Abstract
Background
Several lines of evidence in both human and animal studies suggest that variation in neuropeptide Y (NPY) or its receptor genes (NPY1R, NPY2R and NPY5R) is associated with alcohol dependence as well as alcohol withdrawal symptoms. Additional studies suggest cocaine may affect NPY expression.
Methods
A total of 39 SNPs were genotyped across NPY and its 3 receptor genes in a sample of 1,923 subjects from 219 multiplex alcoholic families of European American descent recruited as part of the Collaborative Studies on the Genetics of Alcoholism (COGA) study. Family-based association analysis was performed to test the primary hypothesis that variation in these genes is associated with alcohol dependence. Secondary analyses evaluated whether there was an association of these SNPs with symptoms of alcohol withdrawal, cocaine dependence, or comorbid alcohol and cocaine dependence.
Results
Although variations in NPY itself were not associated with these phenotypes, variations in two NPY-receptor genes were. SNPs in NPY2R provided significant evidence of association with alcohol dependence, alcohol withdrawal symptoms, comorbid alcohol and cocaine dependence, and cocaine dependence (all p<0.03). Haplotype analyses strengthened the evidence for these phenotypes (global 0.005<p<0.0004). SNPs in NPY5R demonstrated significant association with alcohol withdrawal characterized by seizures (p<0.05).
Conclusion
These results indicate that sequence variations in NPY receptor genes are associated with alcohol dependence, particularly a severe subtype of alcohol dependence characterized by withdrawal symptoms, comorbid alcohol and cocaine dependence or cocaine dependence.
Keywords: Alcoholism, withdrawal, cocaine dependence, NPY, genetic association
Introduction
Alcohol dependence is a common disorder affecting 4–5% of the United States population at any given time, (Li et al., 2007) with a lifetime prevalence of 12.5% (Hasin et al., 2007). Family, twin and adoption studies have consistently demonstrated a substantial genetic contribution to disease etiology (Cloninger et al., 1981; Heath et al., 1997; Kendler et al., 1994; McGue, 1999; Pickens et al., 1991;). Recent human studies have identified several genes associated with alcohol dependence, including GABRA2 (Covault et al., 2004; Edenberg et al., 2004; Fehr et al., 2006; Lappalainen et al., 2005), ADH4 (Edenberg et al., 2006; Guindalini et al., 2005; Luo et al., 2005b), GABRG3 (Dick et al., 2004), CHRM2 (Luo et al., 2005a; Wang et al., 2004), NFKB1 (Edenberg et al., 2007), OPRK1 and PDYN (Edenberg et al 2008; Xuei et al 2006) and TAS2R16 (Hinrichs et al., 2006).
A complementary approach to the identification of genes contributing to the risk for human alcoholism is the analysis of alcohol-related phenotypes in animal models. For example, the alcohol-preferring (P) and -nonpreferring (NP) rats have been shown to be an animal model of alcohol dependence (Files et al., 1992; Li et al., 1991). The P rats voluntarily consume large amounts of alcohol for its pharmacological effects, work hard to obtain alcohol, and demonstrate tolerance when allowed to drink freely (Carr et al., 1998; Li et al., 1993; Murphy et al., 2002). Using data from this animal model, strong evidence of linkage for alcohol preference was found on rat chromosome 4, in a region which included several positional candidate genes including SNCA (Carr et al., 1998; Liang and Carr, 2006; Liang et al., 2003). Subsequent human studies have demonstrated that variation in SNCA does not contribute to the overall risk of alcohol dependence, but is associated with the phenotype of alcohol craving that may be related to the preference in rats (Bonsch et al., 2004; Bonsch et al., 2005a; Bonsch et al., 2005b; Bonsch et al., 2005c; Foroud et al., 2007;).
The same region on rat chromosome 4 also includes the candidate gene Npy (Neuropeptide Y; Spence et al., 2005). Numerous studies in animal models have shown that NPY plays a role in alcohol preference and consummatory behavior (Badia-Elder et al., 2001; Badia-Elder et al., 2003; Badia-Elder et al., 2007; Caberlotto et al., 2001; Cowen et al., 2004; Ehlers et al., 1998; Kimpel et al., 2007; Pandey et al., 2003; Schroeder et al., 2003; Spence et al., 2005; Tecott and Heberlein, 1998; Thiele and Badia-Elder, 2003; Thiele et al., 2004a; Thorsell, 2007). For example, infusion of NPY reduces ethanol intake in P rats (Gilpin et al., 2003), and Npy-deficient mice consume more alcohol than wild-type mice (Thiele et al., 1998). It has also been shown that cocaine administered in Sprague-Dawley rats reduced Npy mRNA in the prefrontal cortex, and reduced NPY-like (NPY-LI) immunoreactivity in the cingulated cortex and nucleus accumbens (Wahlestedt et al., 1991). In the same rat strain NPY-LI immunoreactivity was found to be expressed in the dentate gyrus, a region of the hippocampus where this expression is not typically found, after a cocaine-induced seizure (Goodman and Slovieter, 1993). These results suggest that NPY may also play a role in response to cocaine.
NPY is a highly-conserved 36 amino-acid peptide (de Quidt and Emson, 1986; Sundler et al., 1986) and has multiple functions including anxiolytic regulation (Heilig and Thorsell, 2002), food intake stimulation (Clark et al., 1984; Jolicoeur et al., 1991; Zarjevski et al., 1993), and neuronal excitability (Woldbye et al., 1996). NPY is abundant in the cortex, striatum, nucleus accumbens, amygdala, and hypothalamus (Badia-Elder et al., 2007; Gray and Morley, 1986; Heilig and Widerlov, 1995; Spence et al., 2005).
A functional single nucleotide polymorphism (SNP) in NPY, Leu7Pro (rs16139) (Karvonen et al., 1998), has been extensively studied for its association with alcohol dependence, consumption, and withdrawal symptoms; however, results have been inconsistent. Independent studies have found that the Pro7 allele is more frequent in alcohol dependent individuals than in controls (Kauhanen et al., 2000; Lappalainen et al., 2002), more common in individuals with late onset of alcohol dependence than in those with early onset (Mottagui-Tabar et al., 2005), and more common in alcoholics experiencing severe withdrawal symptoms and higher daily alcohol consumption (Koehnke et al., 2002). In Finnish men, the same allele appeared weakly associated with higher weekly consumption of alcohol (Kauhanen et al., 2000). In contrast, the Pro7 allele when found in heterozygous form was less common among alcoholics than social drinkers (Ilveskoski et al., 2001). Other studies found no significant differences in allele frequency between alcohol dependent individuals and controls of Finnish, Swedish or German origin (Hu et al., 2005; Mottagui-Tabar et al., 2005; Zhu et al., 2003; Zill et al, 2008). Studies of human alcoholics drinking more than 80 g of ethanol per day for most of their adult lives demonstrated a decrease in NPY immunoreactivity in the amygdala (Pluzarev and Crews, 2007) and decreased gene expression of NPY in the frontal and motor cortices (Mayfield et al., 2002).
NPY has also been associated with withdrawal from alcohol. Koehnke et al (2002) reported that the Pro7 allele is more common in alcohol dependent individuals with delirium tremors or who have experienced withdrawal with seizures than in alcoholic dependent individuals with mild withdrawal symptoms. Okubo and Harada (2001) reported association of the 5671C/T polymorphism (rs5574) in NPY with alcohol dependent individuals who experienced withdrawal with seizures. Several animal models have also demonstrated that withdrawal from ethanol reduces NPY expression (Bison and Crews, 2003; Thiele and Badia-Elder, 2003, Thiele et al., 2004b; Thorsell, 2007). For example it has been shown that ethanol withdrawal produced significant reduction in NPY protein levels in the central and medial nuclei of the amygdale, cortical, and hypothalamic structures in rats (Roy and Pandey, 2002). Further evidence shows that intracerebroventricular administration of NPY in Wistar rats in withdrawal significantly decreased the withdrawal scores of the rats (Woldbye et al., 2002).
