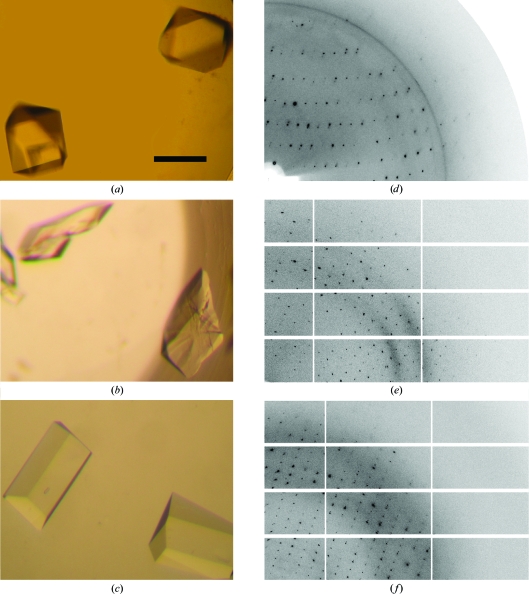
Two C-terminal fragments of the RNA helicase Hera have been crytallized in three crystal forms, one of which was phased by MAD using a single selenium site.
Keywords: Hera, Thermus thermophilus, DEAD-box RNA helicases, ATPases
Abstract
Heat-resistant RNA-dependent ATPase (Hera) from Thermus thermophilus is a DEAD-box RNA helicase. Two constructs encompassing the second RecA-like domain and the C-terminal domain of Hera were overproduced in Escherichia coli and purified to homogeneity. Single crystals of both Hera constructs were obtained in three crystal forms. A tetragonal crystal form belonged to space group P41212, with unit-cell parameters a = 65.5, c = 153.0 Å, and contained one molecule per asymmetric unit. Two orthorhombic forms belonged to space group P212121, with unit-cell parameters a = 62.8, b = 70.9, c = 102.3 Å (form I) and a = 41.6, b = 67.6, c = 183.5 Å (form II). Both orthorhombic forms contained two molecules per asymmetric unit. All crystals diffracted X-rays to beyond 3 Å resolution, but the tetragonal data sets displayed high Wilson B values and high mean |E 2 − 1| values, indicating potential disorder and anisotropy. The tetragonal crystal was phased by MAD using a single selenium site.
1. Introduction
Structural changes in RNA or RNA–protein complexes during (viral) replication, transcription, mRNA splicing, translation and ribosome assembly are mediated by RNA helicases (Pyle, 2008 ▶). These enzymes use the chemical energy of ATP to reorganize their substrates. The DEAD-box helicases constitute the largest family within helicase superfamily 2 and are found in all kingdoms of life. RNA helicases are organized as two RecA-like domains that carry signature motifs for ATP and RNA binding and RNA remodelling. In addition, many helicases contain flanking C-terminal domains that contribute to specific or nonspecific high-affinity RNA binding (Diges & Uhlenbeck, 2001 ▶; Tsu et al., 2001 ▶; Kossen et al., 2002 ▶; Mohr et al., 2008 ▶). It has been suggested that these C-terminal domains may also position the RNA substrate with respect to the helicase core (Mohr et al., 2008 ▶; Halls et al., 2007 ▶; Huang et al., 2005 ▶). The opening and closing of a cleft formed by the RecA domains has been found to be crucial for unwinding (Theissen et al., 2008 ▶). Thus, nucleic acid binding and release cycles require large conformational changes in RNA helicases, rendering them flexible and difficult to crystallize in the apo form. The crystal structures of several RNA helicase–RNA complexes in the closed conformation have been determined (Sengoku et al., 2006 ▶; Andersen et al., 2006 ▶; Bono et al., 2006 ▶) and show pronounced similarity, while the open forms of the helicases tend to differ more strongly from each other. Structural information on ancillary RNA-binding domains is limited to the isolated C-terminal domain of the DEAD-box helicase YxiN, which is involved in ribosome biogenesis. The YxiN C-terminal domain forms a classical RNA-recognition motif that mediates specific binding to a hairpin in the 23S ribosomal RNA (Wang et al., 2006 ▶; Kossen et al., 2002 ▶).
The heat-resistant RNA-dependent ATPase Hera was cloned from Thermus thermophilus and some of its biochemical properties have been assessed (Linden et al., 2008 ▶; Morlang et al., 1999 ▶). Hera contains a C-terminal domain that bears no sequence similarity to the C-terminal domains of other RNA helicases. However, it contains a short sequence with a weak similarity to a conserved motif in the RNase P protein component. It has been shown that the C-terminal domain of Hera, but not the putative RNase P motif, is required for RNA binding (Linden et al., 2008 ▶). In addition, gel-permeation chromatography has shown that Hera forms a stable dimer in solution (Linden et al., 2008 ▶), raising the question as to how dimerization might be linked to RNA binding and helicase function.
