A 2.0 Å resolution neutron diffraction data set has been collected from a D2O-soaked γ-chymotrypsin crystal at low pH on the Institute Laue–Langevin LADI-III beamline.
Keywords: γ-chymotrypsin, neutron diffraction
Abstract
The crystal preparation and preliminary neutron diffraction analysis of γ-chymotrypsin are presented. Large hydrogenated crystals of γ-chymotrypsin were exchanged into deuterated buffer via vapor diffusion in a capillary and neutron Laue diffraction data were collected from the resulting crystal to 2.0 Å resolution on the LADI-III diffractometer at the Institut Laue–Langevin (ILL) at room temperature. The neutron structure of a well studied protein such as γ-chymotrypsin, which is also amenable to ultrahigh-resolution X-ray crystallography, represents the first step in developing a model system for the study of H atoms in protein crystals.
1. Introduction
Perhaps the most important atom in a protein is the smallest one, the one that is almost impossible to see: hydrogen. Its significance may be witnessed through its central role in the fields of mechanistic enzymology, protein folding, protein engineering and computational (or rational) drug design. Enzyme mechanisms can often be determined through careful kinetic analyses; however, the assignment of mechanistic roles to individual side chains within the active site is almost never straightforward. This is partially a consequence of difficulties in assigning protonation states (hydrogen sites) in enzyme active sites, as it is well established that the pK a value of an amino acid in the environment of an enzyme active site can differ significantly from that in solution (Johnson et al., 1981 ▶; Parsons & Raftery, 1972 ▶; Schmidt & Westheimer, 1971 ▶).
In addition to yielding mechanistic information about enzymes, knowledge of the presence or absence of H atoms on ionizable side chains plays an important role in drug discovery. It has been shown that significant improvements in the scoring functions in computational ligand docking are possible when the correct protonation state is used (Ferrara et al., 2004 ▶; Zou et al., 1999 ▶).
In each of the aforementioned research areas, three-dimensional protein structures are critically important for success; yet, protein structures are almost exclusively presented without H atoms. This is not surprising, as even in ultrahigh-resolution structures (<1.2 Å) many H atoms remain invisible, especially those attached to heteroatoms, which are generally the H atoms of greatest interest. A further complication in structural biology arises from the fact that the vast majority of X-ray crystal structures are solved at cryogenic temperatures. Artifacts arising from cryoprotection, such as the alteration of pK a values, have not been examined (Halle, 2004 ▶; Hazemann et al., 2005 ▶). The overarching goal of this research project is to develop a means of improving protein models with respect to H-atom placement, such that they might be better used in various computational methods that depend critically upon accurate and precise placement of H atoms.
Neutron structures for 13 proteins (Bennett et al., 2006 ▶; Blakeley et al., 2008 ▶; Bon et al., 1999 ▶; Borah et al., 1985 ▶; Chatake et al., 2003 ▶, 2004 ▶; Cheng & Schoenborn, 1991 ▶; Coates et al., 2001 ▶; Habash et al., 2000 ▶; Katz et al., 2006 ▶; Kossiakoff, 1984 ▶; Kovalevsky et al., 2008 ▶; Wlodawer et al., 1989 ▶) are currently available from the RCSB Protein Data Bank (http://www.rcsb.org; Berman et al., 2000 ▶); however, none of these studies has explicitly addressed the specific issues described here. Early neutron studies involving the determination of protonation states in proteins relied on NMR spectroscopy to identify changes over a given pH range (Borah et al., 1985 ▶). NMR studies such as these require selective labeling of the side chains of interest for peak assignment and do not provide an overall model of the protein at different pH values. Instead of relying on NMR spectroscopic data, we propose to unambiguously identify the positions of H atoms for a small set of proteins at a variety of pH values and temperatures by directly observing changes in protonation states in the protein crystal. This can only be accomplished directly via neutron diffraction, which is now possible with the recent increased availability of neutron sources. Since D atoms are readily visible at moderate resolution in neutron experiments (2–2.5 Å), a protein crystal exchanged into deuterated mother liquor will provide data showing hydrogen (deuterium) positions at even modest resolution. Furthermore, using ultrahigh-resolution X-ray crystallography, we seek to determine atomic resolution structures for these proteins, under the same conditions, in order to correlate inter-heavy-atom distances (Coates et al., 2001 ▶) (and possibly H-atom positions) with protonation states determined from neutron diffraction analyses. These experiments will yield information not only about the direct positions of H atoms in proteins at varying pH values, but will also provide insight into possible changes in or perturbations of protonation states in protein molecules upon cryoprotection/cryocooling or upon exposure to X-rays. This information will be used to validate computational methods of H-atom placement, which currently rely on small-molecule studies (Hooft et al., 1996 ▶) and protein pK a side-chain calculations, thus improving the accuracy of structural models of proteins.