Three G protein-coupled NPY receptor genes, NPY1R, NPY2R, and NPY5R have been shown to be associated in animals with alcohol preference (Eva et al., 2006; Thiele and Badia-Elder, 2003; Thorsell and Heilig, 2002) and withdrawal (Bison and Crews, 2003; Thiele et al., 2004b; Thorsell et al., 2007; Valdez and Koob, 2004; Woldbye et al., 2002). These three genes are located on chromosome 4q31-q32 (Lutz et al., 1997; Wraith et al., 2000), near the edge of a broad linkage peak for the risk for alcohol dependence identified in the Collaborative Study on the Genetics of Alcoholism (COGA) sample (Reich et al., 1998; Reich, 1996; Williams et al., 1999).
Given that NPY modulates consummatory behavior and the positive, rewarding properties associated with alcohol consumption (Thiele et al., 2004b), and the somewhat inconsistent results from human studies, the neuropeptide Y system seems a good candidate to study in a population of densely affected, alcohol dependent families. We have performed a detailed evaluation of the NPY system, including NPY and its receptor genes NPY2R, NPY1R and NPY5R, in relation to alcohol dependence with and without withdrawal symptoms, and cocaine dependence. The latter was included because of evidence that NPY may be involved in cocaine-seeking behavior and in the response to cocaine.
Materials and methods
Association sample
The Collaborative Study on the Genetics of Alcoholism (COGA) is an ongoing multi-site study that has recruited families at centers across the United States. To limit heterogeneity a sample of 1,923 European American subjects from 219 families was used in the present analysis; they were recruited at Indiana University, State University of New York Downstate Medical Center, University of Connecticut, University of Iowa, University of California/San Diego, and Washington University, St. Louis. This study was approved by the institutional review boards of all participating institutions. Each family was ascertained through a proband seeking treatment at an alcohol treatment program (Begleiter et al., 1995; Foroud et al., 2000; Nurnberger, Jr. et al., 2004; Reich et al., 1998).
A poly-diagnostic instrument, the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) (Bucholz et al., 1994; Hesselbrock et al., 1999) was administered to probands and their families. The families that participated in the genetic phase of this study had at least three first degree relatives who met both lifetime DSM-IIIR criteria for alcohol dependence (American Psychiatric Association, 1987) and lifetime Feighner criteria (Feighner et al., 1972) for definite alcoholism. Further details of the ascertainment and assessment can be found elsewhere (Begleiter et al., 1995; Foroud et al., 2000; Reich et al., 1998).
Phenotypes
We initially tested for an association between the four genes and alcohol dependence as defined by DSM-IV criteria (American Psychiatric Association, 1994). Secondary hypotheses based upon both the human and animal literature included alcohol withdrawal symptoms, such as seizures, and cocaine dependence. The number of affected subjects analyzed for each phenotype is shown in Table 1. Using items from the SSAGA, two measures of alcohol withdrawal were analyzed. The first was whether an individual ever experienced any of nine problems (shakes, sleeplessness, anxiety, sweating, fast heart beat, nausea/vomiting, physically weak, headaches, or seeing/hearing things that weren’t there) after having stopped, cut down, or gone without drinking. Subjects were classified as affected if they met three criteria: 1) responded affirmatively to having at least one of the problems; and 2) took any medication/drug to avoid any of these problems (or to make them go away); and 3) were classified as DSM-IV alcohol dependent. The medication requirement was included in order to more closely approximate the “severe” withdrawal of Koehnke et al. (Koehnke et al., 2002). Subjects were coded as unaffected if they were classified as DSM-IV dependent but did not experience any of the nine symptoms. All other subjects were considered unknown (Table 1). This phenotype is referred to as severe withdrawal.
Table 1.
Phenotypic characteristics of genotyped individuals.
| Phenotypes | # Affected (%) |
# Unaffected (%) |
# Unknown (%) |
|
|---|---|---|---|---|
| Alcohol Dependence | 753 (39%) | 1047 (55%) | 123a (6%) | |
| Withdrawal-Severe |
alcohol dependence +withdrawal symptoms and medication |
176 (9%) | 292 (15%) | 1455 (76%) |
| Withdrawal-Seizures |
alcohol dependence + withdrawal with seizures |
105 (5%) | 648 (34%) | 1170 (61%) |
| Cocaine Dependence | with or without alcohol dependence | 255 (13%) | 1545 (80%) | 123 (6%) |
|
Alcohol Dependence Without Cocaine Dependence |
alcohol dependence no cocaine dependence |
545 (28%) | 1000 (52%) | 378 (20%) |
|
Alcohol Dependence With Cocaine Dependence |
alcohol dependence plus cocaine dependence |
208 (11%) | 1000 (52%) | 715 (37%) |
|
Cocaine Dependence Without Alcohol Dependenceb |
cocaine dependence no alcohol dependence |
47 (2%) | 1000 (52%) | 876 (46%) |
There are 123 individuals who did not complete a SSAGA interview and therefore are classified as unknown for all phenotypes.
Not analyzed for lack of power
The second alcohol withdrawal phenotype classified as affected those subjects who met criteria for DSM-IV alcohol dependence and also responded affirmatively to at least one of two questions: (1) “When you stopped, cut down, or went without drinking, did you have fits, seizures, or convulsions, where you lost consciousness, fell to the floor, and had difficulty remembering what happened;” or (2) “Did you have the DT’s, where you were very confused, extremely shaky, felt very frightened or nervous, or saw things that weren’t really there when you stopped, cut down, or went without drinking?” Subjects who were classified as DSM-IV and responded negatively to both questions were considered unaffected. All other subjects were considered unknown (Table 1). This phenotype is termed withdrawal with seizures.
Because of the reported relationship between NPY-LI expression and cocaine-seeking behavior (Boutrel et al., 2005; Menyhert et al., 2007; Wahlestedt et al., 1991), we tested for an association with cocaine dependence, defined by DSM-IIIR criteria. Due to the large number of cocaine dependent individuals who are comorbid for alcohol dependence (Table 1), we also analyzed individuals who met criteria for both DSM-IV alcohol dependence and cocaine dependence. Individuals who were neither alcohol dependent nor cocaine dependent were classified as unaffected for this phenotype. All other individuals were considered unknown. Since most cocaine dependent individuals were also alcohol dependent (208 out of 255), there was not a sufficient sample to analyze cocaine dependence excluding alcohol dependence (n=47). To avoid confounding cocaine dependence and alcohol dependence, whenever evidence of association was found with both alcohol dependence and cocaine dependence (p<0.05), an additional analysis was performed which included as affected only those individuals who met criteria for DSM-IV alcohol dependence but did not meet DSM-IIIR criteria for cocaine dependence (Table 1). All cocaine dependent individuals who were not alcohol dependent (n=47) were classified as unknown for this alcohol-only phenotype. Thus unaffected individuals were defined as those who were neither alcohol nor cocaine dependent.
SNP genotyping
NPY is located on 7p15.1 and is 7.7 kb in size; NPY2R is on 4q32.1 and is 8.6 kb in size. The other two receptor genes, NPY1R and NPY5R, are only 8kb apart, span 28 kb on 4q32.2 and were analyzed together. SNPs distributed throughout the 4 genes were selected from public databases, primarily dbSNP (http://www.ncbi.nlm.nih.gov/SNP/), based on their spacing and available allele frequencies. At the time some SNPs were selected, allele frequencies were not available. To determine allele frequencies and to test the quality of the assays, SNPs were genotyped in two sets of samples, each consisting of 40 unrelated individuals from the Coriell European- and African-American diversity samples; only SNPs in Hardy Weinberg equilibrium in both test groups were genotyped on the COGA sample.
