Abstract
P210 Bcr-Abl is an activated tyrosine kinase oncogene encoded by the Philadelphia chromosome associated with human chronic myelogenous leukemia (CML). The disease represents a clonal disorder arising in the pluripotent hematopoietic stem cell. During the chronic phase, patients present with a dramatic expansion of myeloid cells and a mild anemia. Retroviral gene transfer and transgenic expression in rodents have demonstrated the ability of Bcr-Abl to induce various types of leukemia. However, study of human CML or rodent models has not determined the direct and immediate effects of Bcr-Abl on hematopoietic cells from those requiring secondary genetic or epigenetic changes selected during the pathogenic process. We utilized tetracycline-regulated expression of Bcr-Abl from a promoter engineered for robust expression in primitive stem cells through multilineage blood cell development in combination with the in vitro differentiation of embryonal stem cells into hematopoietic elements. Our results demonstrate that Bcr-Abl expression alone is sufficient to increase the number of multipotent and myeloid lineage committed progenitors in a dose-dependent manner while suppressing the development of committed erythroid progenitors. These effects are reversible upon extinguishing Bcr-Abl expression. These findings are consistent with Bcr-Abl being the sole genetic change needed for the establishment of the chronic phase of CML and provide a powerful system for the analysis of any genetic change that alters cell growth and lineage choices of the hematopoietic stem cell.
For many human leukemias, precisely defined translocations or inversions can be diagnostic for a particular clinical entity (1, 2). Chronic myelogenous leukemia (CML) patients present with the characteristic translocation t(9:22) known as the Philadelphia (Ph) chromosome, which results in a fused transcription unit producing the P210 Bcr-Abl oncogene with a strongly activated tyrosine kinase critical for cell transformation (3–5). The early or chronic phase of CML presents with a preferential expansion of the myeloid lineage with overproduction of mature granulocytes and associated splenomegaly. Erythropoiesis is ineffective and most patients are mildly anemic. The primary affected cell appears to be the pluripotent hematopoietic stem cell and closely related immature committed progenitors (6). Highly enriched stem cell elements from CML patients are Bcr-Abl-positive at the genomic level by in situ chromosome analysis (7, 8). Analysis of multilineage, myeloid, erythroid, and other colony forming cells demonstrates the Bcr-Abl gene and its chimeric mRNA (9, 10). Without definitive treatment by bone marrow transplantation, most patients evolve into blast crisis phase (11). This can be associated with a wide variety of additional genetic changes, including duplication of the Ph chromosome, various trisomies, other translocations, and loss of function mutations in several tumor suppressors (11, 12).
It is not clear whether specific genetic or epigenetic events are needed for selection of the dominant clone that accompanies the development of chronic phase from the single cell in which the Ph chromosome translocation occurs. High-level expression of Bcr-Abl can have direct and obvious effects in cell culture systems monitoring fibroblast transformation (13), expansion of immature B lineage elements (14), augmentation of multilineage hematopoietic colony forming ability in low growth factor conditions (15), and resistance from apoptosis secondary to growth factor withdrawal from leukemic cell lines (16, 17). However, a variety of transgenic models expressing Bcr-Abl from global or lineage-specific promoters develop leukemia with varying periods of latency and penitrance (18, 19). Introduction of Bcr-Abl by means of retroviral gene transfer into murine bone marrow populations enriched for stem elements followed by transfer to irradiated hosts can result in different types of leukemia, including CML (20–23). These studies combine to support a role for Bcr-Abl in the generation of human and murine leukemia, but do not define the role this oncogene directly plays in the growth and differentiation decisions made by the stem cell.
Several studies have suggested that Bcr-Abl alone is not sufficient to initiate the chronic phase of CML. Fialkow et al. (24) followed patients with myelodysplasia who eventually evolved into frank CML with the Ph chromosome. Using allelic variants of an X chromosome-linked marker enzyme, they defined clonal dominance in the peripheral blood prior to appearance of CML and argued that another genetic event was required before the Ph chromosome in the evolution of the leukemic clone (25). The detection of the Ph chromosome-encoded Bcr-Abl mRNA in apparently normal individuals who do not succumb to CML has been reported (26). Perhaps this translocation occurs at a low frequency in some individuals but does not lead to disease because a second event or correct genetic context does not exist for development of CML.
We needed an in vitro system that reproducibly produced stem cells that was capable of undergoing multilineage differentiation and could be regulated to express Bcr-Abl in 100% of the cells. We have combined an in vitro embryonal stem (ES) cell differentiation system that utilizes stromal cell coculture to direct differentiation into the blood cell lineages (27) with tetracycline-regulated expression (28) of Bcr-Abl employing a promoter engineered for expression throughout hematopoiesis. Our results demonstrate that Bcr-Abl kinase dosage alone can acutely alter the growth rate of immature progenitors and change the balance of myeloid and erythroid differentiation.
Materials and Methods
Cells, Cell Culture, in Vitro ES Cell Differentiation, and Transfection.