To begin answering these questions, the gene coding for T. thermophilus HB27 Hera was cloned from genomic DNA (Linden et al., 2008 ▶) and the protein was recombinantly produced in Escherichia coli. Full-length Hera proved resilient to crystallization, necessitating the generation of the N- and C-terminal domains. The crystal structure of the N-terminal Hera domain in complex with the unusual ligand AMP has been determined previously (Rudolph et al., 2006 ▶), but for a complete picture the structure of the C-terminal domain is required. We crystallized two constructs of the C-terminal domain of Hera in three crystal forms and employed SeMet MAD phasing using a single selenium site to generate initial electron-density maps.
2. Methods
2.1. Protein purification and crystallization
The Hera gene was cloned from genomic DNA of T. thermophilus HB27 and the protein was overproduced in E. coli. Purification of full-length Hera is described in detail elsewhere (Linden et al., 2008 ▶). For the production of C-terminal Hera constructs, two DNA fragments encoding Hera residues 208–510 and 208–419 were generated by PCR and cloned into the NcoI/BamHI sites of pET28a (Novagen), thereby removing the His6 tag of the vector. The plasmid coding for Hera_208–510 was transformed into E. coli BL21 (DE3) RIL (Stratagene) and protein production in LB medium was induced at an OD600 of 0.4 with 0.1 mM IPTG for 4 h at 303 K. Similarly, the plasmid coding for Hera_208–419 was transformed into E. coli BL21 (DE3) Rosetta (Stratagene) and protein production was performed in autoinducing medium (Studier, 2005 ▶). Cells were harvested by centrifugation, resuspended in 50 mM Tris–HCl pH 7.5, 100 mM NaCl, 0.1 mM PMSF and disintegrated in a fluidizer (Microfluidics). The supernatant after centrifugation was discarded and the pellet was extracted with 50 mM Tris–HCl pH 7.5, 1 M NaCl. The Hera constructs were precipitated from the supernatant by the addition of 80%(w/v) ammonium sulfate and were redissolved in 50 mM Tris–HCl pH 7.5, 500 mM NaCl. Dialysis against 50 mM Tris–HCl pH 7.5, 200 mM NaCl, 4 M urea was followed by anion-exchange chromatography on Q-Sepharose in the dialysis buffer to remove nucleic acids. The flowthrough was precipitated with 60%(w/v) ammonium sulfate and the pellet was dissolved in 20 mM HEPES–NaOH pH 7.4, 500 mM NaCl for gel-permeation chromatography on Superdex-200 (GE Healthcare) in the same buffer. The proteins were concentrated to 5–15 mg ml−1 by ultrafiltration. Production of the selenomethionine-substituted protein was performed in minimal medium supplemented with amino acids and it was purified identically to the unsubstituted protein. Generation of the Hera_208–510 triple mutant L272M, L281M, L315M was performed using site-directed mutagenesis (QuikChange, Stratagene).
Initial crystallization conditions for Hera_208–510 were obtained from Crystal Screen (Hampton Research) using vapour diffusion and were refined using the microbatch-under-oil setup. 9.3 mg ml−1 protein in 20 mM HEPES–NaOH pH 7.4, 500 mM NaCl was mixed in a 1:2 volume ratio with precipitant consisting of 0.1 M MES–NaOH pH 6.5, 15–18% ethylene glycol, 0.3–0.5 M NaI, 3–6% PEG 20 000. The final drop volumes were varied from 2 to 6 µl as an additional parameter, with larger drop volumes typically resulting in larger crystals. Tetragonal bipyramidal crystals appeared after 1–4 d at 277 K (Fig. 1 ▶ a). Two rhomboid-shaped crystal forms of Hera_208–419 were obtained using the Index I Screen (Hampton Research). These conditions were refined in the microbatch-under-oil setup at 293 K by mixing 10 mg ml−1 protein with precipitant in a 1:2 volume ratio. One form was obtained from 0.1 M Tris–HCl pH 8.5, 0.2 M ammonium sulfate, 23% PEG 3350 (Fig. 1 ▶ b), while another form grew from 0.1 M Tris–HCl pH 7.0, 0.2 M ammonium sulfate, 25% PEG 3350, 5–10%(w/v) glucose or sucrose (Fig. 1 ▶ c).