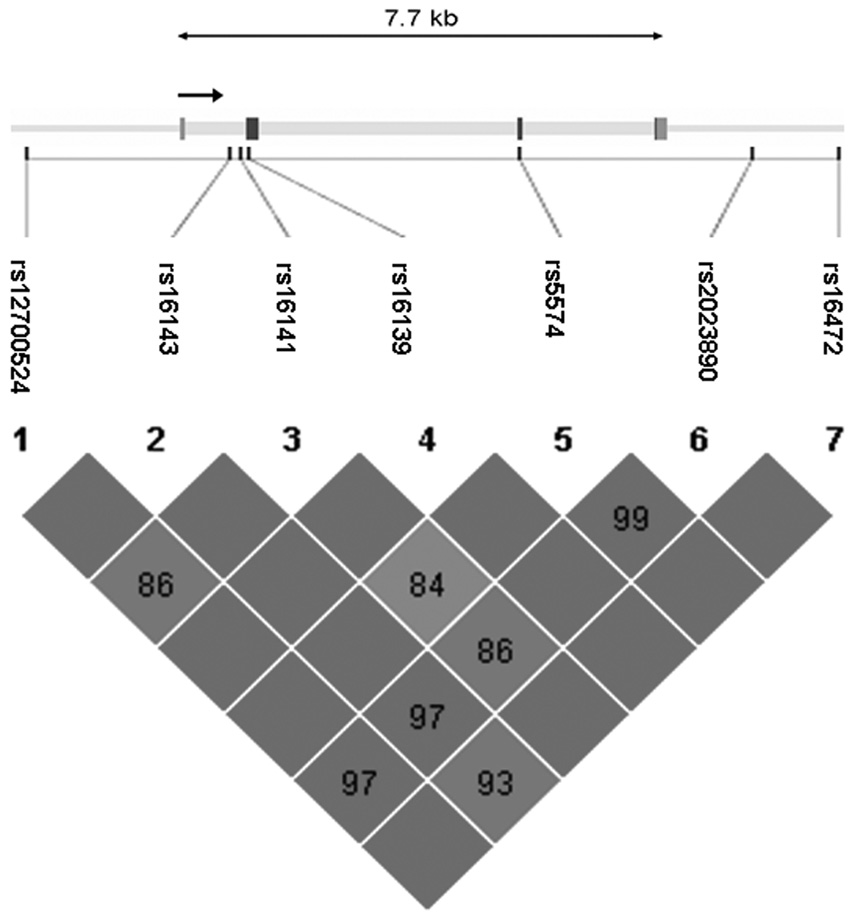
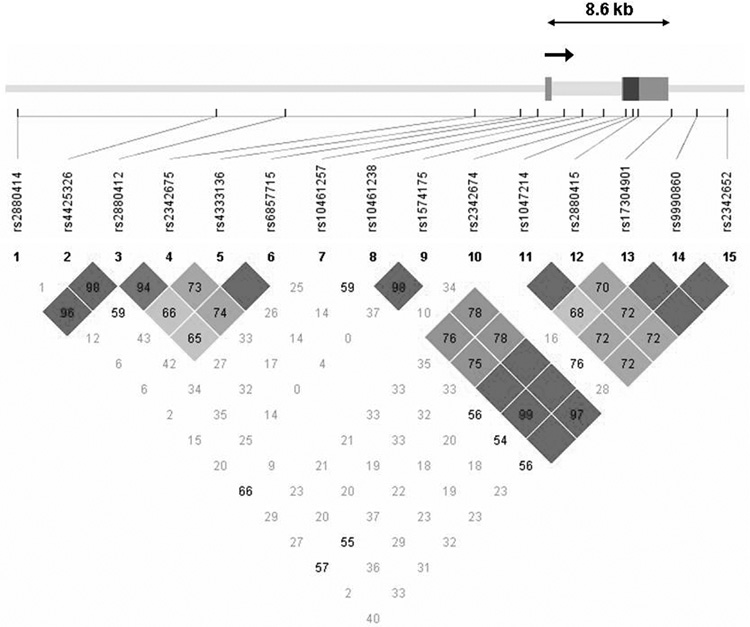
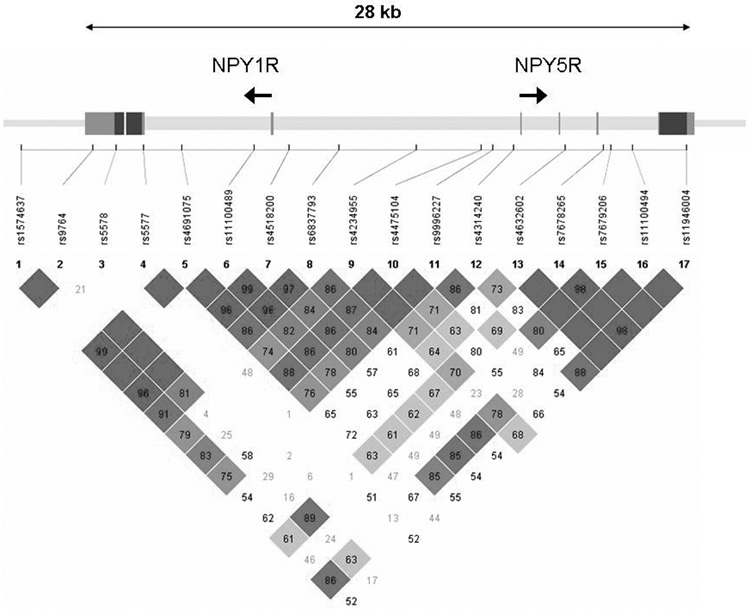
A total of 39 SNPs were genotyped in the sample, including the known coding SNP Leu7Pro in NPY (rs16139) located in exon 2. Genotyping was done using a modified single nucleotide extension reaction, with allele detection by mass spectrometry (Sequenom MassArray Sysem; Sequenom, San Diego, CA). All SNP genotypes were checked for Mendelian inheritance using PEDCHECK (O'Connell and Weeks, 1998). Marker allele frequencies and heterozygosities were computed using USERM13 (Boehnke, 1991). Markers were tested for Hardy-Weinberg equilibrium using Haploview (Barrett et al., 2005). No marker deviated significantly (p<0.01) from Hardy-Weinberg equilibrium. Linkage disequilibrium measured by D’ is depicted for NPY, NPY2R, and the NPY1R/NPY5R cluster in Figures 1A, 1B, and 1C.
Figure 1.
Figure 1A Genomic structure of NPY. The direction of transcription and the exons are indicated in arrow and rectangular block, respectively. Pairwise linkage disequilibrium (LD) estimates, genotyped in the COGA sample, is given as D’. Darkly shaded boxes have strong evidence of LD, defined as a pair of SNPs with the 1-sided upper 95% confidence bound on D’ of 0.98 and the lower bound above 0.70. Lightly shaded boxes have lower LD.
Figure 1B Genomic structure of NPY2R. The direction of transcription and the exons are indicated in arrow and rectangular block, respectively. Pairwise linkage disequilibrium (LD) estimates, genotyped in the COGA sample, is given as D’. Symbols as in Figure 1A.
Figure 1C Genomic structure of NPY1R/NPY5R. The direction of transcription and the exons are indicated in arrow and rectangular block, respectively. Pairwise linkage disequilibrium (LD) estimates, genotyped in the COGA sample, is given as D’. Symbols as in Figure 1A
Statistical analysis
To ensure that the genotyped SNPs adequately covered the genes under consideration, linkage disequilibrium (LD) was computed using the program Haploview (Barrett et al., 2005). An independent evaluation was performed using the program Tagger (de Bakker et al., 2005) to calculate the fraction of all SNPs in each region analyzed by HapMap (with MAF > 0.10) that were in LD (r2≥ 0.80) with the SNPs we genotyped.
The Pedigree Disequilibrium Test (PDT) (Martin et al., 2001), as implemented in the program UNPHASED (Dudbridge, 2003), was used to test whether the qualitative phenotypes in the extended, multiplex COGA pedigrees were associated with the genotyped SNPs. The PDT-average statistic, which weighs each family equally in computing the overall test statistic, was the statistic of interest for each phenotype.
To reduce the scope of hypothesis testing, multi-SNP haplotypes were constructed only when two or more phenotypes were significant (p≤0.05) within any one gene. To avoid constructing haplotypes based on SNPs providing redundant information, only SNPs with low pairwise LD (r2<0.50) were used in haplotype analysis. Each haplotype was then examined to determine whether significant association results were due to the overtransmission of a particular haplotype to affected individuals or to the differential transmission of particular haplotypes to siblings discordant for the phenotype. Except for the severe withdrawal phenotype, haplotypes were estimated using phase-certain genotyped individuals in the program UNPHASED (Dudbridge, 2003). Due to the small number of both affected and unaffected subjects for the severe withdrawal phenotype, missing haplotypes were estimated using the EM algorithm. All haplotypes with a frequency less than 0.05 were omitted from association analyses.