E14tg2a ES cells and OP9 stromal cells were maintained as described previously (27, 29). For differentiation induction, ES cells were seeded onto confluent OP9 cell layers on six-well plates at a density of 104 cell per well in the condition of 20% FBS, α-MEM, and no leukemia inhibitory factor (LIF). On day 5 of differentiation, these cells were harvested by 0.25% trypsin-EDTA (GIBCO/BRL) and replated for 30 min to remove adherent cells. Nonadherent cells were reseeded with new OP9 layers at a density of 105 cells per well of 6-well plate or 7–8 × 105 cells per 10-cm dish. On day 7 or day 8 of differentiation, nonadherent cells were collected to obtain hematopoietic cells. In some experiments, these cells were transferred to another new OP9 cell layer and were cultured until day 14 or 15. The electroporation procedures were carried out as described previously (29).
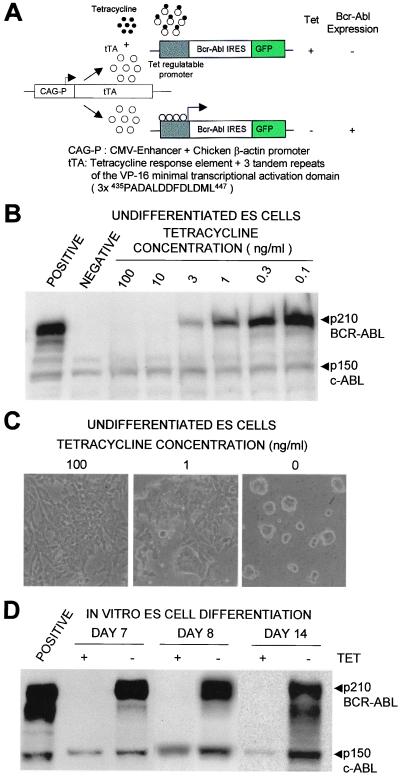
The tetracycline (Tet) regulatory system was used to obtain Bcr-Abl regulatable ES clones. The expression of Bcr-Abl was driven by Tet response promoter (tetO-CMV) (28) and suppressed by the addition of Tet (Tet-off system; see Fig. 1). The Tet-regulated transactivator (tTA) (30) expression vector, pCAG 20-1 (31), was constructed by inserting a tTA cDNA to the downstream of the CAG promoter (Fig. 1). We used a modified-tTA gene containing tetracycline response element fused at its carboxyl terminus with three minimal transcriptional domains of VP-16, FFF; 3× 435PADALDDFDLDML447 (30). The Tet-regulatable Bcr-Abl construct, pUHD 10-3 Bcr-Abl IRES GFP, was generated by inserting p210 Bcr-Abl (14), internal ribosomal entry site (IRES) (32), and green fluorescence protein (GFP) (33) genes to EcoRI and BamHI sites downstream of tetO-CMV promoter in pUHD10-3 vector (28) provided by H. Bujard (Heidelberg University, Heidelberg). The pUHD10-3 puro vector was constructed by ligating puromycin resistance gene to BamHI site downstream of the tetO-CMV promoter in pUHD10-3.
Figure 1.
Inducible expression of Tet-regulated expression of Bcr-Abl in undifferentiated and differentiated ES cells. (A) The constructs and strategy for the regulated expression of Bcr-Abl in ES cells. The CAG promoter drives the cDNA of the tTA (31). This tTA consisted of Tet response element and three tandem repeats of the VP-16 minimal transcriptional transactivation domatins, 3× 435PADALDDFDLDML447 (30). The CAG promoter showed a stable and high level of expression in undifferentiated ES cells. The expression of p210 Bcr-Abl and IRES-linked GFP are driven by a Tet-regulated promoter that is suppressed by the addition of tetracycline (Tet-off-system). (B) Inducible expression of Bcr-Abl protein by the removal of Tet in undifferentiated ES cells. POSITIVE, fibroblast cell line expression P210 Bcr-Abl constitutively. NEGATIVE, normal ES cells. A total of 106 cells were extracted as described in Materials and Methods and analyzed by anti-Abl Western blot. (C) The effect of Bcr-Abl expression on undifferentiated ES cells. Undifferentiated ES cells were cultured in the presence of LIF with 100, 1, or 0 ng/ml Tet. Two days later, the ES cells expressing Bcr-Abl showed alteration of colony shape (0 ng/ml) from normal ES colonies (100 ng/ml). Photographs are taken at ×100 magnification. (D) Inducible expression of Bcr-Abl during in vitro ES cell differentiation. Inducible Bcr-Abl ES cells were differentiated when cultured with OP9 in the presence of Tet for 5 days then split into dishes with (+) Tet (100 ng/ml) or without (−) Tet. Samples harvested on day 7, 8, or 14 were analyzed by Western blot with anti-Abl antibody as described in Materials and Methods. POSITIVE, fibroblast cell line constitutively expression of p210 Bcr-Abl.