Figure 1.
(a) Tetragonal Hera_208–510 crystals. (b) Orthorhombic Hera_208–419 crystals (form I). (c) Orthorhombic Hera_208–419 crystals (form II). The scale bar in (a) is 0.2 mm and applies to all crystal images. (d), (e) and (f) show diffraction patterns of the crystals in (a), (b) and (c), respectively. Resolution limits: (d) 3.7 Å (top) and 2.6 Å (right); (e) 3.0 Å (top) and 2.4 Å (right); (f) 3.0 Å (top) and 2.0 Å (right).
2.2. Data collection and analysis
Crystals were initially obtained by vapour diffusion but grew very rapidly, sometimes within a matter of seconds after setup, and did not diffract X-rays beyond 4 Å resolution. Crystals from microbatch under oil grew much more slowly and tended to exhibit better order as judged by X-ray diffraction. Crystals were cryoprotected by replacing the oil from the microbatch experiment with dry paraffin oil, dragging the crystal through the oil layer using a loop and hyperquenching in liquid nitrogen (Warkentin et al., 2006 ▶). Cystals were mounted in an arbitrary orientation and data were collected on BESSY beamline BL-1 using a MAR CCD detector (tetragonal form) and on SLS beamline X06SA using a PILATUS detector (orthorhombic forms). Data reduction of the tetragonal crystal form was performed with the HKL suite of programs (Otwinowski & Minor, 1997 ▶) and data from the orthorhombic forms were integrated with XDS (Kabsch, 1993 ▶) and scaled with SADABS (Bruker). Data statistics for the tetragonal MAD data sets and the native orthorhombic data sets are collected in Tables 1 ▶ and 2 ▶, respectively. Indexing of the diffraction pattern of the tetragonal crystals was consistent with a primitive lattice of Laue group 4/mmm containing one molecule per asymmetric unit (Fig. 1 ▶ d). Analysis of the systematic absences allowed space group P41212 or its enantiomorph. The diffraction patterns of the orthorhombic crystals (Figs. 1 ▶ e and 1 ▶ f) indexed in a primitive lattice. Systematically absent reflections established the space group as P212121 in both cases but with different unit-cell parameters. There are two molecules per asymmetric unit in the orthorhombic crystal forms.
Table 1. MAD X-ray data-collection statistics for the tetragonal crystal form.
Values in parentheses are for the highest resolution shell.
| Data set | Peak | High remote | Low remote | Inflection |
|---|---|---|---|---|
| Wavelength (Å) | 0.9797 | 0.9770 | 0.9832 | 0.9799 |
| Unit-cell parameters (Å) | a = 65.5, c = 153.0 | |||
| Space group | P41212 | |||
| Wilson B value (Å2) | 92 | |||
| Mean |E2 − 1| | 0.80 | |||
| Resolution range (Å) | 49.8–2.9 (3.0–2.9) | 29.5–2.9 (3.0–2.9) | 29.5–2.6 (2.7–2.6) | 29.5–2.9 (3.0–2.9) |
| Unique reflections† | 13014 | 13443 | 19299 | 13037 |
| Completeness† (%) | 92.5 (61.4) | 94.6 (71.8) | 99.0 (92.8) | 92.0 (64.8) |
| Multiplicity† | 6.2 (1.9) | 6.5 (3.0) | 8.3 (3.9) | 6.5 (2.4) |
| Mean I/σ(I)† | 21.7 (1.9) | 22.6 (2.9) | 24.4 (2.8) | 22.2 (2.3) |
| Rmeas‡ | 0.078 (0.620) | 0.063 (0.635) | 0.082 (0.805) | 0.057 (0.660) |
For anomalous data.
The redundancy-independent R factor on intensities R meas was calculated according to Diederichs & Karplus (1997 ▶).
Table 2. X-ray data-collection statistics for the orthorhombic crystal forms.
Values in parentheses are for data in the highest resolution shell.