Results
Coverage of variation in the genes
To determine how well the genotyped SNPs represented the known variation (from the HapMap CEU database) in the regions of interest, we applied the program Tagger (de Bakker et al., 2005). Seven SNPs were genotyped across the 13 kb region containing NPY, extending 4 kb beyond each end of the gene (Table 2). The average r2 of these 7 SNPs with all of the 27 known HapMap SNPs (MAF≥0.10) in the 13 kb region was 0.96; r2 > 0.8 for 96% of the SNPs. The Leu/Pro7 polymorphism in NPY (rs16139) had a minor allele frequency (MAF) of 0.05.
Table 2.
Association of NPY and NPY receptor genes and study phenotypes.
| Gene | Id | SNP | Positiona | Locationb | MAFc | Alcohol Dependenced |
Withdrawal Severed |
Withdrawal Seizuresd |
Cocaine Dependenced |
|---|---|---|---|---|---|---|---|---|---|
| NPY | 1 | rs12700524 | 24,287,939 | Upstream | 0.13 | 0.79 | 0.77 | 0.52 | 0.81 |
| 2 | rs16143 | 24,291,113 | Intron 1 | 0.25 | 0.70 | 0.80 | 0.80 | 0.20 | |
| 3 | rs16141 | 24,291,284 | Intron 1 | 0.49 | 0.26 | 0.41 | 0.46 | 0.19 | |
| 4 | rs16139 | 24,291,404 | Exon 2, Pro7Leu | 0.05 | 0.84 | 0.68 | 0.41 | 0.91 | |
| 5 | rs5574 | 24,295,658 | Exon 3, Ser68 | 0.49 | 0.39 | 0.73 | 0.73 | 0.42 | |
| 6 | rs2023890 | 24,299,272 | Downstream | 0.23 | 0.73 | 0.80 | 0.41 | 0.63 | |
| 7 | rs161416472 | 24,300,594 | Downstream | 0.09 | 0.31 | 0.82 | 0.14 | 0.25 | |
| NPY2R | 1 | rs2880414 | 156,313,101 | Upstream | 0.29 | 0.27 | 0.89 | 0.37 | 0.38 |
| 2 | rs4425326* | 156,326,686 | Upstream | 0.36 | 0.03 | 0.03 | 0.35 | 0.02 | |
| 3 | rs2880412 | 156,331,429 | Upstream | 0.26 | 0.60 | 0.88 | 0.52 | 0.15 | |
| 4 | rs2342675 | 156,344,373 | Upstream | 0.35 | 0.53 | 0.58 | 0.46 | 0.03 | |
| 5 | rs4333136 | 156,347,499 | Upstream | 0.38 | 0.02 | 0.08 | 0.60 | 0.004 | |
| 6 | rs6857715* | 156,348,632 | Promoter | 0.37 | 0.03 | 0.03 | 0.92 | 0.0005 | |
| 7 | rs10461257 | 156,350,454 | Intron 1 | 0.28 | 0.35 | 0.31 | 0.37 | 0.93 | |
| 8 | rs10461238 | 156,351,666 | Intron 1 | 0.40 | 0.71 | 0.41 | 0.32 | 0.08 | |
| 9 | rs1574175 | 156,353,162 | Intron 1 | 0.12 | 0.83 | 0.85 | 0.94 | 0.28 | |
| 10 | rs2342674 | 156,354,700 | Exon 2, Leu53 | 0.01 | 0.38 | 1.00 | 0.32 | 0.06 | |
| 11 | rs1047214 | 156,355,126 | Exon 2, Ile195 | 0.46 | 0.34 | 0.69 | 0.78 | 0.40 | |
| 12 | rs2880415 | 156,355,477 | Exon 2, Ile312 | 0.47 | 0.28 | 0.49 | 0.81 | 0.18 | |
| 13 | rs17304901 | 156,357,822 | Downstream | 0.27 | 0.35 | 0.18 | 0.31 | 0.005 | |
| 14 | rs9990860 | 156,359,515 | Downstream | 0.22 | 0.72 | 0.82 | 0.28 | 0.21 | |
| 15 | rs2342652 | 156,361,5662 | Downstream | 0.48 | 0.81 | 0.12 | 0.61 | 0.76 | |
| NPY1R | 1 | rs1574637 | 164,461,542 | Downstream | 0.11 | 0.16 | 0.65 | 0.45 | 0.55 |
| NPY1R | 2 | rs9764 | 164,464,855 | 3’ UTR | 0.26 | 0.28 | 0.19 | 0.73 | 0.47 |
| NPY1R | 3 | rs5578 | 164,465,939 | Exon 3, Thr374Lys | 0.01 | 0.89 | 0.32 | 0.32 | 0.78 |
| NPY1R | 4 | rs5577 | 164,467,193 | 5’ UTR | 0.02 | 0.87 | 0.94 | 0.65 | 0.79 |
| NPY1R | 5 | rs4691075 | 164,468,935 | Intron 1 | 0.12 | 0.15 | 0.63 | 0.38 | 0.67 |
| NPY1R | 6 | rs11100489 | 164,472,262 | Intron 1 | 0.10 | 0.25 | 0.85 | 0.54 | 0.75 |
| NPY1R | 7 | rs4518200 | 164,473,872 | Promoter | 0.11 | 0.21 | 0.63 | 0.73 | 0.90 |
| 8 | rs6837793 | 164,476,185 | Intergenic | 0.11 | 0.24 | 0.51 | 0.56 | 0.73 | |
| 9 | rs4234955 | 164,479,726 | Intergenic | 0.26 | 0.45 | 0.87 | 0.97 | 0.51 | |
| 10 | rs4475104 | 164,482,736 | Intergenic | 0.11 | 0.67 | 0.33 | 0.05 | 0.56 | |
| NPY5R | 11 | rs9996227 | 164,483,251 | Promoter | 0.11 | 0.29 | 0.17 | 0.06 | 0.82 |
| NPY5R | 12 | rs4314240 | 164,484,226 | Promoter | 0.11 | 0.87 | 0.20 | 0.05 | 0.34 |
| NPY5R | 13 | rs4632602 | 164,486,579 | Intron 2 | 0.12 | 0.42 | 0.33 | 0.008 | 0.75 |
| NPY5R | 14 | rs7678265 | 164,488,364 | Intron 3 | 0.09 | 0.22 | 0.55 | 0.02 | 0.33 |
| NPY5R | 15 | rs7679206 | 164,488,684 | Intron 3 | 0.23 | 0.09 | 0.21 | 0.36 | 0.86 |
| NPY5R | 16 | rs11100494 | 164,489,703 | Intron 3 | 0.07 | 0.89 | 0.38 | 0.75 | 0.81 |
| NPY5R | 17 | rs11946004 | 164,492,153 | Exon 4, Gly426 | 0.12 | 0.13 | 0.60 | 0.06 | 0.71 |
Position in nucleotides, from NCBI Human Genome Assembly (version 36.2)
position within or near gene.
Minor allele frequency calculated from the COGA dataset.
Pedigree Disequilibrium test average statistic p-value. Bolded are significant (p≤0.05) association SNPs.
SNPs used in haplotype analysis.
NPY, Neuropeptide Y; COGA, Collaborative Study on the Genetics of Alcoholism; SNP, Single Nucleotide Polymorphism
Fifteen SNPs were genotyped across the 48 kb region containing NPY2R, extending 34 kb on the 5’ end and 4 kb on the 3’ end (Table 2). The average r2 of the 8 HapMap SNPs among these with all of the 83 known HapMap SNPs (MAF≥0.10) in the 48 kb NPY2R region was 0.85; r2 > 0.8 for 81% of the SNPs. Because 7 of the 15 SNPs we genotyped were not in the HapMap database, this is the lower boundary of coverage. NPY2R has weak LD along the 3’ end and stronger LD between SNPs 8–15 on the 5’ end of the gene, as can be seen in Figure 2.
Seventeen SNPs were genotyped across the 31 kb region containing NPY1R and NPY5R, extending 3 kb on the 3’ end of NPY1R. The average r2 of 11 SNPs of the 17 SNPs genotyped on NPY1R/NPY5R with all of the 21 known HapMap SNPs (MAF≥0.10) in the 31 kb region was 0.80; r2 > 0.5 for 81% of the SNPs and r2 > 0.8 for 67% of the SNPs (Table 2). Again, this is the lower bound of coverage because we genotyped 6 additional SNPs that could not be evaluated.
