Abstract
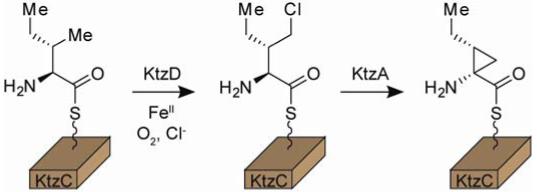
The biosynthetic gene cluster for the kutzneride family of hexapeptidolactones includes the four-gene cassette ktzABCD postulated to generate a nonproteinogenic amino acid. Encoded by this cassette are the nonheme FeII-dependent halogenase KtzD and the acyl-CoA dehydrogenase-like flavoprotein KtzA, proposed to work in conjunction with adenylating protein KtzB and carrier protein KtzC. In the present work, we report the in vitro reconstitution of this four-protein system and identify the final product as (1S, 2R)-allocoronamic acid bound in thioester linkage to KtzC. Further analysis of KtzD and KtzA support a biosynthetic pathway that involves KtzD-mediated generation of a γ-chloroisoleucyl intermediate which is cyclized to the final product by KtzA without redox participation of the bound flavin cofactor. This work introduces a new monomer for potential incorporation into nonribosomal peptides and validates the unique strategy for its biosynthesis.
Nonribosomal peptides constitute a diverse class of secondary metabolites with important biological activities.1 In addition to the twenty proteinogenic amino acids, nonribosomal peptide synthetases make use of a large number of modified amino acid and hydroxy acid building blocks.2 Understanding the chemical pathways and mechanisms used in the biosyntheses of these monomers is a first step towards expanding the pool of new molecules available through metabolic engineering.
Towards this end, we recently used halogenase-directed probes to clone the gene cluster3 responsible for the biosynthesis of a series of hexadepsipeptides, kutznerides, isolated from the soil actinomycete kutzneria4,5 (Figure S1). These molecules contain a number of unusual monomers whose biosyntheses have not been described. Bioinformatic analysis of the gene cluster identified three potential halogenases. KtzQ and KtzR are shown in separate work to be flavin-dependent tryptophan halogenases which can generate the 6,7-dichlorotryptophan residue postulated in kutzneride biosynthesis.6 KtzD was predicted to be an FeII-dependent halogenase capable of chlorinating an unactivated carbon center.
The four-protein cassette KtzABCD could potentially function to generate a novel amino acid via a cryptic chlorination pathway. KtzC and KtzB were assigned roles as carrier protein and adenylation protein, respectively. KtzA is a flavoprotein homologous to members of the acyl-CoA dehydrogenase (ACAD) family of enzymes. We initially hypothesized that these four proteins produce the nonproteinogenic amino acid 2-(1-methylcyclopropyl)glycine (MecPG),3,7 a component of all kutzernide molecules reported to date. Herein, we report the biochemical characterization of KtzABCD and demonstrate that they instead function to produce an alternative cyclopropyl-containing amino acid, (1S, 2R)-allocoronamic acid (alloCMA), tethered to KtzC. To our knowledge, this is the first identification of alloCMA in a natural source.
To study the role of the KtzABCD cassette, the individual genes were cloned into E. coli expression vectors as His-tag fusions. Overexpression and Ni-NTA purification provided soluble protein for each construct. The carrier protein KtzC was isolated in apo form and phosphopantetheinylated via incubation with coenzyme A and Sfp. KtzA was isolated with FAD bound at 50% occupancy. After initial purification of the halogenase KtzD, the His tag was proteolytically cleaved and the enzyme purified by gel filtration and reconstituted anaerobically with FeII and α-ketoglutarate to obtain the active form of the protein.
In accordance with its previously described adenylating activity,3 KtzB efficiently activates and loads both l-Ile and l-allo-Ile to holo-KtzC as the phosphopantetheinyl thioesters. The protein-tethered amino acids can be identified by enzymatic hydrolysis with the Type II thioesterase TycF8 and conversion to readily detectable isoindole derivatives using established procedures.9 Incubation of l-Ile-S-KtzC with KtzD for 1 hour results in complete consumption of starting material and production of a new aminoacyl thioester product (Figure 1A). MS analysis on the released isoindole derivative shows this product results from a single chlorination event. Treatment of l-allo-Ile-S-KtzC with KtzD does not lead to the formation of a new product (Figure S3), illustrating the substrate specificity of the halogenase.
Figure 1.