To consider an enzyme for the study proposed here, several important criteria must be met. Firstly, the protein should be able to form large diffraction-quality crystals. When considering hydrogenated crystals, a size of 1 mm3 or greater is generally required (Cooper & Myles, 2000 ▶). Secondly, given current detector limitations, the protein must have a relatively small unit cell, with the longest axis less than 100 Å in order to avoid a high percentage of spot overlaps on the detector. Thirdly, the protein must be amenable to ultrahigh-resolution X-ray crystallography. H-atom electron density is extremely difficult to observe even in the highest resolution X-ray structures. Atomic resolution X-ray data are necessary for at best the direct visualization of H atoms and at worst the accurate measurement of heavy-atom distances that may reflect the protonation state. Finally, the ideal protein should be of biological importance. Based on these factors, γ-chymotrypsin is an excellent choice for a model system for which to study H-atom positions.
2. Methods
2.1. Crystallization
γ-Chymotrypsin (Sigma–Aldrich, St Louis, Missouri, USA) was used as supplied without further purification. Crystals of γ-chymotrypsin failed to form using previously published conditions (Stoddard et al., 1990 ▶). Further screening using Additive Screen (Hampton Research, Aliso Viejo, California, USA) demonstrated that supplementation of the previously reported condition (Stoddard et al., 1990 ▶) with sodium iodide was sufficient to produce large crystals of γ-chymotrypsin (Moulin et al., 2007 ▶). Crystal size was further increased using a modified sitting-drop technique in which an uncapped 0.5 dram vial was nested in a tightly sealed scintillation vial. This setup allowed the use of very large volumes of protein plus mother liquor. Large γ-chymotrypsin crystals were obtained after several months in this fashion. Briefly, a drop composed of 150 µl of 30 mg ml−1 γ-chymotrypsin in water, 120 µl of a solution containing 45% saturated ammonium sulfate, 10 mM sodium cacodylate pH 5.6, 0.75% saturated cetyltrimethylammonium bromide and 30 µl 1 M sodium iodide was equilibrated against 2 ml well solution that contained only 65% saturated ammonium sulfate. The largest hydrogenated crystals were capillary-mounted in 2.0 mm quartz capillaries. Deuterated mother-liquor plugs at pH*1 5.0 and of about 2 mm in length were added to one side of the crystal and deuteration of the protein proceeded via vapor diffusion for a period of approximately 1 year.
Smaller γ-chymotrypsin crystals for X-ray structure determination were obtained using identical solutions but utilizing the hanging-drop format (Moulin et al., 2007 ▶). These crystals were transferred stepwise into deuterated ammonium sulfate solution and capillary-mounted with deuterated plugs.
2.2. X-ray and neutron data collection and processing
Room-temperature neutron data were collected using the LADI-III instrument on the H142 cold neutron guide at the Institut Laue–Langevin (ILL) using a nickel/titanium bandpass filter (Δλ/λ = 25%) centered at 3.5 Å and extending from 3.2 to 4.2 Å. Using an exposure time of 18 h and a step separation of 7°, seven contiguous Laue diffraction images were recorded in one orientation, with an additional five images collected in a second orientation to maximize data completeness. At the beginning of the collection of the second orientation, the crystal became wet and moved during the first exposure. This may have been caused by liquid from the deuterated plug located at the top of the capillary sliding down into the crystal. The crystal was transferred to a new 1.5 mm capillary and a single deuterated plug was used at the bottom of the capillary. The crystal diffracted similarly and the five frames of the second orientation were collected with the new setup.