| Data set | Form I | Form II |
|---|---|---|
| Wavelength (Å) | 1.0 | 1.0 |
| Space group | P212121 | P212121 |
| Unit-cell parameters (Å) | a = 62.8, b = 70.9, c = 102.3 | a = 41.6, b = 67.6, c = 183.5 |
| Matthews coefficient (Å3 Da−1) | 2.42 | 2.75 |
| Solvent content (%) | 49.3 | 55.2 |
| Wilson B value (Å2) | 58 | 46 |
| Mean |E2 − 1| | 0.80 | 0.93 |
| Resolution range (Å) | 47.0–2.6 (2.78–2.60) | 45.9–2.24 (2.38–2.24) |
| Unique reflections | 17245 (2026) | 25476 (2630) |
| Completeness (%) | 93.4 (77.8) | 87.1 (64.6) |
| Multiplicity | 6.4 (6.6) | 6.0 (4.5) |
| Mean I/σ(I) | 17.6 (2.6) | 21.7 (2.4) |
| Rmeas | 0.075 (0.778) | 0.045 (0.593) |
3. Results and discussion
3.1. Phasing using MAD on a single selenium site
An anomalous difference Patterson map at Harker section w = 0.25 calculated to 3 Å resolution showed a significant peak of >5σ, indicating successful SeMet substitution (Fig. 2 ▶). The substructure consisting of a single selenium site was determined with SOLVE (Terwilliger & Berendzen, 1999 ▶). This solution displayed an overall figure of merit (FOM) of 0.34 at 2.9 Å resolution but did not result in an interpretable electron-density map.
Figure 2.
Anomalous difference Patterson map at section w = 0.25 calculated to a resolution of 3 Å showing the single peak at u = 0.42, v = 0.30 for SeMet242 with fractional coordinates (0.776, 0.138, 0.116). Contours are in steps of 1σ and are coloured differently for each level.
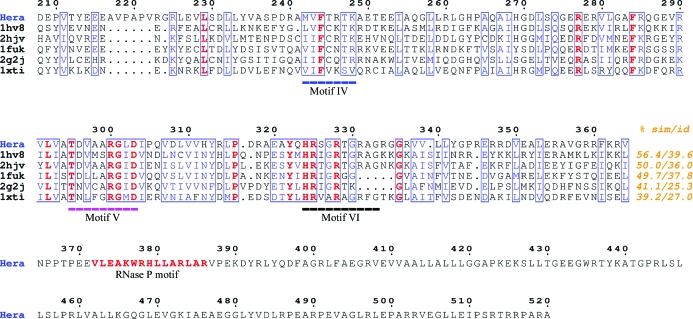
In order to improve the experimental phases, two main strategies were pursued: the exploitation of spurious anomalous signal from halide ions and the incorporation of further Se sites by site-directed mutagenesis. The presence of iodide in the crystallization mother liquor (calculated f′ = −2.3 e−, f′′ = 3.1 e− at the peak wavelength) could in principle allow a mixed Se/I substructure. Searches using SOLVE or SHELXD (Sheldrick, 2008 ▶) did not result in additional sites, although substructures of different chemical composition have previously been used for phasing (Roeser et al., 2005 ▶). In contrast, crystals containing high concentrations of iodide exhibited severe radiation damage, with the crystals turning red after long exposure times, presumably owing to oxidation of iodide to iodine. Substitution of bromide for iodide during crystallization and repetition of the MAD experiment was unsuccessful and radiation damage at the peak wavelengths for Br and Se prevailed. A SIRAS experiment using an iodide data set as a heavy-atom derivative for a bromide data set was also unsuccessful, although the data sets were isomorphous as judged by similar unit-cell parameters (data not shown). Based on multiple sequence alignment of Hera with other DEAD-box helicases (Fig. 3 ▶), three leucine residues (272, 281 and 315) in Hera_208–510 were replaced by methionine in order to increase the number of Se atoms in the substructure. However, while the crystallization conditions remained unchanged, these crystals did not diffract X-rays beyond 3.5 Å resolution and did not result in interpretable electron-density maps (data not shown). A strategy combining molecular replacement, using models derived from homologous RNA helicases, with MAD phasing is currently being used.
Figure 3.
Multiple sequence alignment of C-terminal RecA-like domains from related RNA helicases with Hera_208–510. Conserved residues are in bold, similar residues are boxed and helicase signature motifs as defined in Linder (2006 ▶) are underlined. The percentage similarity and identity for the displayed sequence region are given at the end of the alignment. The sequences are labelled with their PDB codes. 1hv8, M. jannaschii RNA helicase (Story et al., 2001 ▶); 2hjv, B. subtilis YxiN (Caruthers et al., 2006 ▶); 1fuk, yeast initiation factor 4A (Caruthers et al., 2000 ▶); 2g2j, human DDX25 RNA helicase (L. Lehtio et al., unpublished work); 1xti, human UAP56 (Shi et al., 2004 ▶). None of these structures proved useful as a molecular-replacement model. The figure was created using ESPript (Gouet et al., 2003 ▶).