To establish a reliable primary ES cell line expressing tTA, both constructs (pCAG 20–1 and pUHD10-3 Puro) were transfected into ES cells by electroporation (34). When the Tet-regulatory system is working, the expression of the puromycin resistance gene is suppressed by the presence of Tet and the cells will not survive puromycin selection. The cells were grown with 1 μg/ml puromycin in Tet-free medium. A total of 48 puromycin resistant colonies were picked and split into parallel 24-well plates either in the presence or absence of Tet. The six parental clones proliferating in Tet-free medium but dying in the presence of Tet were selected as primary parental Tet-regulatory ES cells. The two parental ES clones were electroporated with pUHD 10-3 Bcr-Abl IRES GFP vector and neomycin plasmid, pMC1NEO (Stratagene). The cells were maintained in the presence of 1 μg/ml Tet and 200 μg/ml G418. Multiple neomycin resistant colonies from each parental clone were picked and split into parallel 24-well plates either in the presence or absence of Tet. Clones tightly regulated by Tet were initially analyzed for GFP expression by FACScan (Becton Dickinson). The GFP expressing clones were subsequently examined for Bcr-Abl expression by Western blotting in Tet+ and Tet− conditions. Finally, a total of 11 Tet-regulatable Bcr-Abl clones were obtained.
Flow Cytometric Analysis, Western Blotting, and Cycle Analysis.
The phycoerythrin (PE)-conjugated TER119 (erythroid lineage marker), M1/70 (anti-CD11b/Mac-1), anti-BrdUrd antibody, biotin-conjugated anti-c-kit, and CD34 monoclonal antibodies were purchased from PharMingen. Cells were stained with the combination of the antibodies as described previously (35). For biotin staining, we used streptavidin-tricolor. Cells were analyzed by FACScan (Becton Dickinson). Anti-ABL Western blots were probed with 21–24 mouse monoclonal antibody (36). A total of 1 × 106 cells were resuspended in 100 μl of boiling lysis buffer containing 100 mM Tris (pH 6.8), 20% glycerol, 2% SDS, 5% 2-ME, and 0.1% bromophenol blue. Twenty microliters of each sample were separated on 4–12% gradient SDS Tris⋅Glysine Gel (NOVEX, San Diego). Proteins were transferred onto nitrocellulose membrane (Micron Separations, Westboro, MA) and visualized with an enhanced chemiluminescence detection kit (Amersham Pharmacia). Goat anti-mouse IgG antibody conjugated by HRP (Bio-Rad) was used as a secondary antibody. For cell cycle analysis, hematopoietic cells on day 7 and 8 were incubated in medium supplemented with 10 μM BrdUrd (Sigma) for 1 or 2 h at 37°C. After fixing and denaturing, the cells were stained with anti-BrdUrd antibody as described previously (37). Samples were analyzed by FACScan. For apoptosis analysis, cell were harvested from cultures and washed in PBS twice. A total of 2 × 105 cells were resuspended in 100 μl of binding buffer, 10 mM Hepes/NaOH (pH 7.4), 140 mM NaCl, 2.5 mM CaCl2, and then PE-conjugated annexin-V (PharMingen) and 7-amino-actinomycin D (PharMingen) were added at 5% final concentration. These samples were incubated for 15 min at room temperature in the dark. Samples were analyzed by FACScan.
Results
Multilineage Development of Blood Cells from ES Lines and Regulation of Bcr-Abl Expression.
Previous work has defined an ES cell cocultivation strategy with the OP9 cell line as a highly reproducible way to model hematopoiesis in vitro (38, 39). OP9 is a marrow stromal cell line derived from a mouse strain deficient in macrophage colony-stimulating factor (27). It provides a favorable environment for the differentiation of ES cells removed from LIF into primitive hemagioblasts (day 4–5), which produce immature hematopoietic stem and progenitor cells (day 7–8) and eventually mature blood cell elements (day 10–14). In the presence of low amounts of FCS, but without added growth factors, both primitive and definitive erythroid cells, granulocytes, and other myeloid lineages predominate by 2 wk.
We needed to utilize a promoter system that was capable of expression in primitive ES cells, hematopoietic stem and progenitor cells, and throughout all the different intermediate and mature hematopoietic cell types produced in these cultures. Previous data had defined that a composite control element comprised of the chicken actin promoter with a CMV enhancer (CAG; Fig. 1) could drive expression in undifferentiated ES cells (31). In preliminary experiments (data not shown), we established multiple independent ES clones constitutively expressing Bcr-Abl from this composite promoter and demonstrated excellent expression throughout the differentiation of ES cells into blood cells of multiple lineages. We observed that variation in the dose of Bcr-Abl in different clones of ES cells was a critical factor in determining the efficiency of conversion into hematopoiesis and production of mature cell forms (data not shown).
To control the expression of Bcr-Abl, we utilized the CAG promoter to drive the tetracycline transactivator, which was modified with synthetic activator sequences (Fig. 1A) chosen to enhance expression in the ES/OP9 differentiation system, and placed Bcr-Abl under the control of the Tet operator sequence. Expression of Bcr-Abl is suppressed in the presence of Tet and activated when the drug is removed. About 50 ES clones were established by transfection and double drug selection (see Materials and Methods) and analyzed for their ability to express Bcr-Abl (Fig. 1B) and their variation in cellular morphology (Fig. 1C) as drug concentration was lowered. Each clone was evaluated for its ability to induce Bcr-Abl expression during the course of hematopoietic differentiation (Fig. 1D). The Tet control system worked efficiently at all times during the culture development.
The Dosage of Bcr-Abl Directly Regulates the Rate of Proliferation of Immature Multilineage Progenitors.