Although peaks were observed to ∼1.8 Å resolution, because of spot overlaps neutron data could only be indexed and integrated to a d min of 2.0 Å using the LAUEGEN software suite (Campbell et al., 1998 ▶). The LSCALE program (Arzt et al., 1999 ▶) was used to derive the normalization curve for using the intensities of symmetry-equivalent reflections measured at different wavelengths. Data were scaled and merged using the SCALA program (Collaborative Computational Project, Number 4, 1994 ▶). The final data set for γ-chymotrypsin consists of 12 051 unique reflections and had an R merge of 13.5%. Neutron data-processing statistics are listed in Table 1 ▶.
Table 1. Data-processing statistics for the D2O-soaked γ-chymotrypsin neutron Laue data set.
| dmin (Å) | Rmerge | Rcum | 〈I/σ(I)〉 | Completeness (%) | Cumulative completeness (%) | Multiplicity |
|---|---|---|---|---|---|---|
| 6.32 | 0.079 | 0.079 | 20.2 | 96.3 | 96.3 | 4.7 |
| 4.47 | 0.098 | 0.091 | 21.3 | 96.8 | 96.6 | 7.0 |
| 3.65 | 0.110 | 0.100 | 18.4 | 94.1 | 95.5 | 6.6 |
| 3.16 | 0.134 | 0.108 | 12.8 | 89.5 | 93.4 | 5.8 |
| 2.83 | 0.163 | 0.115 | 8.7 | 79.9 | 89.6 | 5.1 |
| 2.58 | 0.184 | 0.121 | 6.5 | 74.0 | 85.8 | 4.9 |
| 2.39 | 0.196 | 0.127 | 5.3 | 71.4 | 82.9 | 4.7 |
| 2.24 | 0.201 | 0.131 | 4.0 | 67.4 | 80.1 | 3.9 |
| 2.11 | 0.201 | 0.134 | 3.3 | 62.2 | 77.2 | 3.2 |
| 2.00 | 0.200 | 0.135 | 2.6 | 53.0 | 73.6 | 2.4 |
| Total | 0.135 | 9.2 | 73.6 | 4.8 |
X-ray diffraction data were collected from small deuterated crystals at room temperature using a Xenocs GeniX Cu High Flux generator equipped with a MAR 345 image-plate detector. 180 frames of data were collected using 2 min exposure times and an oscillation sweep of 1.0°. Data were collected to 2.0 Å resolution and were indexed and integrated using MOSFLM (Leslie, 1992 ▶). SCALA was used to scale and merge the data (Collaborative Computational Project, Number 4, 1994 ▶). X-ray data-processing statistics are shown in Table 2 ▶.
Table 2. Data-processing statistics for the D2O-soaked γ-chymotrypsin X-ray data set.
| dmin (Å) | Rmerge | Rcum | 〈I/σ(I)〉 | Completeness (%) | Cumulative completeness (%) | Multiplicity |
|---|---|---|---|---|---|---|
| 6.28 | 0.027 | 0.027 | 58.5 | 99.2 | 99.2 | 11.8 |
| 4.44 | 0.030 | 0.028 | 59.4 | 100.0 | 99.7 | 13.5 |
| 3.62 | 0.037 | 0.033 | 54.4 | 100.0 | 99.8 | 13.8 |
| 3.14 | 0.053 | 0.038 | 42.4 | 100.0 | 99.9 | 14.0 |
| 2.81 | 0.081 | 0.044 | 31.3 | 100.0 | 99.9 | 14.0 |
| 2.56 | 0.118 | 0.052 | 23.1 | 100.0 | 99.9 | 13.9 |
| 2.37 | 0.159 | 0.059 | 17.2 | 100.0 | 100.0 | 13.6 |
| 2.22 | 0.194 | 0.067 | 13.6 | 100.0 | 100.0 | 13.3 |
| 2.09 | 0.279 | 0.076 | 9.9 | 100.0 | 100.0 | 13.2 |
| 1.98 | 0.377 | 0.084 | 6.0 | 91.4 | 98.7 | 10.9 |
| Total | 0.084 | 25.7 | 98.7 | 13.2 |
2.3. X-ray and neutron structure determination
The coordinates from PDB entry 1gct (Dixon & Matthews, 1989 ▶) were used for rigid-body refinement of the X-ray data using PHENIX (Adams et al., 2002 ▶). Several rounds of refinement using the phenix.refine module of PHENIX (Adams et al., 2002 ▶) and model building with Coot (Emsley & Cowtan, 2004 ▶) yielded an X-ray model of D2O-soaked γ-chymotrypsin with R cryst and R free values of 0.141 and 0.189, respectively. H and D atoms (where exchangeable) were added to this model using the phenix.ready_set utility (Adams et al., 2002 ▶). A consistent set of test reflections for both the neutron and X-ray data sets was generated with PHENIX (Adams et al., 2002 ▶) and initial joint X-ray/neutron refinement with phenix.refine (Adams et al., 2002 ▶) using our deuterated X-ray model yielded R cryst/R free values of 0.278 and 0.298 for the neutron data, respectively, indicating successful phasing of the neutron data. Data reduction and initial refinement statistics for the X-ray and neutron data sets are listed in Table 3 ▶.
