HSA and serum homocystamide solutions are reported in units of mg/mL or μg/mL, assuming complete modification. Mass spectra (Figure S1) and additional discussion are provided in the Supporting Information.
Unless otherwise noted, protein homocystamide standard solutions were prepared by diluting the stock solution with phosphate buffer. Excess biological materials were heat sterilized prior to disposal in an approved biohazard waste container. All appropriate safety apparels such as protective gloves, clothing, and eyewear were worn while handling the potentially biohazardous human serum protein material.
Colorimetric Detection of HTL-Modified Protein Using a Gold Nanoparticle Sensor
Protein homocystamide solutions were immediately treated with the GNP sensor in order to minimize the loss of colorimetric response due to spontaneous intermolecular disulfide bonding between modified proteins and other miscellaneous auto-oxidation processes. The protein homocystamide solutions used in this study were not treated with a disulfide reductant. Unless otherwise indicated, a 100 μL aliquot of the protein homocystamide or control solution was treated with 200 μL of 1.0 mM GNP stock solution at room temperature. Changes in the visible absorption due to the GNPs and the nanoparticle-modified protein complex were monitored immediately at 520 and 620 nm, respectively. The UV absorption associated with the protein was monitored between 220 and 280 nm. Protein–sensor solutions were typically diluted 2-fold prior to measuring UV–vis absorption.
Instrumentation
Absorption measurements were acquired using a Shimadzu, model UV-3101PC UV-vis-NIR spectrophotometer with either quartz (UV–vis) or disposable polystyrene (visible only) reduced volume cells. DLS measurements were performed using a Malvern Zetasizer Nano ZS (Worcestershire, U.K.). TEM images were acquired with a JEOL model 100CX transmission electron microscope using an acceleration voltage of 80 kV. TEM specimens were prepared by adsorbing a thin layer of the sensor–protein solution onto carbon-coated grids followed by air drying. The sample grids were treated with 2% uranyl acetate staining reagent prior to TEM imaging.
Results and Discussion
Evaluation of GNPs as a Serum Protein Sensor
A preliminary assessment of the proposed GNP sensor was performed using the model protein human serum albumin (HSA). HSA was chosen for these studies because it is a major protein constituent in human serum, accounting for approximately one-half of the total serum protein. In addition, it is known that highly stable albumin–GNP conjugates are readily obtained via adsorption interactions under physiological pH and ionic strength conditions.40,47,48 However, it was necessary to evaluate the behavior of the proposed HSA-GNP system under the conditions used for these studies.
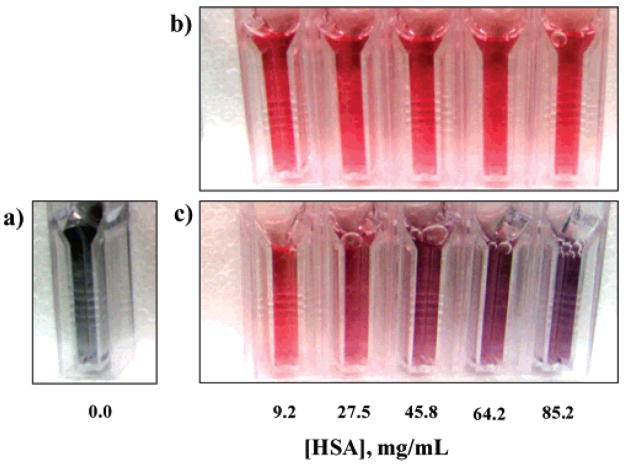
Figure 1 is an image of cuvettes containing equivalent concentrations of the GNP sensor (~0.3 mM) in the presence of either HSA or HSA-homocystamide (0–85.2 mg/mL) dissolved in phosphate buffer (100 mM at pH 7.4). The protein concentration range used for this study encompasses the expected physiological concentration range for HSA in human plasma, ~34–54 mg/mL.49 As expected, the GNPs were only sparingly soluble in the phosphate buffer in the absence of HSA. The resultant metallic gray-colored suspension shown in Figure 1a is slightly opaque due to scattering from salt-induced aggregation and precipitation of the GNPs. This behavior is attributed to the citrate-capped GNPs becoming destabilized by the relatively high ionic strength of the phosphate buffer.50
Figure 1.