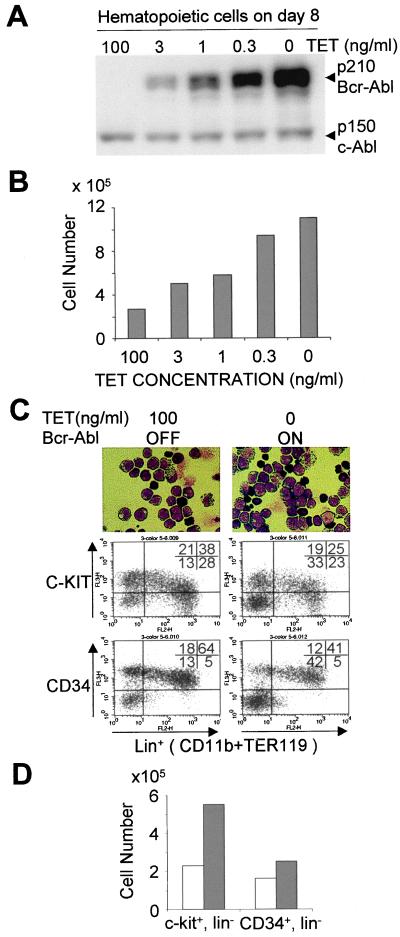
Several reports have shown that growth factor requirements of continuous cell lines or marrow multilineage progenitors' function in colony forming assays can be lessened or overcome by high levels of Bcr-Abl expression (11, 40, 41). We directly evaluated the quantitative effects of Bcr-Abl on the least mature populations of hematopoietic stem and progenitor elements by regulating expression coincident with their appearance in ES/OP9 cultures from day 5–8 post LIF removal (Fig. 2). The results demonstrate that an increase in Bcr-Abl level (Fig. 2A) is directly related to the number of cells that accumulate in the cultures over the 3-day period (Fig. 2B). We observed up to a 4-fold increase in the total cell number recovered over this 3-day period. The developmental phenotype of the cell populations in the presence or absence of Bcr-Abl over this period is quite similar as monitored by morphology and FACS analysis for early hematopoietic markers (CD34 and c-kit) and lineage-specific markers of myeloid (CD11b) or erythroid (Fig. 2C) cell types. One significant difference is that the absolute number of progenitor elements (c-kit+, lin− or CD34+,lin−) was increased when Bcr-Abl was expressed (Fig. 2D). Multiple independent ES clones regulated to produce Bcr-Abl showed this behavior, and control ES clones without Bcr-Abl expression showed no variation in development when treated with Tet over a wide dose range (data not shown).
Figure 2.
A dose-dependent enhancement of immature hematopoietic cells by Bcr-Abl expression. (A) The expression level of Bcr-Abl protein is controlled by alteration of Tet concentration. The Bcr-Abl-inducible ES cells were differentiated by coculturing with OP9 stromal cells in the presence of 100 ng/ml Tet until day 5. The differentiated ES cells were subcultured (7 × 105 cells per 10-cm dish) at various concentrations of Tet (100, 3, 1, 0.3, or 0 ng/ml), and 3 days later the hematopoietic cells that developed on the OP9 cell layers were harvested. Bcr-Abl expression was analyzed by Western blotting (on equivalent cell numbers) using anti-Abl antibody as described in Materials and Methods. (B) The number of hematopoietic cells on day 8 of ES cell differentiation cultured at various Tet concentrations. As described above, 7 × 105 cells at day 5 were seeded on new OP9 layers and cultured at various concentrations of Tet. The hematopoietic cells were harvested on day 8 and counted. The expression level of Bcr-Abl protein at each Tet concentration correspond to those of Fig. 2A. (C) Immature hematopoietic phenotype of day 8 differentiated ES cells. Inducible Bcr-Abl ES cells are cultured in Tet on OP9 cell layers for 5 days. On day 5, one was maintained with Tet (100 ng/ml) to suppress Bcr-Abl expression (Left). Three days later, the clusters of hematopoietic cells were harvested and analyzed by FACScan and Giemsa staining. Both hematopoietic cell populations exhibited an immature morphological phenotype with occasional primitive erythroid cells (Top, ×500 magnification). (Middle and Bottom) The phenotype of cell surface markers in day 8 hematopoietic cells differentiated from the inducible Bcr-Abl ES cells. The expression of the c-kit or CD34 and lineage markers, CD11b (myeloid cells) and TER119 (erythroid cells) are shown. (Left) The Bcr-Abl-off condition with 100 ng/ml Tet added. (Right) Results for day 8 Bcr-Abl-expressing cells. (D) The number of day 8 c-kit+/lin− cells derived from inducible Bcr-Abl ES cells. Open bars show the Tet off-condition with 100 ng/ml Tet, in which Bcr-Abl expression of inducible Bcr-Abl ES cells are suppressed. Filled bars indicate the on-condition of Bcr-Abl expression. The experiment shows the mean of triplicate cultures. The results are plotted as total cell number of the designated phenotype per culture dish. Each experiment was carried out three times using four different clones. A representative result is shown.