Table 3. Data-reduction and initial refinement statistics.
| X-ray | Neutron | |
|---|---|---|
| Source | Cu Kα | ILL |
| Detector | MAR 345 | LADI-III |
| Wavelength (Å) | 1.54 | 3.2–4.2 |
| Space group | P42212 | |
| Unit-cell parameters | a = b = 69.5, c = 97.4 | |
| Reflections, measured/unique | 395507/17127 | 61431/12051 |
| Resolution range (Å) | 29.62–1.98 | 56.80–2.00 |
| Rcryst/Rfree test-set reflections | 15227/1705 | 10725/1071 |
| Rcryst/Rfree | 0.141/0.189 | 0.278/0.298 |
3. Results and discussion
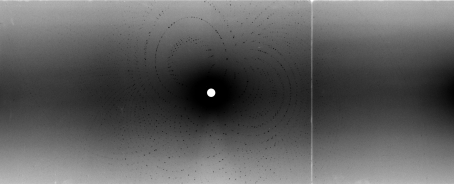
The crystal used for neutron diffraction in this study has dimensions of approximately 1.3 × 1.0 × 1.0 mm and is shown in Fig. 1 ▶. With an 18 h exposure, neutron diffraction data extended to 1.8 Å resolution; however, owing to the high percentage of spot overlaps, the data were only processed to 2.0 Å resolution. Fig. 2 ▶ shows a representative Laue diffraction pattern for γ-chymotrypsin and data-processing statistics are listed in Table 1 ▶. It can be seen that the 〈I/σ(I)〉 for the data set is strong, even out to 2.0 Å resolution, where its value is 2.6. The multiplicity is fair to the 2.0 Å resolution limit, with a value of 2.4 (4.8 overall). The completeness, however, is only 50% in the highest resolution bin. The reason for the lack of completeness is almost certainly loss of data owing to spatial overlaps rather than actual absence of the data, as the crystals visibly diffract beyond 2.0 Å resolution and the signal at this resolution is quite strong. This suggests that a new data-collection strategy might be in order for crystals with a long unit-cell axis: specifically, a good strategy may be to first collect low-resolution data quickly with a large bandwidth of neutrons and then to collect higher resolution data with the crystal in a different orientation using a smaller bandwidth of neutrons. This strategy should help achieve greater completeness and higher resolution as the smaller bandwidth of neutrons will reduce the number of spatial overlaps [Bennett et al., 2005 ▶; Kalb (Gilboa) et al., 2001 ▶].
Figure 1.
Photograph of the γ-chymotrypsin crystal used for this neutron diffraction study on the LADI-III beamline at the ILL. The crystal volume is approximately 1.3 mm3.
Figure 2.
Neutron Laue diffraction pattern from an 18 h exposure of γ-chymotrypsin using the LADI-III beamline at the ILL.
From these preliminary maps there is clear nuclear density in the active site of γ-chymotrypsin for the important catalytic residues and significant nuclear density for D atoms can be visualized on a number of heteroatoms in the protein (data not shown). γ-Chymotrypsin is known to be an acyl-enzyme representative of an intermediate along the chymotrypsin reaction pathway (Hartley, 1960 ▶) and there is nuclear density for a bound peptide linked to serine 195 via an acyl bond. The sequence of the peptide as determined in these experiments is GSWPW, which is a self-cleavage product of chymotrypsin (residues 25–29). The fragment is most likely to be longer than five amino acids, but only these five residues can be resolved in the electron-density and nuclear density maps.