Photographs of the gold nanoparticle (GNP) sensor: (a) in the absence of protein, (b) in the presence of unmodified HSA (9.2–85.2 mg/mL), and (c) in the presence of HSA–homocystamide (9.2–85.2 mg/mL). All samples were prepared in 100 mM phosphate, 0.2 mM EDTA buffer. Each cuvette contains 0.3 mM GNP.
Cuvettes containing unmodified HSA (9.2–85.2 mg/mL) are shown in Figure 1b. The nanoparticles remain stable and dispersed in the presence of unmodified HSA, as indicated by the reddish-pink color of the sensor solutions. Notably, there is no discernible change in the color of the sensor solutions in response to increasing concentrations of unmodified HSA, which suggests that non-specific nanoparticle assembly does not occur in the presence of unmodified HSA. Furthermore, the UV–vis absorption spectra shown in Figure 2a are similar to those previously reported for bovine serum albumin–GNP conjugates dissolved in 10 mM phosphate at pH 7.0.40
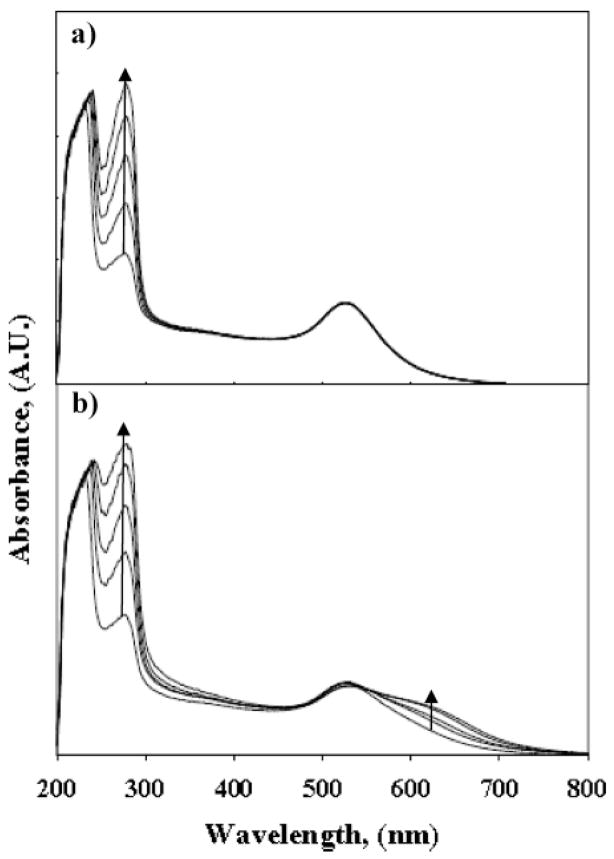
Figure 2.
UV–vis absorption spectra of GNP sensor solutions shown in Figure 1: (a) unmodified HSA and (b) HSA protein homocystamide. Extended plasmon bands are only observed for sensor solutions containing HSA–homocystamide. The arrows indicate the change in absorption with increasing HSA or HSA–homocystamide.
The GNP sensor produced a significant colorimetric signal in response to increasing concentrations of protein homocystamide, which is clearly visible in Figure 1c. The sensor solutions transition from a reddish-pink to a deep purple-violet color, which is easily distinguishable from the highly scattering metallic gray-colored suspension produced in the absence of protein due to salt-induced nanoparticle aggregation.
The corresponding UV–vis absorption spectra for sensor–protein solutions collected approximately 20 min after treatment with the GNPs (Figure 1b, c) are provided in Figure 2. The absorption spectra shown in Figure 2a indicate the absorption maximum (λmax) for the GNP sensor in the presence of unmodified HSA is ~520 nm, which is comparable to absorption of dispersed gold nanoparticles in water. No significant change in the visible absorption spectra was observed in response to increasing concentrations of unmodified HSA over the concentration range investigated in this study. Conversely, the absorption spectra of the sensor in the presence of an equivalent concentration range of HSA–homocystamide have red-shifted extended plasmon bands (Figure 2b). A proportional increase in UV absorption at the protein absorption bands ~220–280 nm was also observed in response to increasing concentrations of both unmodified HSA and HSA–homocystamide in the sensor solutions. A representative plot of the colorimetric response of the GNP sensor detected at 620 nm in the presence of HSA–homocystamide is provided in the Supporting Information (Figure S2). The colorimetric response was found to be linear (r2 = 0.9849) over a range of HSA concentrations (2.8–28 mg/mL). The limit of detection (LOD) and calibration sensitivity for the method were 2.3 mg/mL and 2.7 AU·(μg/mL)−1, respectively. Collectively, these results suggest that the colorimetric signal produced by the GNP sensor is due to HTL modification-directed nanoparticles assembly and not salt-induced nanoparticle aggregation. Moreover, the stability of the GNP sensor in the HSA-phosphate buffer solutions suggests that the GNP sensor will be stable in the presence of human sera (vide infra).