The increase in expansion of stem and progenitor elements between days 5 and 8 could be the result of an increase in proliferation rate, a reduction in cell death, or a combination of both effects. A clear increase (almost 2-fold from 7% to 13%) in the fraction of cells staining positive for BrdUrd uptake was observed on both days 7 and 8, suggesting that the major effect of Bcr-Abl on stem and progenitor elements is likely to be stimulating the entry into active cycle. Analysis of programmed cell death by staining for expression of annexin to mark cells entering the death pathway, and 7-AAD dye to count those that are already dead showed little difference between the Bcr-Abl on or off conditions at day 7 or day 8. A small increase in the percentage of dead and dying cells was observed (11 compared with 14%) when Bcr-Abl was expressed.
Bcr-Abl Expression Changes the Balance of Lineage Development.
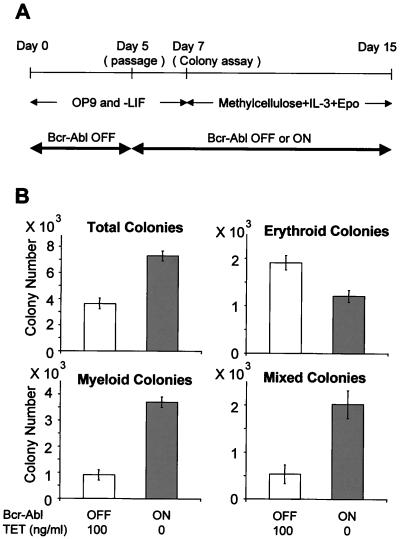
The observation that Bcr-Abl expression increased the percentage and absolute number of very immature progenitor elements between days 5 and 8 during ES/OP9 culture could be solely due to differential proliferative effects or coupled to a partial block in differentiation. The surface phenotypes of such cells cannot predict their eventual developmental outcome, which can be monitored by colony forming assays. We evaluated the efficiency and balance of myeloid and erythroid cell fates by colony forming assays on cells from day 7 of ES/OP9 cultures. Cells were plated in methylcellulose in the presence of the multilineage factor IL-3 and erythropoietin, which favors the growth and development of colonies restricted to the erythroid lineage, but allows growth of myeloid colonies dominated by granulocytic and monocytic cell types (35, 42). The balance of differentiated phenotypes from this culture system resembles those produced by cells from the fetal liver stage of hematopoiesis. The results shown in Fig. 3 demonstrate that Bcr-Abl has two dramatic effects. First, the total number of colony-forming units is increased, and, second, the normal balance of erythroid to myeloid colonies (2:1) is reversed to a dominance of myeloid over erythroid (4:1). In addition, the fraction of colonies showing mixtures of alternative myeloid and erythroid cell types is more than doubled. All of these results are consistent with Bcr-Abl directly and acutely expanding a very immature cell type with multilineage differentiation capabilities and favoring myeloid over erythroid development, even in the presence of erythropoietin.
Figure 3.
Effects of Bcr-Abl on early hematopoietic colony formation. Control and inducible Bcr-Abl ES cells were differentiated on OP9 stromal cell layers in the presence of Tet until day 5. At that point, these cells were divided into two subgroups: one was maintained with Tet, and the other was cultured in the absence of Tet. On day 7, clusters of immature hematopoietic cells had developed on the OP9 cell layers. The hematopoietic cells in these clusters were harvested and analyzed. (A) Experimental design of hematopoietic colony assays in methylcellulose culture. Inducible Bcr-Abl ES cells were differentiated on OP9 stromal cells with Tet (100 ng/ml) in the absence of LIF. On day 5, differentiated ES cells were harvested and seeded onto a new OP9 cell layer with or without Tet. On day 7, the hematopoietic cells developing on the OP9 cell layers were harvested and plated in methylcellulose cultures with recombinant IL-3 (10 ng/ml) and erythropoetin (2 units/ml) present (42). (B) Eight days later, colonies were counted and scored as to differentiated phenotype. Individual colonies were picked, and cytospin specimens were stained with Giemsa solution to confirm the colony types. Colonies containing more than 50 hemoglobinized cells were counted as erythroid. Open bars show the Tet (+) condition, in which the Bcr-Abl protein is suppressed. Filled bars indicate the Tet (−) condition, which results in the strong expression of Bcr-Abl protein. Colony numbers are represented as the calculated total present per culture harvested at day 7.
Reversing Bcr-Abl Expression Allows Completion of Erythroid and Myeloid Development.