Heavy-water molecules, especially those in the active site, also show significant deuterium nuclear density (data not shown). After full refinement, the combination of X-ray and neutron data may facilitate the orientation of heavy-water molecules with strongly conserved positions, as the locations of the O atoms of heavy water can be determined in the electron-density map.
4. Conclusion
In this work, we have described the collection and preliminary nuclear density maps of γ-chymotrypsin to 2.0 Å resolution. Joint X-ray/neutron refinement is under way with both nCNS (Langan & Mustyakimov, 2007 ▶) and PHENIX (Adams et al., 2002 ▶). This work demonstrates the feasibility of using γ-chymotrypsin in the development of a benchmark set of H-atom positions. The future goals of this project are continuing. We are preparing large γ-chymotrypsin crystals at higher pH* values and we are in the process of collecting ultrahigh-resolution X-ray data at both ambient and cryogenic temperatures.
This study also highlights the difficulties of neutron crystallography data collection for crystals with large unit cells. Spatial overlaps of reflections can significantly reduce completeness and resolution. Our situation may be similar to thaumatin (a = b = 57.8, c = 150.1 Å), for which data were collected to 1.8 Å resolution but could only be processed to 2.0 Å resolution (Teixeira et al., 2008 ▶). It remains to be seen whether the data-collection strategy using a narrower bandpass filter, as suggested here and by others [Bennett et al., 2005 ▶; Kalb (Gilboa) et al., 2001 ▶], would be helpful in extending resolution and completeness for crystals containing long unit-cell axes.
Acknowledgments
The authors thank the Institut Laue–Langevin for the generous provision of neutron beam time for testing and data collection (proposals 8-01-271 and 8-01-290, respectively) on the LADI-III diffractometer. We wish to thank Dr Hassan Belrhali for assistance with the collection of X-ray data at EMBL Grenoble and Dr Flora Meilleur and Dr Dean Myles for helpful discussions. DR acknowledges generous support by the Alexander von Humboldt Foundation. This work was supported by the National Institutes of Health Grants GM-32415 to GAP and DR and NRSA 5-F32-GM-075719 to WRPN.
Footnotes
pH* refers to the reading of a D2O solution using an H2O-calibrated pH meter.
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Arzt, S., Campbell, J. W., Harding, M. M., Hao, Q. & Helliwell, J. R. (1999). J. Appl. Cryst.32, 554–562.
- Bennett, B., Langan, P., Coates, L., Mustyakimov, M., Schoenborn, B., Howell, E. E. & Dealwis, C. (2006). Proc. Natl Acad. Sci. USA, 103, 18493–18498. [DOI] [PMC free article] [PubMed]
- Bennett, B. C., Meilleur, F., Myles, D. A. A., Howell, E. E. & Dealwis, C. G. (2005). Acta Cryst. D61, 574–579. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res.28, 235–242. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P., Ruiz, F., Cachau, R., Hazemann, I., Meilleur, F., Mitschler, A., Ginell, S., Afonine, P., Ventura, O. N., Cousido-Siah, A., Haertlein, M., Joachimiak, A., Myles, D. & Podjarny, A. (2008). Proc. Natl Acad. Sci. USA, 105, 1844–1848. [DOI] [PMC free article] [PubMed]
- Bon, C., Lehmann, M. S. & Wilkinson, C. (1999). Acta Cryst. D55, 978–987. [DOI] [PubMed]
- Borah, B., Chen, C. W., Egan, W., Miller, M., Wlodawer, A. & Cohen, J. S. (1985). Biochemistry, 24, 2058–2067. [DOI] [PubMed]
- Campbell, J. W., Hao, Q., Harding, M. M., Nguti, N. D. & Wilkinson, C. (1998). J. Appl. Cryst.31, 496–502.