Association results
Results of association analyses are provided in Table 2. None of the SNPs in NPY, including the Leu7Pro coding SNP rs16139, or in NPY1R were associated with any of the phenotypes.
One SNP in the promoter region of NPY2R (rs6857715) and two SNPs further upstream of the gene (rs4333136 and rs4425326) were associated with alcohol dependence (p<0.03), and two of these with the secondary phenotype of severe withdrawal (Table 2). Five SNPs in this gene, the same three plus rs2342675 and rs17304901, were associated with cocaine dependence (Table 2). The same three SNPs were also associated with comorbid alcohol and cocaine dependence (rs687715, p=0.006; rs4333136, p=0.02; rs4425326, p=0.03; all other SNPs were non-significant (p>0.07), data not shown). To determine whether the significant evidence of association for alcohol dependence was due to the subset of affected individuals who also met criteria for cocaine dependence (208 of the 753; Table 1), the analysis was repeated using only alcohol dependent individuals who were not cocaine dependent; there was no significant association of alcohol-only dependence with any of the SNPs (p>0.08, data not shown).
Due to the consistency of the association results with the same set of three SNPs, we constructed haplotypes using only SNPs rs4425326 and rs6857715 (r2=0.18); the high r2 (0.98) between SNPs rs4333136 and rs6857715 made use of both redundant. The global haplotype test and several individual haplotypes were significantly associated with alcohol dependence, alcohol dependence with severe withdrawal, alcohol dependence in the absence of cocaine dependence, comorbid alcohol and cocaine dependence, and cocaine dependence (all global haplotypes p<0.04; Table 3). For all five phenotypes, the most frequent haplotype, C-C, was overtransmitted to affected individuals. The complementary, second-most frequent haplotype, T-T, was overtransmitted to individuals who did not meet the criteria.
Table 3.
Association analysis of haplotypes in NPY2R, all entries are p-values using the Pedigree Disequilibrium Test, average statistic
| rs4425326 nucleotide |
rs6857715 nucleotide |
Alcohol Dependence |
Severe Withdrawal |
Alcohol Dependence Without Cocaine Dependence |
Comorbid Alcohol with Cocaine dependence |
Cocaine Dependence |
|---|---|---|---|---|---|---|
| C | C | 0.0002 | 0.0006 | 0.01 | 0.002 | 0.002 |
| C | T | 0.32 | 0.27 | 0.50 | 0.28 | 0.25 |
| T | C | 0.19 | 0.15 | 0.29 | 0.52 | 0.62 |
| T | T | 0.006 | 0.009 | 0.08 | 0.01 | 0.02 |
| Global test | 0.0004 | 0.0009 | 0.04 | 0.005 | 0.005 |
Four SNPs in NPY5R were associated with the secondary phenotype of withdrawal with seizures: rs4475104, rs4314240, rs4632602, and rs7678265 (p≤0.05). SNPs rs9996227 and rs11946004 approached significance (p≤0.06).
Discussion
This study is the first extensive examination of the association between the neuropeptide Y system and alcohol dependence, alcohol withdrawal and cocaine dependence. We took a systems approach and analyzed not only the NPY gene but also three of its receptor genes. The strongest association we found was between a haplotype in NPY2R with alcohol dependence as well as with several subphenotypes, including alcohol withdrawal symptoms, alcohol dependence without comorbid cocaine dependence, comorbid alcohol and cocaine dependence, and cocaine dependence. For each of the phenotypes analyzed, the same haplotype was preferentially transmitted to alcohol dependent individuals, with the strongest association being with all alcohol dependent individuals (the numerically largest group) and those suffering from alcohol withdrawal. Due to ascertainment criteria for the sample, we had insufficient power to test for an association of NPY2R and cocaine dependence in subjects who were not also alcohol dependent. Our results suggest that sequence variations in the NPY system, specifically in NPY2R, are associated with alcohol dependence characterized by severe withdrawal, as was reported in humans by Koehnke et al (2002) and Okubo and Harada (Okubo and Harada, 2001) and by numerous findings in the animal literature (for a review, see Thiele et al, 2004).
Neither NPY1R nor NP5YR were associated with alcohol dependence or cocaine dependence. However, NPY5R was associated with the phenotype of alcohol withdrawal with seizures, representing a small but severely affected subset of alcoholics.
We found no association of alcohol dependence with any SNPs in NPY, which is consistent with some previously reported results (Hu et al., 2005; Mottagui-Tabar et al., 2005; Zhu et al., 2003). The bulk of evidence of the evidence for a role of the entire NPY system in alcohol-related phenotypes comes from animal models. While alcohol preference and consumption in rats and mice mimic similar traits of alcohol dependence in humans, their equivalence is still unclear (Crabbe, 2007). Our examination of the secondary phenotype of alcohol withdrawal provides another means to examine the nature of the association of these SNPs in the NPY system with alcohol dependence and provide important insights regarding disease heterogeneity.
Although our primary hypothesis was that genes in the NPY system were associated with alcohol dependence, we analyzed additional phenotypes related to alcoholism that were suggested by the literature. The withdrawal phenotypes each consist of a subset of the alcohol dependent subjects, as does the phenotype of comorbid alcohol and cocaine dependence; thus they are not independent phenotypes, but phenotypes nested within alcohol dependence, analyzed to better understand what aspect of alcoholism is most affected by these genes. The cocaine dependence phenotype was considered a secondary analysis, and while it includes many subjects who also meet criteria for alcohol dependence, it is not a nested subset. Therefore, we are testing two phenotypes (alcohol dependence and cocaine dependence) and we have considered the strength of our association results if we were to apply a conservative Bonferroni correction (0.05/2 = 0.025). Applying this correction, we would still identify one SNP in NPY2R which is significantly associated with alcohol dependence and with comorbid alcohol and cocaine dependence, two SNPs in NPY5R which are significantly associated with withdrawal with seizures and four SNPs in NPY2R that are significantly associated with cocaine dependence. The results from haplotype analyses of NPY2R are even stronger (Table 3).
There are several strengths of this study. The sample is based on 1,923 individuals from 219 extended families, with a wealth of reliable and valid information obtained on each individual through the well-characterized SSAGA (Bucholz et al., 1994; Hesselbrock et al., 1999). This large sample was limited to European-Caucasian, non-Hispanic families, thus limiting heterogeneity of the haplotypes used in analyses. The use of family-based association tests reduced potential confounding from population stratification. Finally 39 SNPs were genotyped across NPY and its three receptor genes on chromosome 4, NPY2R, NPY1R, and NPY5R, all with moderate LD to establish extensive coverage of all genes.
In summary, using a family-based association test in extended alcoholic pedigrees, we found evidence of association of SNPs in the NPY receptor genes NPY2R, and NPY5R with alcohol dependence, comorbid alcohol and cocaine dependence, alcohol withdrawal, and cocaine dependence phenotypes which were identified previously in the animal literature. These results indicate that sequence variations in NPY receptor genes are associated with alcohol dependence, particularly a severe subtype of alcohol dependence characterized by withdrawal symptoms, comorbid alcohol and cocaine dependence or cocaine dependence.
Acknowledgements
The Collaborative Study on the Genetics of Alcoholism (COGA), Co-Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes nine different centers where data collection, analysis, and storage take place. The nine sites and Principal Investigators and Co-Investigators are: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., P.M. Conneally, T. Foroud); University of Iowa (S. Kuperman, R. Crowe); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, A. Goate, J. Rice); University of California at San Diego (M. Schuckit); Howard University (R. Taylor); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy). Zhaoxia Ren serves as the NIAAA Staff Collaborator. This national collaborative study is supported by the NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
Genotyping facilities were provided by the Center for Medical Genomics at Indiana University School of Medicine, supported in part by the Indiana Genomics Initiative (INGEN, supported in part by the Lilly Endowment, Inc.). We thank Gayathri Rajan and Rachel Thowe for their superb technical support on SNP genotyping.
In memory of Henri Begleiter and Theodore Reich, Principal and Co-Principal Investigators of COGA since its inception; we are indebted to their leadership in the establishment and nurturing of COGA, and acknowledge with great admiration their seminal scientific contributions to the field.