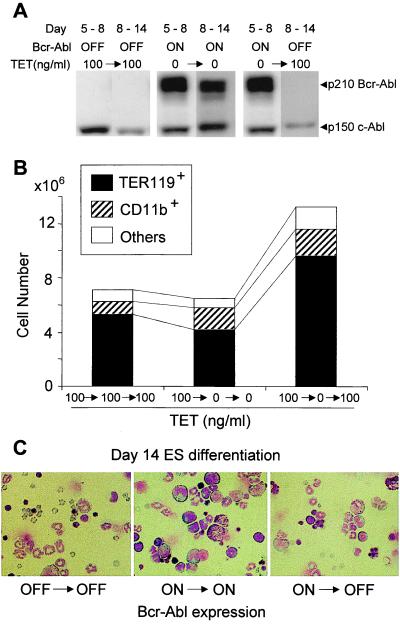
The Tet-regulated expression of Bcr-Abl allowed us to investigate whether the relative block in differentiation and alteration of erythroid/myeloid ratio could be reversed. Control cultures produce mainly mature erythrocytes and granulocytes by the end of the 14-day culture period. Continuous expression of Bcr-Abl results in about the same total cell number, but there are fewer mature cells and a relative increase in immature myeloid cell types of granulocyte and macrophage lineages (Fig. 4). If the expression of Bcr-Abl is shut off (Fig. 4A) during the last phase of culture development (days 8–14), the accumulated multilineage and restricted progenitors can effectively complete their differentiation program and revert to the pattern of erythroid dominance over myeloid cell types (Fig. 4 B and C). The effects of Bcr-Abl are clearly reversible in this in vitro system.
Figure 4.
Reversible effect of Bcr-Abl expression on erythroid differentiation. (A) ES cells were induced to differentiate by removal of LIF and cultured on OP9 layers between days 0 and 5 in the presence of Tet (100 ng/ml) to suppress Bcr-Abl expression. Differentiated cells were harvested, washed, and replated on OP9 layers in three different groups. In the first group, cells were continuously cultured in Tet between days 5 and 8, when a sample was harvested, and between days 8 and 14. In the second group, cells harvested on day 5 were washed and recultured in the absence of Tet between days 5 and 8, when the culture was sampled, and continued between days 8 and 14 without Tet. In the third group, cells from day 5 were washed and cultured between days 5 and 8 without Tet, a sample was harvested, and Tet was added back to suppress Bcr-Abl expression. A portion of each sample was extracted for Western blot analysis with anti-Abl monoclonal as described in Materials and Methods to monitor expression of Bcr-Abl at the indicated time points. (B) Cells harvested on day 14 from each of the groups described above were analyzed by FACScan for the percentage of erythroid (TER119+), myeloid (CD11b+), and other cell types. The total cell number per culture is shown. (C) Morphology of cell preparations harvested and analyzed after Giemsa staining. Magnification is ×500.
Discussion
The combined use of the CAG promoter and Tet regulation system allowed us to tightly control the expression level of P210 Bcr-Abl throughout hematopoietic development and observe its immediate and direct effects on different cell types. The effects occurred on a population-wide basis, and there was insufficient time to allow for random second genetic changes to be selected. The overall data strongly support the idea that the Ph chromosome-encoded P210 Bcr-Abl gene product alone is sufficient to change the growth and differentiation properties of immature progenitors. The strategy employed here should be generally useful for the evaluation of any candidate gene involved in hematopoiesis.
Therapeutic interventions during chronic phase can include the use of IFN-α (43). Recent data evaluating cells pre- or postinterferon treatment suggest that the mode of action may be to induce degradation of the Bcr-Abl mRNA, and hence lower effective protein tyrosine kinase dosage (44). The leukemic clone loses its competitive advantage and normal progenitors can now compete for expansion and differentiation. The system described here should allow more precise evaluation of mixtures of Bcr-Abl-positive and -negative stem cells undergoing development within a common environment.
We observed an increase in proliferative rate and BrdUrd uptake for immature progenitors during days 5–8 of differentiation with minimal effects on cell death rates. Much data in the literature support a role for Bcr-Abl as providing a strong anti-apoptotic signal in various leukemia-derived cell lines stressed by growth factor removal or cellular damage (16). Alternative and synergistic mechanisms including activation of anti-apoptotic cellular genes like Bcl-xl and autocrine production of growth factors has been suggested (45, 46). To date, there has not been a dramatic change in reducing or enhancing the rate of apoptosis under our standard culture conditions.
There is considerable evidence for Bcr-Abl expression leading to discordant hematopoiesis in chronic phase CML (47). Levels of burst-forming unit (BFU)-E and colony-forming unit (CFU)-E are relatively normal, but most patients are mildly anemic (48, 49). Our cultures showed a clear association of Bcr-Abl expression with blockage of erythroid colony development and lower numbers of red cells derived from definitive erythropoiesis pathways. When Bcr-Abl expression was turned off during the later stages of culture development (days 8–14), the accumulated immature and intermediate cells of erythroid lineage could proceed through development. Inhibition of P210 Bcr-Abl activity with specific tyrosine kinase inhibitors in the CML-derived leukemic cell line K562 allows erythroid development to proceed as monitored by accumulation of hemoglobinized cells, suggesting a role in suppression of globin gene expression (50). These combined observations suggest that Bcr-Abl may antagonize erythroid development at several stages.
An important effect we have observed is the Bcr-Abl-mediated enhancement of immature progenitors with the surface antigen phenotypes of c-kit+, lin−, and CD34+,lin− associated with multilineage and myeloid development. Similar cell types with the Ph chromosome are enhanced in human marrow from CML patients (51). After multiparameter FACS separation, cells with the surface phenotype of pluripotential stem cells isolated from CML marrow show the presence of the Bcr-Abl rearrangement at the chromosomal level by in situ chromosome analysis (7, 8). Reverse transcription–PCR analysis on small numbers of such cells produces Bcr-Abl junction products (9, 52). However, it is not clear if the number of such stem cells is increased and if there is expression of functional mRNA, protein, and kinase activity for Bcr-Abl within human hematopoietic stem cells. This effect of Bcr-Abl on pluripotential stem cells can be approached in future studies with the ES differentiation system.