Effect of Temperature on GNP Sensor-Protein Interactions
Many GNP sensors reported in the literature are designed such that nanoparticle assembly is achieved via reversible noncovalent interactions.51–53 Consequently, the mechanisms for nanoparticle assembly are often loosely bound and environment dependent. For example, Mirkin et al. demonstrated temperature-dependent reversible nanoparticle assembly over a decade ago using DNA-functionalized GNPs.54 This system exploits noncovalent DNA hybridization interactions to produce a colorimetric signal that could be effectively “switched on and off” by cycling the solution temperature between 0 and 80 °C, the latter of which is well above the dissociation temperature of the DNA–nanoparticle complex. Additionally, a temperature- and pH-sensitive Hcy GNP sensor was recently developed by Lim et al.33 This novel GNP sensor utilizes reversible zwitterionic-type interactions to generate a colorimetric response.
A distinguishing characteristic of the proposed GNP sensor for protein homocystamide is that it was designed to exhibit essentially irreversible binding behavior via thiol binding interactions. In principle, a tightly bound nanoparticle–protein assembly would allow for highly stable and long-lived colorimetric detection. Consequently, it was found that sensor solutions containing HSA–homocystamide incubated in an 80 °C water bath for 20 min exhibited remarkable stability as indicated by a persistent colorimetric signal. Only a very slight increase in colorimetric signal was observed after exposing the sensor solutions to 80 °C for ~10 min (data not shown). This result is likely caused by additional GNP binding sites being exposed due to thermal denaturation of the modified protein.
Confirmation of Modification-Directed Nanoparticle Assembly
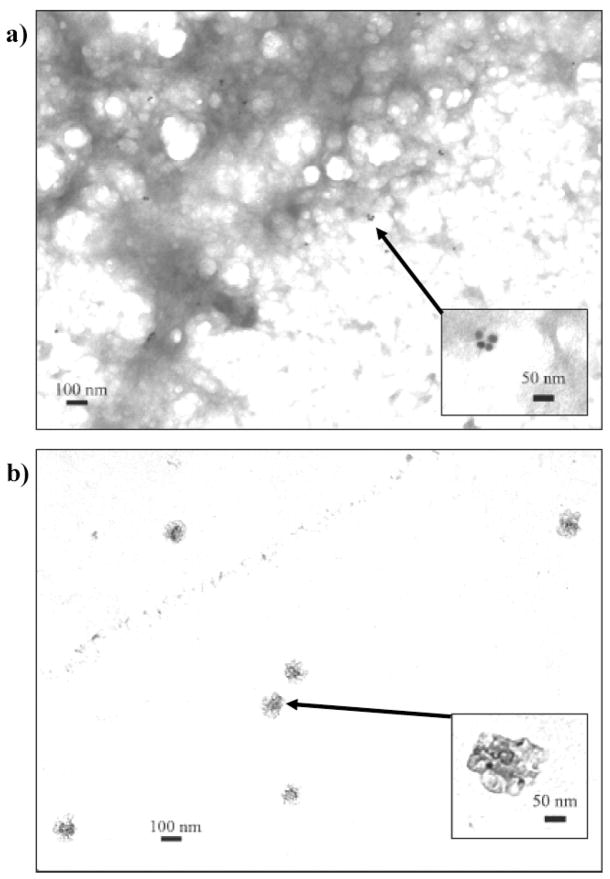
Transmission electron microscopy (TEM) was used to image the GNP sensor in the presence of HSA and HSA homocystamide. As expected, the purple-colored GNP sensor–protein homocystamide solutions contained oligomeric HSA, which appears as a dark sponge-like film in the TEM image (33 000× magnification) shown in Figure 3a. The GNPs appear as clearly defined black dots in the TEM images. Further examination of Figure 3a reveals an assortment of nanoparticle clusters within the oligomeric protein network. The multiplicity of shapes and sizes of the nanoparticle assemblies in the sensor specimen is consistent with the broad extended plasmon bands observed at 620 nm (Figure 2b).55,56 The inset of Figure 3a (100 000× magnification) shows an example of modification-directed nanoparticle assembly, a 4-nanoparticle cluster within the oligomeric protein network. Other regions within the oligomeric protein network containing GNP clusters are highlighted in Figure S3 in the Supporting Information. These data suggest that both modification-induced protein oligomerization and modification-directed nanoparticle assembly occur concomitantly in the GNP sensor solutions containing protein homocystamide.