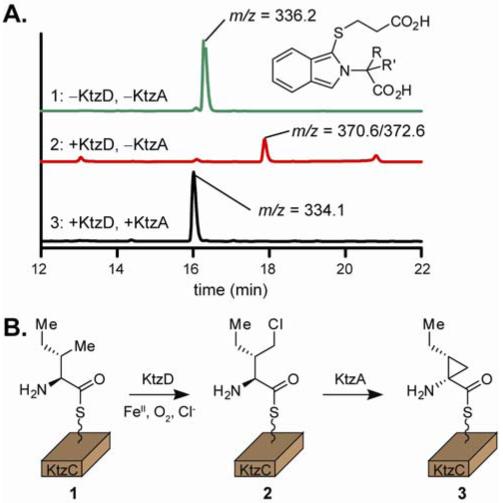
A. HPLC and MS analysis of cleaved reaction products as isoindole derivatives. Treatment of L-Ile-S-KtzC (1) with KtzD yields a chlorinated intermediate (2) which is dehydrochlorinated by KtzA (3). B. The biosynthetic pathway for allocoronamic acid tethered in thioester linkage to KtzC. A detailed mechanism is presented in scheme S1.
Exposure of the chlorinated intermediate to KtzA again leads to complete conversion to a new product. MS analysis shows the released amino acid product to have a m/z ratio consistent with dehydrochlorination of the intermediate and net two-electron oxidation of the starting isoleucine. Supplementing the reaction with excess FAD had no effect on product formation (data not shown). To confirm the structure of the final product, the isoindole derivative was purified by HPLC and analyzed by 1H- and COSY-NMR. Additionally, 13C6-Ile was used as the starting material, and the isotope-enriched product analyzed by HMQC and 1H-NMR spectroscopy. The spectral data support the atom connectivity of alloCMA (Figures S7/S8, Table S1).
The stereochemistry of the product was deduced from two observations. First, the observed product fails to co-elute with an authentic sample of coronamic acid under the typical HPLC conditions (Figure S9). Second, the γ-CH2 protons show 1H-NMR resonances at 1.47 and -0.14 ppm. The unusual upfield resonance is best explained by strong shielding of the proton as a result of its location directly above the aromatic ring of the isoindole group. For this to occur on the rigid cyclopropyl skeleton, the ethyl side chain and isoindole group must be in a cis arrangement. Based on the observed alloCMA structure, we propose a biosynthetic scheme which involves γ-CH3 chlorination of l-Ile-S-KtzC followed by KtzA-mediated α,γ cyclization to alloCMA-S-KtzC via an intermediate Cα-carbanion (Figure 1B).
To confirm the regiochemistry of γ-chlorination by KtzD, we synthesized substrates deuterated at the γ-CH3 position (Ile-d3) or at the β-CH,γ-CH2, and δ-CH3 positions (Ile-d6). Treatment of Ile-d6-S-KtzC with KtzD produces a chlorinated product that retains all deuterium labels. Identical treatment of Ile-d3-S-KtzC produces a chlorinated product which retains only two deuterium labels (Figure 2). Additionally, analysis of the reaction at partial conversion shows that the rate of chlorination of Ile-d3 is retarded relative to Ile and Ile-d6 (Figure S4). This kinetic isotope effect for H/D abstraction is consistent with the isotope effect observed in stopped flow studies of the related halogenase CytC3.10
Figure 2.
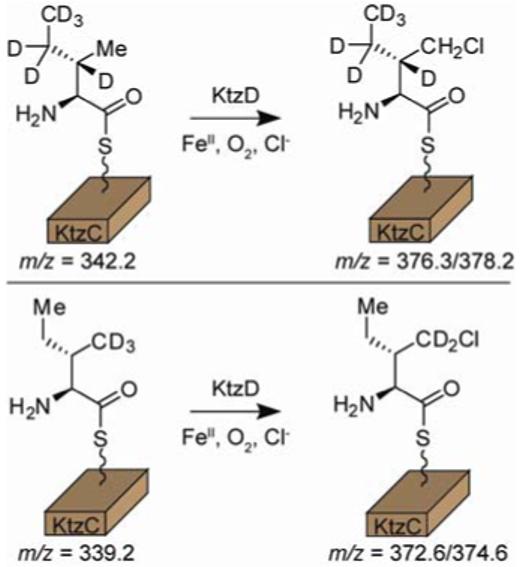
Treatment of deuterated Ile-S-KtzC isotopologues with KtzD indicates regioselective chlorination at the γ-CH3 position.
Cyclization of 2 to alloCMA-S-KtzC is mediated by the ACAD-like flavoprotein KtzA. ACADs catalyze dehydrogenation of acyl/aminoacyl CoA thioesters by abstraction of the acidic α- proton and transfer of a β-hydride to the FAD cofactor to yield the α,β-unsaturated thioester and FADH2.11 In contrast, KtzA need only abstract the alpha proton of 2 to generate a stabilized thioester enolate which can collapse by intramolecular attack at Cγ to form the cyclopropane. Loss of Cl- constitutes the two-electron oxidation of substrate and avoids formation of the FADH2 oxidation state.12 To verify that KtzA can catalyze reversible α-proton abstraction as the initial event in cyclization, unlabeled Ile-S-KtzC was incubated with KtzA in the presence of D2O. Recovery of the amino acid and analysis by HPLC and MS showed incorporation of a single deuterium atom (Figures S5/S6). The yield is similar to that for untreated Ile-S-KtzC indicating that there is no significant loss of amino acid through hydrolysis of a dehydrogenated intermediate. Attempts to generate apo-KtzA by treatment of Ni-bound protein with 2M KBr and 2M urea gave negligible yields of protein; thus it is possible the flavin cofactor is needed to maintain the tertiary structure of KtzA.