Acknowledgments
We thank Anke Henne for providing T. thermophilus HB27 genomic DNA, Ines Hertel, Kathrin Gasow and Andreas Schmidt for technical assistance and the staff at BESSY beamline BL-1 and SLS beamline X06SA for guidance during data collection. We would also like to thank Gérard Bricogne and Clemens Vonrhein for a pre-release of BUSTER-TNT and George Sheldrick for a conversion program from XDS to SADABS reflection-data format. This work was supported by the German Research Foundation (MGR), the VolkswagenStiftung (DK) and the Swiss National Science Foundation (DK).
References
- Andersen, C. B., Ballut, L., Johansen, J. S., Chamieh, H., Nielsen, K. H., Oliveira, C. L., Pedersen, J. S., Seraphin, B., Le Hir, H. & Andersen, G. R. (2006). Science, 313, 1968–1972. [DOI] [PubMed]
- Bono, F., Ebert, J., Lorentzen, E. & Conti, E. (2006). Cell, 126, 713–725. [DOI] [PubMed]
- Caruthers, J. M., Hu, Y. & McKay, D. B. (2006). Acta Cryst. F62, 1191–1195. [DOI] [PMC free article] [PubMed]
- Caruthers, J. M., Johnson, E. R. & McKay, D. B. (2000). Proc. Natl Acad. Sci. USA, 97, 13080–13085. [DOI] [PMC free article] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Diges, C. M. & Uhlenbeck, O. C. (2001). EMBO J.20, 5503–5512. [DOI] [PMC free article] [PubMed]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res.31, 3320–3323. [DOI] [PMC free article] [PubMed]
- Halls, C., Mohr, S., Del Campo, M., Yang, Q., Jankowsky, E. & Lambowitz, A. M. (2007). J. Mol. Biol.365, 835–855. [DOI] [PMC free article] [PubMed]
- Huang, H. R., Rowe, C. E., Mohr, S., Jiang, Y., Lambowitz, A. M. & Perlman, P. S. (2005). Proc. Natl Acad. Sci. USA, 102, 163–168. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Kossen, K., Karginov, F. V. & Uhlenbeck, O. C. (2002). J. Mol. Biol.324, 625–636. [DOI] [PubMed]
- Linden, M. H., Hartmann, R. K. & Klostermeier, D. (2008). Nucleic Acids Res.36, 5800–5811. [DOI] [PMC free article] [PubMed]
- Linder, P. (2006). Nucleic Acids Res.34, 4168–4180. [DOI] [PMC free article] [PubMed]
- Mohr, G., Del Campo, M., Mohr, S., Yang, Q., Jia, H., Jankowsky, E. & Lambowitz, A. M. (2008). J. Mol. Biol.375, 1344–1364. [DOI] [PMC free article] [PubMed]
- Morlang, S., Weglohner, W. & Franceschi, F. (1999). J. Mol. Biol.294, 795–805. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pyle, A. M. (2008). Annu. Rev. Biophys.37, 317–336. [DOI] [PubMed]
- Roeser, D., Dickmanns, A., Gasow, K. & Rudolph, M. G. (2005). Acta Cryst. D61, 1057–1066. [DOI] [PubMed]
- Rudolph, M. G., Heissmann, R., Wittmann, J. G. & Klostermeier, D. (2006). J. Mol. Biol.361, 731–743. [DOI] [PubMed]
- Sengoku, T., Nureki, O., Nakamura, A., Kobayashi, S. & Yokoyama, S. (2006). Cell, 125, 287–300. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shi, H., Cordin, O., Minder, C. M., Linder, P. & Xu, R. M. (2004). Proc. Natl Acad. Sci. USA, 101, 17628–17633. [DOI] [PMC free article] [PubMed]
- Story, R. M., Li, H. & Abelson, J. N. (2001). Proc. Natl Acad. Sci. USA, 98, 1465–1470. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif.41, 207–234. [DOI] [PubMed]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed]
- Theissen, B., Karow, A. R., Kohler, J., Gubaev, A. & Klostermeier, D. (2008). Proc. Natl Acad. Sci. USA, 105, 548–553. [DOI] [PMC free article] [PubMed]
- Tsu, C. A., Kossen, K. & Uhlenbeck, O. C. (2001). RNA, 7, 702–709. [DOI] [PMC free article] [PubMed]
- Wang, S., Hu, Y., Overgaard, M. T., Karginov, F. V., Uhlenbeck, O. C. & McKay, D. B. (2006). RNA, 12, 959–967. [DOI] [PMC free article] [PubMed]
- Warkentin, M., Berejnov, V., Husseini, N. S. & Thorne, R. E. (2006). J. Appl. Cryst.39, 805–811. [DOI] [PMC free article] [PubMed]