Acknowledgments
We thank Drs. Hermann Bujard (Heidelberg University), Kiwamu Akagi (Saitama Cancer Institute, Japan), and Takeshi Yagi (National Institute for Physiological Sciences, Japan) for providing plasmids for the Tet system; Drs. Austin Smith (University of Edinburgh), Toru Nakano, and Hitoshi Niwa (Osaka University) for the gift of ES and OP9 stromal cells; J. C. White for the preparation of the manuscript; and Irv Weissman and Charles Sawyers for critical review of this manuscript. T.E. is a postdoctoral associate, and O.N.W. is an Investigator of the Howard Hughes Medical Institute. This work was partially supported by Grant CA 76204 (to O.N.W.) from the National Cancer Institute.
Abbreviations
- ES
embryonal stem
- CML
chronic myelogenous leukemia
- Ph chromosome
Philadelphia chromosome
- LIF
leukemia inhibitory factor
- Tet
tetracycline
- tTA
Tet-regulated transactivator
References
- 1.Lowenberg B, Downing J R, Burnett A. N Engl J Med. 1999;341:1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- 2.DeClue J E, Lowy D R. In: The Molecular Basis of Blood Diseases. Stamatoyannopoulos G, Nienhuis A, Majerus P, Varmus H, editors. Philadelphia: Saunders; 1994. pp. 789–834. [Google Scholar]
- 3.Groffen J, Stephenson J R, Heisterkamp N, de Klein A, Bartram C R, Grosveld G. Cell. 1984;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 4.Konopka J B, Watanabe S M, Witte O N. Cell. 1984;37:1035–1042. doi: 10.1016/0092-8674(84)90438-0. [DOI] [PubMed] [Google Scholar]
- 5.Shtivelman E, Lifshitz B, Gale R P, Roe B A, Canaani E. Nature (London) 1985;315:550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- 6.Clarkson B D, Strife A, Wisniewski D, Lambek C, Carpino N. Leukemia. 1997;11:1404. doi: 10.1038/sj.leu.2400751. [DOI] [PubMed] [Google Scholar]
- 7.Irving J A, Lennard A, Storey N, Conn J, Dunn J, Oliver K, Proctor S J, Dickinson A M. Leukemia. 1999;13:944–949. doi: 10.1038/sj.leu.2401435. [DOI] [PubMed] [Google Scholar]
- 8.Tkachuk D C, Westbrook C A, Andreeff M, Donlon T A, Cleary M L, Suryanarayan K, Homge M, Redner A, Gray J, Pinkel D. Science. 1990;250:559–562. doi: 10.1126/science.2237408. [DOI] [PubMed] [Google Scholar]
- 9.Maguer-Satta V, Petzer A L, Eaves A C, Eaves C J. Blood. 1996;88:1796–1804. [PubMed] [Google Scholar]
- 10.Diamond J, Goldman J M, Melo J V. Blood. 1995;85:2171–2175. [PubMed] [Google Scholar]
- 11.Sawyers C L. N Engl J Med. 1999;340:1330–1340. doi: 10.1056/NEJM199904293401706. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein R. Semin Hematol. 1988;25:20–34. [PubMed] [Google Scholar]
- 13.Lugo T G, Pendergast A, Muller A J, Witte O N. Science. 1990;247:1079–1082. doi: 10.1126/science.2408149. [DOI] [PubMed] [Google Scholar]
- 14.McLaughlin J, Chianese E, Witte O N. Proc Natl Acad Sci USA. 1987;84:6558–6562. doi: 10.1073/pnas.84.18.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gishizky M L, Witte O N. Science. 1992;256:836–839. doi: 10.1126/science.1375394. [DOI] [PubMed] [Google Scholar]
- 16.Kabarowski J H, Allen P B, Wiedemann L. EMBO J. 1994;13:5887–5895. doi: 10.1002/j.1460-2075.1994.tb06934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daley G Q, Baltimore D. Proc Natl Acad Sci USA. 1988;85:9312–9316. doi: 10.1073/pnas.85.23.9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voncken J W, Griffiths S, Greaves M F, Pattengale P K, Heisterkamp N, Groffen J. Cancer Res. 1992;52:4534–4539. [PubMed] [Google Scholar]
- 19.Honda H, Oda H, Suzuki T, Takahashi T, Witte O N, Ozawa K, Ishikawa T, Yazaki Y, Hirai H. Blood. 1998;91:2067–2075. [PubMed] [Google Scholar]
- 20.Pear W S, Miller J P, Xu L, Pui J C, Soffer B, Quackenbush R C, Pendergast A M, Bronson R, Aster J C, Scott M L, et al. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 21.Kelliher M A, McLaughlin J, Witte O N, Rosenberg N. Proc Natl Acad Sci USA. 1990;87:6649–6653. doi: 10.1073/pnas.87.17.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daley G Q, Van Etten R A, Baltimore D. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 23.Gishizky M L, Johnson-White J, Witte O N. Proc Natl Acad Sci USA. 1993;90:3755–3759. doi: 10.1073/pnas.90.8.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fialkow P J, Martin P J, Najfeld V, Penfold G K, Jacobson R J, Hansen J A. Blood. 1981;58:158–163. [PubMed] [Google Scholar]