Figure 3.
Representative transmission electron microscopy (TEM) images of the GNP sensor (a) in the presence of HSA–homocystamide at 33 000× magnification (inset shows 100 000× magnification of a 4-nanoparticle cluster assembly within the oligomeric protein network) and (b) in the presence of unmodified HSA (inset shows 100 000× magnification of unassembled GNPs surrounded by unmodified HSA). The arrows indicate the location of the insets in lower magnification images.
In contrast, neither nanoparticle assembly nor protein oligomerization is observed in a specimen prepared from the red-colored GNP sensor solution containing unmodified HSA (Figure 3b). The unmodified protein appears as dark semitransparent spots in the TEM images. Closer examination of the sensor specimens containing unmodified HSA reveals that the spherical GNPs are randomly dispersed throughout the specimen within individual protein spots. The inset of Figure 3b shows GNPs in the presence of unmodified HSA at 100 000× magnification. Nanoparticle assembly is not induced by unmodified HSA, presumably due to the lack of protein homocystamide binding sites. The absence of significant nanoparticle assembly in the unmodified protein sample is consistent with the absorption spectra shown in Figure 2a and consistent with the expected results for this detection scheme.
Proposed Dectection Scheme for Protein Homocystamide in Human Serum
Biochemical sensor design requires careful consideration of the intended sample matrix. Blood and urine samples have been traditionally collected from patients for use in generic clinical diagnostics. Urine analysis is a versatile medical screening procedure that has been used historically for the in vitro detection of a number of physiologically relevant analytes including illicit drugs, steroids, and numerous other analytes. The detection of HTL in urinary excretions has been reported.57 However, the existence of substantial amounts of protein homocystamide in urinary excretions is not expected because of the relatively low protein content of urine. In contrast, human serum is a more ideal sample medium for the protein sensor because it is relatively protein rich and thus more likely to contain detectable levels of protein homocystamide. Another potential advantage of working with human serum is that it is already routinely screened for other cardiovascular disease risk factors such as cholesterol.
Unlike the purified HSA used in the previous studies, human serum is a complex medium that consists of an assortment of proteins and small molecules, including carbohydrates, drugs, hormones, etc., which may exist as free or protein-bound species. Human serum also contains various thiol-containing species that could interfere with the GNP sensor. For example, cysteine (Cys),56 glutathione,56 and Hcy33 are typically found at micromolar concentrations in human sera. Fortunately, these low molecular weight species can be easily removed from sera by use of a molecular cutoff filter device prior to treatment with the GNP sensor.
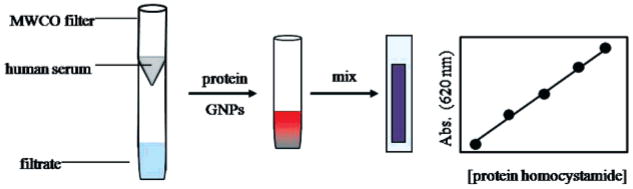
Frozen whole human sera collected and pooled from healthy adult donors were purchased from Sigma-Aldrich (St. Louis, MO) for use in this component of our study. The sera solutions were faintly yellow in color and slightly turbid due to sparingly soluble components. The protein content of the sera was determined by a gravimetric assay of lyophilized 1 mL aliquots after washing with deionized water. The average protein concentration of the aliquots was 52.4 ± 6.7 mg/mL (n = 4). Sera were typically diluted with phosphate buffer to obtain ~65% (v/v) serum solution in order to decrease the viscosity and turbidity. The diluted serum was post-translationally modified with HTL in vitro as previously described. Scheme 2 depicts the proposed scheme for detecting serum protein homocystamide using the GNP sensor. First, the protein modification mixture is purified by use of centrifugal 10 kDa molecular cutoff (MWCO) filters. Next, the modified protein homocystamide retained by the MWCO filter is promptly reconstituted with phosphate buffer in order to obtain a stock solution. The resultant protein homocystamide is then immediately treated with the GNP sensor. Finally, the resultant GNP–protein homocystamide solution is diluted and transferred to a cuvette for monitoring of the colorimetric response.