Reference List
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders DSM-III-R. 3rd ed. Washington, DC: American Psychiatric Association; 1987. [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders DSM-IV. 4th ed. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- Badia-Elder NE, Gilpin NW, Stewart RB. Neuropeptide Y modulation of ethanol intake: effects of ethanol drinking history and genetic background. Peptides. 2007;28:339–344. doi: 10.1016/j.peptides.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Badia-Elder NE, Stewart RB, Powrozek TA, Murphy JM, Li TK. Effects of neuropeptide Y on sucrose and ethanol intake and on anxiety-like behavior in high alcohol drinking (HAD) and low alcohol drinking (LAD) rats. Alcohol Clin Exp Res. 2003;27:894–899. doi: 10.1097/01.ALC.0000071929.17974.DA. [DOI] [PubMed] [Google Scholar]
- Badia-Elder NE, Stewart RB, Powrozek TA, Roy KF, Murphy JM, Li TK. Effect of neuropeptide Y (NPY) on oral ethanol intake in Wistar, alcohol-preferring (P), and - nonpreferring (NP) rats. Alcohol Clin Exp Res. 2001;25:386–390. [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B, Wang W. Event-related brain potentials differentiate priming and recognition to familiar and unfamiliar faces. Electroencephalogr Clin Neurophysiol. 1995;94:41–49. doi: 10.1016/0013-4694(94)00240-l. [DOI] [PubMed] [Google Scholar]
- Bison S, Crews F. Alcohol withdrawal increases neuropeptide Y immunoreactivity in rat brain. Alcohol Clin Exp Res. 2003;27:1173–1183. doi: 10.1097/01.ALC.0000075827.74538.FE. [DOI] [PubMed] [Google Scholar]
- Boehnke M. Allele frequency estimation from data on relatives. Am J Hum Genet. 1991;48:22–25. [PMC free article] [PubMed] [Google Scholar]
- Bonsch D, Greifenberg V, Bayerlein K, Biermann T, Reulbach U, Hillemacher T, Kornhuber J, Bleich S. Alpha-synuclein protein levels are increased in alcoholic patients and are linked to craving. Alcohol Clin Exp Res. 2005a;29:763–765. doi: 10.1097/01.alc.0000164360.43907.24. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Lederer T, Reulbach U, Hothorn T, Kornhuber J, Bleich S. Joint analysis of the NACP-REP1 marker within the alpha synuclein gene concludes association with alcohol dependence. Hum Mol Genet. 2005b;14:967–971. doi: 10.1093/hmg/ddi090. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Lenz B, Kornhuber J, Bleich S. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005c;16:167–170. doi: 10.1097/00001756-200502080-00020. [DOI] [PubMed] [Google Scholar]
- Bonsch D, Reulbach U, Bayerlein K, Hillemacher T, Kornhuber J, Bleich S. Elevated alpha synuclein mRNA levels are associated with craving in patients with alcoholism. Biol Psychiatry. 2004;56:984–986. doi: 10.1016/j.biopsych.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Boutrel B, Kenny PJ, Specio SE, Martin-Fardon R, Markou A, Koob GF, de LL. Role for hypocretin in mediating stress-induced reinstatement of cocaine-seeking behavior. Proc Natl Acad Sci USA. 2005;102:19168–19173. doi: 10.1073/pnas.0507480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Caberlotto L, Thorsell A, Rimondini R, Sommer W, Hyytia P, Heilig M. Differential expression of NPY and its receptors in alcohol-preferring AA and alcohol-avoiding ANA rats. Alcohol Clin Exp Res. 2001;25:1564–1569. [PubMed] [Google Scholar]
- Carr LG, Foroud T, Bice P, Gobbett T, Ivashina J, Edenberg H, Lumeng L, Li TK. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998;22:884–887. [PubMed] [Google Scholar]
- Clark JT, Kalra PS, Crowley WR, Kalra SP. Neuropeptide Y and human pancreatic polypeptide stimulate feeding behavior in rats. Endocrinology. 1984;115:427–429. doi: 10.1210/endo-115-1-427. [DOI] [PubMed] [Google Scholar]
- Cloninger CR, Bohman M, Sigvardsson S. Inheritance of alcohol abuse. Cross-fostering analysis of adopted men. Arch Gen Psychiatry. 1981;38:861–868. doi: 10.1001/archpsyc.1981.01780330019001. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2004;129:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Crabbe J. Beyond preference: efforts to improve rodent oral self-administration models; Presentation at the 30th Annual Scientific Meeting of the Research Society on Alcoholism; July 7–12, 2007; Chicago IL. 2007. [Google Scholar]
- Cowen MS, Chen F, Lawrence AJ. Neuropeptides: implications for alcoholism. Neurochem. 2004;89:273–185. doi: 10.1111/j.1471-4159.2004.02394.x. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- de Quidt ME, Emson PC. Distribution of neuropeptide Y-like immunoreactivity in the rat central nervous system--II. Immunohistochemical analysis. Neuroscience. 1986;18:545–618. doi: 10.1016/0306-4522(86)90057-6. [DOI] [PubMed] [Google Scholar]
- Dick DM, Edenberg HJ, Xuei X, Goate A, Kuperman S, Schuckit M, Crowe R, Smith TL, Porjesz B, Begleiter H, Foroud T. Association of GABRG3 with alcohol dependence. Alcohol Clin Exp Res. 2004;28:4–9. doi: 10.1097/01.ALC.0000108645.54345.98. [DOI] [PubMed] [Google Scholar]
- Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr, O'Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Wang J, Tian H, Pochareddy S, Xuei X, Wetherill L, Goate A, Hinrichs T, Kuperman S, Nurnberger JI, Jr, Schuckit M, Tischfield JA, Foroud T. A regulatory variation in OPRK1, the gene encoding the {kappa}-opioid receptor, is associated with alcohol dependence. Hum Mol Genet epub. 2008 doi: 10.1093/hmg/ddn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Wetherill LF, Bierut L, Bucholz K, Dick DM, Hesselbrock V, Kuperman S, Porjesz B, Schuckit MA, Tischfield JA, Almasy LA, Nurnberger JI, Jr, Foroud T. Association of NFKB1, which encodes a subunit of the transcription factor NF-{kappa}B, with Alcohol Dependence. Hum Mol Genet Dec 12. 2007 doi: 10.1093/hmg/ddm368. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Li TK, Lumeng L, Hwang BH, Somes C, Jimenez P, Mathe AA. Neuropeptide Y levels in ethanol-naive alcohol-preferring and nonpreferring rats and in Wistar rats after ethanol exposure. Alcohol Clin Exp Res. 1998;22:1778–1782. [PubMed] [Google Scholar]
- Eva C, Serra M, Mele P, Panzica G, Oberto A. Physiology and gene regulation of the brain NPY Y1 receptor. Front Neuroendocrinol. 2006;27:308–339. doi: 10.1016/j.yfrne.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, Dahmen N, Schmidt LG, Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- Feighner JP, Robins E, Guze SB, Woodruff RA, Jr, Winokur G, Munoz R. Diagnostic criteria for use in psychiatric research. Arch Gen Psychiatry. 1972;26:57–63. doi: 10.1001/archpsyc.1972.01750190059011. [DOI] [PubMed] [Google Scholar]
- Files FJ, Andrews CM, Samson HH, Lumeng L, Li TK. Alcohol self-administration in a nonrestricted access situation with alcohol-preferring (P) rats. Alcohol Clin Exp Res. 1992;16:751–756. doi: 10.1111/j.1530-0277.1992.tb00672.x. [DOI] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, Crowe R, Schuckit M, Porjesz B, Begleiter H, Reich T. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–945. [PubMed] [Google Scholar]
- Foroud T, Wetherill LF, Dick DM, Hesselbrock V, Nurnberger JI, Jr, Kramer J, Tischfield J, Schuckit M, Bierut LJ, Xuei X, Edenberg HJ. Lack of association of alcohol dependence and habitual smoking with catechol-O-methyltransferase (COMT) Alcohol Clin Exp Res. 2007;31:1773–1779. doi: 10.1111/j.1530-0277.2007.00505.x. [DOI] [PubMed] [Google Scholar]
- Gilpin NW, Stewart RB, Murphy JM, Li TK, Badia-Elder NE. Neuropeptide Y reduces oral ethanol intake in alcohol-preferring (P) rats following a period of imposed ethanol abstinence. Alcohol Clin Exp Res. 2003;27:787–794. doi: 10.1097/01.ALC.0000065723.93234.1D. [DOI] [PubMed] [Google Scholar]