- Chatake, T., Kurihara, K., Tanaka, I., Tsyba, I., Bau, R., Jenney, F. E., Adams, M. W. W. & Niimura, N. (2004). Acta Cryst. D60, 1364–1373. [DOI] [PubMed]
- Chatake, T., Mizuno, N., Voordouw, G., Higuchi, Y., Arai, S., Tanaka, I. & Niimura, N. (2003). Acta Cryst. D59, 2306–2309. [DOI] [PubMed]
- Cheng, X. D. & Schoenborn, B. P. (1991). J. Mol. Biol.220, 381–399. [DOI] [PubMed]
- Coates, L., Erskine, P. T., Wood, S. P., Myles, D. A. A. & Cooper, J. B. (2001). Biochemistry, 40, 13149–13157. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cooper, J. B. & Myles, D. A. A. (2000). Acta Cryst. D56, 246–248. [DOI] [PubMed]
- Dixon, M. M. & Matthews, B. W. (1989). Biochemistry, 28, 7033–7038. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Ferrara, P., Gohlke, H., Price, D. J., Klebe, G. & Brooks, C. L. III (2004). J. Med. Chem.47, 3032–3047. [DOI] [PubMed]
- Habash, J., Raftery, J., Nuttall, R., Price, H. J., Wilkinson, C., Kalb (Gilboa), A. J. & Helliwell, J. R. (2000). Acta Cryst. D56, 541–550. [DOI] [PubMed]
- Halle, B. (2004). Proc. Natl Acad. Sci. USA, 101, 4793–4798. [DOI] [PMC free article] [PubMed]
- Hartley, B. S. (1960). Annu. Rev. Biochem.29, 45–72. [DOI] [PubMed]
- Hazemann, I., Dauvergne, M. T., Blakeley, M. P., Meilleur, F., Haertlein, M., Van Dorsselaer, A., Mitschler, A., Myles, D. A. A. & Podjarny, A. (2005). Acta Cryst. D61, 1413–1417. [DOI] [PubMed]
- Hooft, R. W. W., Sander, C. & Vriend, G. (1996). Proteins, 26, 363–376. [DOI] [PubMed]
- Johnson, F. A., Lewis, S. D. & Shafer, J. A. (1981). Biochemistry, 20, 44–48. [DOI] [PubMed]
- Kalb (Gilboa), A. J., Myles, D. A. A., Habash, J., Raftery, J. & Helliwell, J. R. (2001). J. Appl. Cryst.34, 454–457.
- Katz, A. K., Li, X., Carrell, H. L., Hanson, B. L., Langan, P., Coates, L., Schoenborn, B. P., Glusker, J. P. & Bunick, G. J. (2006). Proc. Natl Acad. Sci. USA, 103, 8342–8347. [DOI] [PMC free article] [PubMed]
- Kossiakoff, A. A. (1984). Basic Life Sci.27, 281–304. [DOI] [PubMed]
- Kovalevsky, A. Y., Chatake, T., Shibayama, N., Park, S.-Y., Ishikawa, T., Mustyakimov, M., Fisher, S. Z., Langan, P. & Morimoto, Y. (2008). Acta Cryst. F64, 270–273. [DOI] [PMC free article] [PubMed]
- Langan, P. & Mustyakimov, M. (2007). Computational Tools For Macromolecular Neutron Crystallography. http://mnc.lanl.gov.
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Moulin, A., Bell, J. H., Pratt, R. F. & Ringe, D. (2007). Biochemistry, 46, 5982–5990. [DOI] [PMC free article] [PubMed]
- Parsons, S. M. & Raftery, M. A. (1972). Biochemistry, 11, 1623–1629. [DOI] [PubMed]
- Schmidt, D. E. Jr & Westheimer, F. H. (1971). Biochemistry, 10, 1249–1253. [DOI] [PubMed]
- Stoddard, B. L., Bruhnke, J., Porter, N., Ringe, D. & Petsko, G. A. (1990). Biochemistry, 29, 4871–4879. [DOI] [PubMed]
- Teixeira, S. C. M., Blakeley, M. P., Leal, R. M. F., Mitchell, E. P. & Forsyth, V. T. (2008). Acta Cryst. F64, 378–381. [DOI] [PMC free article] [PubMed]
- Wlodawer, A., Savage, H. & Dodson, G. (1989). Acta Cryst. B45, 99–107. [DOI] [PubMed]
- Zou, X. Q., Sun, Y. X. & Kuntz, I. D. (1999). J. Am. Chem. Soc.121, 8033–8043.