Scheme 2.
Proposed Scheme for Detection of Protein Homocystamide in Human Sera
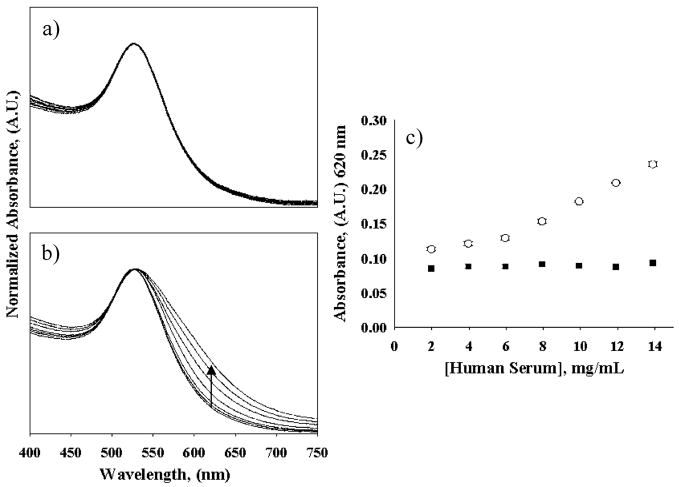
Representative absorption plots of the sensor under various experimental conditions are shown in Figure 4. Similar to the HSA model studies, unmodified sera were found to be reasonably stable and did not produce appreciable shifts in the plasmon resonance band over the sera concentration range investigated in this study (Figures 4a). Consistent with the HSA model studies, the sensor also produces extended plasmon resonance bands in the presence of serum protein homocystamide (Figure 4b). Figure 4c is a comparison plot of sensor signal versus serum protein concentration. It is apparent that the colorimetric signal detected at 620 nm increases in response to increasing concentrations of serum protein homocystamide but remains relatively constant in the presence of equivalent concentrations of unmodified sera. Notably, the sensor response deviates from linearity at modified serum concentrations lower than ~5 mg/mL, giving a background signal of ~0.12 AU. This effect is partially attributed to the GNPs being sparingly soluble in the phosphate buffer at low protein concentrations, which produces increased sample turbidity and light scattering. The colorimetric response in the range of 6.0–14 mg/mL modified serum protein provides excellent linearity (r2 = 0.9994) and a calibration sensitivity of 13.6 AU· (μg/mL)−1. The calculated LOD of the serum method is 5.2 mg/mL.
Figure 4.
Visible absorption spectra of the GNP sensor in response to human sera: (a) in the presence of unmodified human sera and (b) in the presence of human serum homocystamide. (c) Sensor response at 620 nm in the presence of modified (○) and unmodified (■) sera versus human serum concentration (2–14 mg/mL). The arrow indicates the change in signal at 620 nm with increasing concentrations of human serum homocystamide.
Human plasma can contain from 0.1 to 13 μM protein homocystamide, which is comparable to the mg/mL protein concentration range.58 Accordingly, a solution containing 2.3 mg/mL sera homocystamide and 0.7 mM GNP was allowed to react as previously described. The resultant solution containing the HSA–homocystamide–GNP complex was serially diluted with ultrapure water in order to evaluate the dilutional stability of the protein homocystamide–GNP assembly at concentrations well below the physiologically relevant range. The resultant curve exhibited excellent linearity (r2 = 0.9899), which is expected for a tightly bound colorimetric complex (Figure S4, Supporting Information). The sensor also provided an exceptional calibration sensitivity, 0.30 AU·(ng/mL)−1. These data further support the hypothesis of modification-directed nanoparticle assembly via chemisorption and intermolecular disulfide bonding interactions.