- Goodman JH, Sloviter RS. Cocaine neurotoxicity and altered neuropeptide Y immunoreactivity in the rat hippocampus; a silver degeneration and immunocytochemcial study. Brain Research. 1993;616:263–272. doi: 10.1016/0006-8993(93)90217-b. [DOI] [PubMed] [Google Scholar]
- Gray TS, Morley JE. Neuropeptide Y: anatomical distribution and possible function in mammalian nervous system. Life Sci. 1986;38:389–401. doi: 10.1016/0024-3205(86)90061-5. [DOI] [PubMed] [Google Scholar]
- Guindalini C, Scivoletto S, Ferreira RG, Breen G, Zilberman M, Peluso MA, Zatz M. Association of genetic variants in alcohol dehydrogenase 4 with alcohol dependence in Brazilian patients. Am J Psychiatry. 2005;162:1005–1007. doi: 10.1176/appi.ajp.162.5.1005. [DOI] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch Gen Psychiatry. 2007;64:830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Heilig M, Thorsell A. Brain neuropeptide Y (NPY) in stress and alcohol dependence. Rev Neurosci. 2002;13:85–94. doi: 10.1515/revneuro.2002.13.1.85. [DOI] [PubMed] [Google Scholar]
- Heilig M, Widerlov E. Neurobiology and clinical aspects of neuropeptide Y. Crit Rev Neurobiol. 1995;9:115–136. [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94:1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Hinrichs AL, Wang JC, Bufe B, Kwon JM, Budde J, Allen R, Bertelsen S, Evans W, Dick D, Rice J, Foroud T, Nurnberger J, Tischfield JA, Kuperman S, Crowe R, Hesselbrock V, Schuckit M, Almasy L, Porjesz B, Edenberg HJ, Begleiter H, Meyerhof W, Bierut LJ, Goate AM. Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence. Am J Hum Genet. 2006;78:103–111. doi: 10.1086/499253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Oroszi G, Chun J, Smith TL, Goldman D, Schuckit MA. An expanded evaluation of the relationship of four alleles to the level of response to alcohol and the alcoholism risk. Alcohol Clin Exp Res. 2005;29:8–16. doi: 10.1097/01.alc.0000150008.68473.62. [DOI] [PubMed] [Google Scholar]
- Ilveskoski E, Kajander OA, Lehtimaki T, Kunnas T, Karhunen PJ, Heinala P, Virkkunen M, Alho H. Association of neuropeptide y polymorphism with the occurrence of type 1 and type 2 alcoholism. Alcohol Clin Exp Res. 2001;25:1420–1422. doi: 10.1097/00000374-200110000-00003. [DOI] [PubMed] [Google Scholar]
- Jolicoeur FB, Michaud JN, Rivest R, Menard D, Gaudin D, Fournier A, St-Pierre S. Neurobehavioral profile of neuropeptide Y. Brain Res Bull. 1991;26:265–268. doi: 10.1016/0361-9230(91)90237-e. [DOI] [PubMed] [Google Scholar]
- Karvonen MK, Pesonen U, Koulu M, Niskanen L, Laakso M, Rissanen A, Dekker JM, Hart LM, Valve R, Uusitupa MI. Association of a leucine(7)-to-proline(7) polymorphism in the signal peptide of neuropeptide Y with high serum cholesterol and LDL cholesterol levels. Nat Med. 1998;4:1434–1437. doi: 10.1038/4027. [DOI] [PubMed] [Google Scholar]
- Kauhanen J, Karvonen MK, Pesonen U, Koulu M, Tuomainen TP, Uusitupa MI, Salonen JT. Neuropeptide Y polymorphism and alcohol consumption in middle-aged men. Am J Med Genet. 2000;93:117–121. doi: 10.1002/1096-8628(20000717)93:2<117::aid-ajmg7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. A twin-family study of alcoholism in women. Am J Psychiatry. 1994;151:707–715. doi: 10.1176/ajp.151.5.707. [DOI] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring and - non-preferring rats in five brain regions. Alcohol. 2007;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehnke MD, Schick S, Lutz U, Willecke M, Koehnke AM, Kolb W, Gaertner I. Severity of alcohol withdrawal symptoms and the T1128C polymorphism of the neuropeptide Y gene. J Neural Transm. 2002;109:1423–1429. doi: 10.1007/s00702-002-0752-1. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Kranzler HR, Malison R, Price LH, Van DC, Rosenheck RA, Cramer J, Southwick S, Charney D, Krystal J, Gelernter J. A functional neuropeptide Y Leu7Pro polymorphism associated with alcohol dependence in a large population sample from the United States. Arch Gen Psychiatry. 2002;59:825–831. doi: 10.1001/archpsyc.59.9.825. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, Somberg LK, Covault J, Kranzler HR, Krystal JH, Gelernter J. Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcohol Clin Exp Res. 2005;29:493–498. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- Li TK, Hewitt BG, Grant BF. The Alcohol Dependence Syndrome, 30 years later: a commentary. The 2006 H. David Archibald lecture. Addiction. 2007;102:1522–1530. doi: 10.1111/j.1360-0443.2007.01911.x. [DOI] [PubMed] [Google Scholar]
- Li TK, Lumeng L, Doolittle DP. Selective breeding for alcohol preference and associated responses. Behav Genet. 1993;23:163–170. doi: 10.1007/BF01067421. [DOI] [PubMed] [Google Scholar]
- Li TK, Lumeng L, Doolittle DP, Carr LG. Molecular associations of alcohol-seeking behavior in rat lines selectively bred for high and low voluntary ethanol drinking. Alcohol Alcohol Suppl. 1991;1:121–124. [PubMed] [Google Scholar]
- Liang T, Carr LG. Regulation of alpha-synuclein expression in alcohol-preferring and-non preferring rats. J Neurochem. 2006;99:470–482. doi: 10.1111/j.1471-4159.2006.04111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Spence J, Liu L, Strother WN, Chang HW, Ellison JA, Lumeng L, Li TK, Foroud T, Carr LG. alpha-Synuclein maps to a quantitative trait locus for alcohol preference and is differentially expressed in alcohol-preferring and -nonpreferring rats. Proc Natl Acad Sci USA. 2003;100:4690–4695. doi: 10.1073/pnas.0737182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Blumberg HP, Gelernter J. CHRM2 gene predisposes to alcohol dependence, drug dependence and affective disorders: results from an extended case-control structured association study. Hum Mol Genet. 2005a;14:2421–2434. doi: 10.1093/hmg/ddi244. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence: results from family controlled and population-structured association studies. Pharmacogenet Genomics. 2005b;15:755–768. doi: 10.1097/01.fpc.0000180141.77036.dc. [DOI] [PubMed] [Google Scholar]
- Lutz CM, Richards JE, Scott KL, Sinha S, Yang-Feng TL, Frankel WN, Thompson DA. Neuropeptide Y receptor genes mapped in human and mouse: receptors with high affinity for pancreatic polypeptide are not clustered with receptors specific for neuropeptide Y and peptide YY. Genomics. 1997;46:287–290. doi: 10.1006/geno.1997.5024. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Kaplan NL. Correcting for a potential bias in the pedigree disequilibrium test. Am J Hum Genet. 2001;68:1065–1067. doi: 10.1086/319525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- McGue M. Phenotyping alcoholism. Alcohol Clin Exp Res. 1999;23:757–758. doi: 10.1111/j.1530-0277.1999.tb04180.x. [DOI] [PubMed] [Google Scholar]
- Menyhert J, Wittmann G, Lechan RM, Keller E, Liposits Z, Fekete C. Cocaine- and amphetamine-regulated transcript (CART) is colocalized with the orexigenic neuropeptide Y and agouti-related protein and absent from the anorexigenic alpha-melanocyte-stimulating hormone neurons in the infundibular nucleus of the human hypothalamus. Endocrinology. 2007;148:4276–4281. doi: 10.1210/en.2007-0390. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S, Prince JA, Wahlestedt C, Zhu G, Goldman D, Heilig M. A novel single nucleotide polymorphism of the neuropeptide Y (NPY) gene associated with alcohol dependence. Alcohol Clin Exp Res. 2005;29:702–707. doi: 10.1097/01.alc.0000164365.04961.b1. [DOI] [PubMed] [Google Scholar]