- 25.Raskind W H, Ferraris A M, Najfeld V, Jacobson R J, Moohr J W, Fialkow P J. Leukemia. 1993;7:1163–1167. [PubMed] [Google Scholar]
- 26.Biernaux C, Loos M, Sels A, Huez G, Stryckmans P. Blood. 1995;86:3118–3122. [PubMed] [Google Scholar]
- 27.Nakano T, Kodama H, Honjo T. Science. 1994;265:1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- 28.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa H, Burdon T, Chambers I, Smith A. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baron U, Gossen M, Bujard H. Nucleic Acids Res. 1997;25:2723–2729. doi: 10.1093/nar/25.14.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 32.Hellen C U, Wimmer E. Curr Top Immunol. 1995;203:31–63. doi: 10.1007/978-3-642-79663-0_2. [DOI] [PubMed] [Google Scholar]
- 33.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 34.Chambard J C, Pognonec P. Nucleic Acids Res. 1998;26:3443–3444. doi: 10.1093/nar/26.14.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suwabe N, Takahashi S, Nakano T, Yamamoto M. Blood. 1998;92:4108–4118. [PubMed] [Google Scholar]
- 36.Schiff-Maker L, Burns M C, Konopka J B, Clark S, Witte O N, Rosenberg N. J Virol. 1986;57:1182–1186. doi: 10.1128/jvi.57.3.1182-1186.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartley S B, Cooke M P, Fulcher D A, Harris A W, Cory S, Basten A, Goodnow C C. Cell. 1993;72:325–335. doi: 10.1016/0092-8674(93)90111-3. [DOI] [PubMed] [Google Scholar]
- 38.Era, T., Takagi, T., Takahashi, T., Bories, J.-C. & Nakano, T. (2000) Blood, in press. [PubMed]
- 39.Nakano T, Kodama H, Honjo T. Science. 1996;272:722–724. doi: 10.1126/science.272.5262.722. [DOI] [PubMed] [Google Scholar]
- 40.Van Etten R A. The Molecular Pathogenesis of the Philadelphia-Positive Leukemias: Implications for Diagnosis and Therapy. Norwood: Klumer Academic; 1993. [DOI] [PubMed] [Google Scholar]
- 41.Faderl S, Talpaz M, Estrov Z, O'Brien S, Kurzrock R, Kantarjian H M. N Engl J Med. 1999;341:164–172. doi: 10.1056/NEJM199907153410306. [DOI] [PubMed] [Google Scholar]
- 42.Era T, Takahashi T, Sakai K, Kawamura K, Nakano T. Blood. 1997;89:1207–1213. [PubMed] [Google Scholar]
- 43.Silver R T, Woolf S H, Hehlmann R, Appelbaum F R, Anderson J, Bennett C, Goldman J M, Guilhot F, Kantarjian H M, Lichtin A E, et al. Blood. 1999;94:1517–1536. [PubMed] [Google Scholar]
- 44.Pane F, Mostarda I, Selleri C, Salzano R, Raiola A M, Luciano L, Saglio G, Rotoli B, Salvatore F. Blood. 1999;94:2200–2207. [PubMed] [Google Scholar]
- 45.Jiang X, Lopez A, Holyoake T, Eaves A, Eaves C. Proc Natl Acad Sci USA. 1999;96:12804–12809. doi: 10.1073/pnas.96.22.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amarante-Mendes G P, McGahon A J, Nishioka W K, Afar D E, Witte O N, Green D R. Oncogene. 1998;16:1383–1390. doi: 10.1038/sj.onc.1201664. [DOI] [PubMed] [Google Scholar]
- 47.Clarkson B, Strife A. Leukemia. 1993;7:1683–1721. [PubMed] [Google Scholar]
- 48.Goldman J M, Shiota F, Th'ng K H, Orchard K H. Br J Haematol. 1980;46:7–13. doi: 10.1111/j.1365-2141.1980.tb05929.x. [DOI] [PubMed] [Google Scholar]
- 49.Marley S B, Lewis J L, Goldman J M, Gordon M Y. Br J Haematol. 1996;93:878–883. doi: 10.1046/j.1365-2141.1996.d01-1738.x. [DOI] [PubMed] [Google Scholar]
- 50.Anafi M, Gazit A, Zehavi A, Ben-Neriah Y, Levitzki A. Blood. 1993;82:3524–3529. [PubMed] [Google Scholar]
- 51.Martin-Henao G A, Quiroga R, Sureda A, Garcia J. Am J Hematol. 1999;61:178–186. doi: 10.1002/(sici)1096-8652(199907)61:3<178::aid-ajh4>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 52.Bedi A, Zehnbauer B A, Collector M I, Barber J P, Zicha M S, Sharkis S J, Jones R J. Blood. 1993;81:2898–2902. [PubMed] [Google Scholar]