Overall, the proposed GNP sensor exhibited exceptional performance for the detection of the in vitro modified serum protein species used in these studies. This is largely because freshly prepared protein homocystamide was promptly treated with the sensor, thereby minimizing the effects of auto-oxidation in the protein reaction mixture. However, essentially all of the protein homocystamide produced in vivo exists in an oxidized state and would thus be unavailable for binding with the sensor. Therefore, the issue of reducing the oxidized protein homocystamide species must be addressed prior to applying this method to screening clinical samples containing in vivo synthesized serum protein homocystamide.
Conclusions
We demonstrated a straightforward colorimetric method for the detection of serum protein homocystamide, an emergent cardiovascular disease biomarker. GNP assembly was observed only in the presence of serum protein homocystamide. TEM images of the GNP sensor in the presence of unmodified and HTL-modified serum protein are consistent with the spectroscopy data. The TEM images are also indicative of modification-directed nanoparticle assembly as well as modification-induced protein oligomerization. The nanoparticle–protein homocystamide complex exhibited excellent stability and irreversible binding behavior under the condition used in this study. Studies are underway to adapt this model GNP biochemical sensor for use in clinical sera samples. On the basis of our studies to date, we believe that the approach reported here will lead to the development of a GNP sensing scheme that could aid in the rapid detection and diagnosis of cardiovascular disease.
Supplementary Material
Additional experimental data including TEM images, mass spectra for unmodified and post-translationally modified HSA, as well as calibration and dilutional stability curves for the GNP sensor solutions. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
A.T.G. is the recipient of fellowships from the Procter & Gamble Company and the Dow Chemical Company through the National Organization for the Professional Advancement of Black Chemist and Chemical Engineers (NOBCChE). R.M.S. acknowledges support from the National Institutes of Health (NIH) award RO1 EB002044. I.M.W. acknowledges the National Science Foundation award 0616824, NIH award RO1 GM079670, and the Philip W. West Endowment for their support of this work.
References
- 1.Clarke R, Daly L, Robinson K, Naughten E, Cahalane S, Fowler B, Graham I. N Engl J Med. 1991;324:1149–1155. doi: 10.1056/NEJM199104253241701. [DOI] [PubMed] [Google Scholar]
- 2.Taylor LM, Defrang RD, Harris EJ, Porter JM. J Vasc Surg. 1991;13:128–136. doi: 10.1067/mva.1991.24913. [DOI] [PubMed] [Google Scholar]
- 3.Clarke R, Smith AD, Jobst KA, Refsum H, Sutton L, Ueland PM. Arch Neurol. 1998;55:1449–1455. doi: 10.1001/archneur.55.11.1449. [DOI] [PubMed] [Google Scholar]
- 4.Mercie P, Garnier O, Lascoste L. Apoptosis. 2000;5:403–411. doi: 10.1023/a:1009652011466. [DOI] [PubMed] [Google Scholar]
- 5.Jakubowski H, Ambrosius WT, Pratt JH. FEBS Lett. 2001;491:35–39. doi: 10.1016/s0014-5793(01)02143-3. [DOI] [PubMed] [Google Scholar]
- 6.Yang X, Gao Y, Zhou J. Clin Chim Acta. 2006;364:230–234. doi: 10.1016/j.cccn.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Jakubowski H. Biomed Pharmacother. 2001;55:443–447. doi: 10.1016/s0753-3322(01)00085-3. [DOI] [PubMed] [Google Scholar]