The rerouting of a carbanion intermediate in a flavoprotein active site to intramolecular elimination rather than two-electron transfer to the FAD coenzyme has previously been observed for the flavoprotein D-alanine oxidase when presented with a non-natural substrate.13 In that case, β-chloroalanine can be partitioned between αβ-dehydrochlorination to dehydroalanine and Cα oxidation to β-chloro-iminopyruvate. In KtzA catalysis, the net removal of Hα and Clγ appears to be the physiological reaction. We have previously reported a related four protein pathway in coronamic acid biosynthesis where l-allo-Ile is comparably converted to (1S,2S)-coronamic acid via the same cryptic γ-chlorination strategy, but in that case the cyclization catalyst is the Zn-dependent enzyme CmaC.9,14 KtzA and CmaC thus represent two distinct enzymatic strategies for generation of the thioester enolate needed to initiate cyclopropane formation.
In conclusion, we have characterized the activities of the KtzABCD proteins and identified the novel metabolite alloCMA as the protein-tethered product of these enzymes. Deuterium-labeling studies confirm the regioselective chlorination of l-Ile-S-KtzC at the γ-CH3 position by KtzD and support the unusual role of the flavoprotein KtzA as the catalytic base which induces cyclization without redox participation of bound FAD. This cassette provides a biosynthetic route to a novel amino acid monomer that is a stereochemical complement to the previously described coronamic acid. At the same time, the results suggest future directions for research on kutzneria. Identifying additional nonribosomal peptides which may contain alloCMA and locating the gene cassette which provides for MecPG biosynthesis are now key avenues for metabolomic and genomic approaches.
Supplementary Material
Acknowledgement
We thank Dr. Danica Galonic Fujimori for technical assistance and helpful discussions. We thank Dr. Alex Koglin and Dr. Dominique Frueh for assistance in obtaining HMQC spectra. This work was supported by NIH grant GM20011 to C.T.W.
REFERENCES
- (1).Felnagle EA, Jackson EE, Chan YA, Podevels AM, Berti AD, McMahon MD, Thomas MG. Mol. Pharm. 2008;5:191–211. doi: 10.1021/mp700137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Döhren H. v. In: Biochemistry of peptide antibiotics. Döhren H. v., Kleinkauf H., editors. de Gruyter; Berlin, New York: 1990. pp. 411–506. [Google Scholar]
- (3).Fujimori DG, Hrvatin S, Neumann CS, Strieker M, Marahiel MA, Walsh CT. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16498–16503. doi: 10.1073/pnas.0708242104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Broberg A, Menkis A, Vasiliauskas R. J. Nat. Prod. 2006;69:97–102. doi: 10.1021/np050378g. [DOI] [PubMed] [Google Scholar]
- (5).Pohanka A, Menkis A, Levenfors J, Broberg A. J. Nat. Prod. 2006;69:1776–1781. doi: 10.1021/np0604331. [DOI] [PubMed] [Google Scholar]
- (6).Heemstra JR, Walsh CT. J. Am. Chem. Soc. 2008;130 doi: 10.1021/ja806467a. ####-####. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Neumann CS, Fujimori DG, Walsh CT. Chem. Biol. 2008;15:99–109. doi: 10.1016/j.chembiol.2008.01.006. [DOI] [PubMed] [Google Scholar]
- (8).Yeh E, Kohli RM, Bruner SD, Walsh CT. Chembiochem. 2004;5:1290–3. doi: 10.1002/cbic.200400077. [DOI] [PubMed] [Google Scholar]
- (9).Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- (10).Fujimori DG, Barr EW, Walsh CT, Bollinger JM, Krebs C. Nat. Chem. Biol. 2007;3:113–116. doi: 10.1038/nchembio856. [DOI] [PubMed] [Google Scholar]
- (11)(a).Ghisla S, Thorpe C. Eur. J. Biochem. 2004;271:494–508. doi: 10.1046/j.1432-1033.2003.03946.x. [DOI] [PubMed] [Google Scholar]; (b) Mansoorabadi SO, Thibodeaux CJ, Liu H. J. Org. Chem. 2007;72:6329–6342. doi: 10.1021/jo0703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).While not common, redox inactive flavoproteins are known. For examples, see ref 11(b) and:Bornemann S. Nat. Prod. Rep. 2002;19:761–772. doi: 10.1039/b108916c.
- (13).Walsh CT, Schonbrunn A, Abeles RH. J. Biol. Chem. 1971;246:6855–6866. [PubMed] [Google Scholar]
- (14).Kelly WL, Boyne MT, Yeh E, Vosburg DA, Galonic DP, Kelleher NL, Walsh CT. Biochemistry. 2007;46:359–368. doi: 10.1021/bi061930j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