- Murphy JM, Stewart RB, Bell RL, Badia-Elder NE, Carr LG, McBride WJ, Lumeng L, Li TK. Phenotypic and genotypic characterization of the Indiana University rat lines selectively bred for high and low alcohol preference. Behav Genet. 2002;32:363–388. doi: 10.1023/a:1020266306135. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Wiegand R, Bucholz K, O'onnor S, Meyer ET, Reich T, Rice J, Schuckit M, King L, Petti T, Bierut L, Hinrichs AL, Kuperman S, Hesselbrock V, Porjesz B. A family study of alcohol dependence: coaggregation of multiple disorders in relatives of alcohol-dependent probands. Arch Gen Psychiatry. 2004;61:1246–1256. doi: 10.1001/archpsyc.61.12.1246. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo T, Harada S. Polymorphism of the neuropeptide Y gene: an association study with alcohol withdrawal. Alcohol Clin Exp Res. 2001;25:59S–62S. doi: 10.1097/00000374-200106001-00014. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Carr LG, Heilig M, Ilveskoski E, Thiele TE. Neuropeptide y and alcoholism: genetic, molecular, and pharmacological evidence. Alcohol Clin Exp Res. 2003;27:149–154. doi: 10.1097/01.ALC.0000052706.21367.0E. [DOI] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, Lykken DT, Heston LL, Clayton PJ. Heterogeneity in the inheritance of alcoholism. A study of male and female twins. Arch Gen Psychiatry. 1991;48:19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- Pluzarev O, Crews FT. NPY immunoreactivity is decreased in the amygdala of human alcoholics; Presentation at the 30th Annual Scientific Meeting of the Research Society on Alcoholism; July 7–12, 2007; Chicago IL. 2007. [Google Scholar]
- Reich T. A genomic survey of alcohol dependence and related phenotypes: results from the Collaborative Study on the Genetics of Alcoholism (COGA) Alcohol Clin Exp Res. 1996;20:133A–137A. doi: 10.1111/j.1530-0277.1996.tb01763.x. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van EP, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Roy A, Pandey SC. The decreased cellular expression of neuropeptide Y protein in rat brain structures during ethanol withdrawal after chronic ethanol exposure. Alcohol Clin Exp Res. 2005;26:796–803. [PubMed] [Google Scholar]
- Schroeder JP, Iller KA, Hodge CW. Neuropeptide-Y Y5 receptors modulate the onset and maintenance of operant ethanol self-administration. Alcohol Clin Exp Res. 2003;27:1912–1920. doi: 10.1097/01.ALC.0000098873.80433.BA. [DOI] [PubMed] [Google Scholar]
- Spence JP, Liang T, Habegger K, Carr LG. Effect of polymorphism on expression of the neuropeptide Y gene in inbred alcohol-preferring and -nonpreferring rats. Neuroscience. 2005;131:871–876. doi: 10.1016/j.neuroscience.2004.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundler F, Hakanson R, Ekblad E, Uddman R, Wahlestedt C. Neuropeptide Y in the peripheral adrenergic and enteric nervous systems. Int Rev Cytol. 1986;102:243–269. doi: 10.1016/s0074-7696(08)61277-2. [DOI] [PubMed] [Google Scholar]
- Tecott LH, Heberlein U. Y do we drink? Cell. 1998;95:733–735. doi: 10.1016/s0092-8674(00)81695-5. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Marsh DJ, Ste Marie L, Bernstein IL, Palmiter RD. Ethanol consumption and resistance are inversely related to neuropeptide Y levels. Nature. 1998;396:313–314. doi: 10.1038/24614. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Badia-Elder NE. A role for neuropeptide Y in alcohol intake control: evidence from human and animal research. Physiol Behav. 2003;79:95–101. doi: 10.1016/s0031-9384(03)00109-4. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Naveilhan P, Ernfors P. Assessment of ethanol consumption and water drinking by NPY Y(2) receptor knockout mice. Peptides. 2004a;25:975–983. doi: 10.1016/j.peptides.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Sparta DR, Hayes DM, Fee JR. A role for neuropeptide Y in neurobiological responses to ethanol and drugs of abuse. Neuropeptides. 2004b;38:235–243. doi: 10.1016/j.npep.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Thorsell A. Neuropeptide Y (NPY) in alcohol intake and dependence. Peptides. 2007;28:480–483. doi: 10.1016/j.peptides.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Thorsell A, Heilig M. Diverse functions of neuropeptide Y revealed using genetically modified animals. Neuropeptides. 2002;36:182–193. doi: 10.1054/npep.2002.0897. [DOI] [PubMed] [Google Scholar]
- Thorsell A, Repunte-Canonigo V, O'Dell LE, Chen SA, King AR, Lekic D, Koob GF, Sanna PP. Viral vector-induced amygdala NPY overexpression reverses increased alcohol intake caused by repeated deprivations in Wistar rats. Brain. 2007;130:1330–1337. doi: 10.1093/brain/awm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez GR, Koob GF. Allostasis and dysregulation of corticotropin-releasing factor and neuropeptide Y systems: implications for the development of alcoholism. Pharmacol Biochem Behav. 2004;79:671–689. doi: 10.1016/j.pbb.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Wahlestedt C, Karoum F, Jaskiw G, Wyatt RJ, Larhammar D, Ekman R, Reis DJ. Cocaine-induced reduction of brain neuropeptide Y synthesis dependent on medial prefrontal cortex. Proc Natl Acad Sci U S A. 1991;88:2078–2082. doi: 10.1073/pnas.88.6.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Hinrichs AL, Stock H, Budde J, Allen R, Bertelsen S, Kwon JM, Wu W, Dick DM, Rice J, Jones K, Nurnberger JI, Jr, Tischfield J, Porjesz B, Edenberg HJ, Hesselbrock V, Crowe R, Schuckit M, Begleiter H, Reich T, Goate AM, Bierut LJ. Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Hum Mol Genet. 2004;13:1903–1911. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, Van EP, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet. 1999;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldbye DP, Greisen MH, Bolwig TG, Larsen PJ, Mikkelsen JD. Prolonged induction of c-fos in neuropeptide Y- and somatostatin-immunoreactive neurons of the rat dentate gyrus after electroconvulsive stimulation. Brain Res. 1996;720:111–119. doi: 10.1016/0006-8993(96)00158-8. [DOI] [PubMed] [Google Scholar]
- Woldbye DP, Ulrichsen J, Haugbol S, Bolwig TG. Ethanol withdrawal in rats is attenuated by intracerebroventricular administration of neuropeptide Y. Alcohol Alcohol. 2002;37:318–321. doi: 10.1093/alcalc/37.4.318. [DOI] [PubMed] [Google Scholar]
- Wraith A, Tornsten A, Chardon P, Harbitz I, Chowdhary BP, Andersson L, Lundin LG, Larhammar D. Evolution of the neuropeptide Y receptor family: gene and chromosome duplications deduced from the cloning and mapping of the five receptor subtype genes in pig. Genome Res. 2000;10:302–310. doi: 10.1101/gr.10.3.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarjevski N, Cusin I, Vettor R, Rohner-Jeanrenaud F, Jeanrenaud B. Chronic intracerebroventricular neuropeptide-Y administration to normal rats mimics hormonal and metabolic changes of obesity. Endocrinology. 1993;133:1753–1758. doi: 10.1210/endo.133.4.8404618. [DOI] [PubMed] [Google Scholar]
- Zhu G, Pollak L, Mottagui-Tabar S, Wahlestedt C, Taubman J, Virkkunen M, Goldman D, Heilig M. NPY Leu7Pro and alcohol dependence in Finnish and Swedish populations. Alcohol Clin Exp Res. 2003;27:19–24. doi: 10.1097/01.ALC.0000050642.62233.44. [DOI] [PubMed] [Google Scholar]
- Zill P, Preuss UW, Koller G, Bondy B, Soyka M. Analysis of single nucleotide polymorphisms and haplotypes in the neuropeptide Y gene: no evidence for association with alcoholism in a German population sample. Alcohol Clin Exp Res. 2008;32:430–434. doi: 10.1111/j.1530-0277.2007.00586.x. [DOI] [PubMed] [Google Scholar]