- 8.Jakubowski H. Biochemistry. 1997;36:11077–11085. doi: 10.1021/bi970589n. [DOI] [PubMed] [Google Scholar]
- 9.Jakubowski H. J Nutr. 2006;136:1741S–1749S. doi: 10.1093/jn/136.6.1741S. [DOI] [PubMed] [Google Scholar]
- 10.Austin RC, Lentz SR, Werstuck GH. Cell Death Differ. 2004;11:S56–S64. doi: 10.1038/sj.cdd.4401451. [DOI] [PubMed] [Google Scholar]
- 11.Jakubowski H. FASEB J. 1999;13:2277–2283. [PubMed] [Google Scholar]
- 12.Jakubowski H. J Nutr. 2000;130:377S–381S. doi: 10.1093/jn/130.2.377S. [DOI] [PubMed] [Google Scholar]
- 13.Hop ECA, Bakhtiar R. Rapid Commun Mass Spectrom. 2002;16:1049–1053. doi: 10.1002/rcm.681. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg A. AAPSJ. 2006;8:E501–E507. doi: 10.1208/aapsj080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang WH, Rusin O, Xiangyang X, Kwang K, Escobedo JO, Fakayode SO, Fletcher KA, Lowry M, Schowalter CM, Lawrence CM, Fronczeck F, Warner IM, Strongin RM. J Am Chem Soc. 2005;127:15949–15958. doi: 10.1021/ja054962n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rusin O, St Luce NN, Agbaria RA, Escobedo JO, Jiang S, Warner IM, Dawan FB, Lian K, Strongin RM. J Am Chem Soc. 2004;126:438–439. doi: 10.1021/ja036297t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escobedo JO, Wang W, Strongin RM. Nat Protocols. 2006;1:2759–2762. doi: 10.1038/nprot.2006.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang WH, Escobedo JO, Lawrence CM, Strongin RM. J Am Chem Soc. 2004;126:3400–3401. doi: 10.1021/ja0318838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pacsial-Ong EJ, McCarley RL, Wang W, Strongin RM. Anal Chem. 2006;78:7577–7581. doi: 10.1021/ac061451q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gates AT, Lowry M, Fletcher KA, Merugeshu A, Rusin O, Robinson JW, Strongin RM, Warner IM. Anal Chem. 2007;79:8249–8256. doi: 10.1021/ac0706731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakubowski H, Zhang L, Bardeguez A, Aviv A. Circ Res. 2000;87:45–51. doi: 10.1161/01.res.87.1.45. [DOI] [PubMed] [Google Scholar]
- 22.Uji Y, Motmiya Y, Hanyu N, Ukaji F, Okabe H. Clin Chem. 2002;48:941–944. [PubMed] [Google Scholar]
- 23.Ferguson E, Parthasarathy S, Joseph J, Kalyanaraman B. J Lipid Res. 1998;39:925–933. [PubMed] [Google Scholar]
- 24.Undas A, Perla J, Lacinski M, Trzeciak W, Kazmierski R, Jakubowski H. Stroke. 2004;35:1299–1304. doi: 10.1161/01.STR.0000128412.59768.6e. [DOI] [PubMed] [Google Scholar]
- 25.Frens G. Nat Phys Sci. 1973;241:20–22. [Google Scholar]
- 26.Grant CD, Schwartzberg AM, Norman TJ, Zhang JZ. J Am Chem Soc. 2002;125:549–553. doi: 10.1021/ja028532y. [DOI] [PubMed] [Google Scholar]
- 27.Bloomfield N. Am J Clin Pathol. 1964;41:15–21. doi: 10.1093/ajcp/41.1.15. [DOI] [PubMed] [Google Scholar]
- 28.Willner I, Baron R, Willner B. Biosens Bioelectron. 2007;22:1841–1852. doi: 10.1016/j.bios.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Liao HW, Nehl CL, Hafner JH. Nanomedicine. 2006;1:201–208. doi: 10.2217/17435889.1.2.201. [DOI] [PubMed] [Google Scholar]
- 30.Chen JR, Miao YQ, He NY, Wu XH, Li SJ. Biotechnol Adv. 2004;22:505–518. [Google Scholar]
- 31.Maxwell DJ, Taylor JR, Nie SM. J Am Chem Soc. 2002;124:9606–9612. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 32.Reynolds AJ, Haines AH, Russell DA. Langmuir. 2007;22:1156–1163. doi: 10.1021/la052261y. [DOI] [PubMed] [Google Scholar]
- 33.Lim IS, Ip W, Crew E, Njoki PN, Mott D, Zhong C, Pan Y, Zhou S. Langmuir. 2007;23:826–833. doi: 10.1021/la062334t. [DOI] [PubMed] [Google Scholar]
- 34.Zhang FX, Han L, Israel LB, Daras JG, Maye MM, Ly NK. Analyst. 2002;127:462–465. doi: 10.1039/b200007e. [DOI] [PubMed] [Google Scholar]
- 35.Liu JW, Lu Y. J Fluoresc. 2004;14:343–354. doi: 10.1023/b:jofl.0000031816.06134.d3. [DOI] [PubMed] [Google Scholar]
- 36.Storhoff JJ, Mirkin CA. Chem Rev. 1999;99:1849–1862. doi: 10.1021/cr970071p. [DOI] [PubMed] [Google Scholar]
- 37.Aubin-Tam ME, Hamad-Schifferli K. Langmuir. 2005;21:12080–12084. doi: 10.1021/la052102e. [DOI] [PubMed] [Google Scholar]
- 38.Chah S, Hammond MR, Zare RN. Chem Biol. 2005;12:323–328. doi: 10.1016/j.chembiol.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharya J, Jasrapuria S, Sarkar T, GhoshMoulick R, Dasgupta AK. Nanomedicine. 2007;3:14–19. doi: 10.1016/j.nano.2006.10.159. [DOI] [PubMed] [Google Scholar]
- 40.Shang L, Wang Y, Jiang J, Dong S. Langmuir. 2007;23:2714–2721. doi: 10.1021/la062064e. [DOI] [PubMed] [Google Scholar]
- 41.Tom RT, Samal AK, Sreeprasad TS, Pradeep T. Langmuir. 2007;23:1320–1325. doi: 10.1021/la061150b. [DOI] [PubMed] [Google Scholar]
- 42.Huang C, Huang Y, Cao Z, Tan W, Chang H. Anal Chem. 2005;77:5735–5741. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- 43.Aslan K, Luhrs CC, Perez-Luna VH. J Phys Chem B. 2004;108:15631–15639. [Google Scholar]
- 44.Schofield CL, Field RA, Russell DA. Anal Chem. 2007;79:1356–1361. doi: 10.1021/ac061462j. [DOI] [PubMed] [Google Scholar]
- 45.Liu JW, Lu Y. Anal Chem. 2004;76:1627–1632. doi: 10.1021/ac0351769. [DOI] [PubMed] [Google Scholar]
- 46.Kimling J, Maier M, Okenve B, Kotaidis V, Ballot H, Plech A. J Phys Chem B. 2006;110:15700–15707. doi: 10.1021/jp061667w. [DOI] [PubMed] [Google Scholar]
- 47.Brewer SH, Glomm WR, Johnson MC, Knag MKSF. Langmuir. 2005;21:9303–9307. doi: 10.1021/la050588t. [DOI] [PubMed] [Google Scholar]
- 48.Liu S, Yang Z, Liu Z, Kong L. Anal Biochem. 2006;353:108–116. doi: 10.1016/j.ab.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 49.Walter H, Schobel B. Hum Genet. 1975:30. doi: 10.1007/BF00275146. [DOI] [PubMed] [Google Scholar]
- 50.Wilcoxon JP, Martin JE. Phys Rev A. 1989;39:2675–2688. doi: 10.1103/physreva.39.2675. [DOI] [PubMed] [Google Scholar]
- 51.Naka K, Itoh H, Chujo Y. Langmuir. 2003;19:5496–5501. [Google Scholar]
- 52.Guo Y, Yudan X, Li J, Yang W. J Phys Chem C. 2007;111:9172–9176. [Google Scholar]
- 53.Li M, Mann S. J Mater Chem. 2004;14:2260–2263. [Google Scholar]
- 54.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 55.Schwartzberg AM, Grant CD, Wilcott A, Talley CE, Huser TR, Bogomolni R, Zhang JZ. J Phys Chem B. 2004;108:19191–19197. [Google Scholar]
- 56.Sudeep PK, Joseph STS, Thomas KG. J Am Chem Soc. 2005;127:6516–6517. doi: 10.1021/ja051145e. [DOI] [PubMed] [Google Scholar]
- 57.Chwatko G, Jakubowski H. Clin Chem. 2005;51:408–415. doi: 10.1373/clinchem.2004.042531. [DOI] [PubMed] [Google Scholar]
- 58.Jakubowski H. CMLS. 2004;61:470–487. doi: 10.1007/s00018-003-3204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional experimental data including TEM images, mass spectra for unmodified and post-translationally modified HSA, as well as calibration and dilutional stability curves for the GNP sensor solutions. This material is available free of charge via the Internet at http://pubs.acs.org.