Abstract
The role of extracellular DNA in the maintenance of biofilms formed by gram-positive and gram-negative bacteria was studied. This study evaluated all the bacterial strains that were tested for the presence of extracellular DNA with an average size of 30 kb in the matrix. Our results indicate changes in community biomass, architecture, morphology, and the numbers of CFU in the presence of DNase. This effect seems to be common to biofilms established by various unrelated gram-positive and gram-negative bacteria. The cleavage of extracellular DNA leads to the formation of an altered biofilm that permits the increased penetration of antibiotics. Thus, the addition of DNase enhances the effect of antibiotics, resulting in decreased biofilm biomass and numbers of CFU.
It is well known that bacteria form biofilms, in which they survive in the presence of high concentrations of antimicrobial agents (13, 23, 28, 51, 52, 55). Antibiotics at concentrations of 102 to 104 times the MIC cause no killing effect on the bacteria in biofilm communities (2, 7, 14, 22, 24, 32, 54). Bacterial survival in biofilms may be determined by multiple different factors. Biofilms are covered by a surface film composed of lipid components similar to those in bacterial membranes, which are a barrier for the penetration of some antibiotics (57, 59). The roles of separate components of the biofilm matrix in the bacterial life have recently been studied (17, 26, 50, 63). It is known that the matrix consists of proteins, polysaccharides, lipids, and nucleic acids, which form an extracellular polymeric substance (EPS) (21, 34, 56, 64). Some suggest that EPS can interact with antibiotics in a manner that leads to a decline in antibacterial activity (5, 8, 29). It was also shown that bacterial survival in biofilms in the presence of antibiotics can be determined by special cells persisters, which are tolerant to various drugs (27, 31, 35, 51). Recently, extracellular DNA has been found in the matrix of Pseudomonas aeruginosa and Neisseria gonorrhoeae biofilms (1, 26, 32, 50, 63). Previously, free circulating DNA was found in human blood plasma (3, 45, 61), and it is also present in marine sediments and soil (6, 15, 16, 44, 60). It was shown that the extracellular DNA in the biofilm matrix could take part in the development of bacterial communities (25, 40, 49, 63). It can be released by live cells, possibly via membrane vesicles composed of bacterial lipids (65), or it may enter the matrix from lysed cells (10, 18, 19, 48, 58, 65). We have recently found cell-free DNA as a component of EPS in biofilms formed by various unrelated gram-positive and gram-negative bacteria and fungi (G. Tetz and V. Tetz, unpublished data). It was previously shown that the destruction of the extracellular DNA of P. aeruginosa and Streptococcus pneumoniae could change the properties of the biofilms formed by these bacteria (30, 40, 63). At the same time, this phenomenon has not been studied with other microorganisms. The role of extracellular DNA in the interaction of bacterial biofilms with environmental factors is also unclear. Thus, the aim of the present study was to evaluate established biofilms for the presence of extracellular DNA and the role of the extracellular DNA in the established biofilms formed by various bacteria and to evaluate whether the antimicrobial effect of antibiotics can be enhanced by destroying this DNA.
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli ATCC 25922, Haemophilus influenzae VT 450-2006, Klebsiella pneumoniae VT 1367, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, Streptococcus pyogenes VT 59, and Acinetobacter baumannii VT 126 were used in this study. The strains designated VT were isolated from patients in the clinics of the St. Petersburg State Pavlov Medical University.
Medium and culture conditions.
The liquid media used for bacterial growth were Luria-Bertani (Becton Dickinson, Sparks, MD), Mueller-Hinton (bioMerieux, Paris, France), Schaedler (bioMerieux), and Haemophilus test medium (Becton Dickinson) broths. The strains were grown at 37°C, and liquid cultures were incubated without shaking. Before use in the biofilm experiments, the cells were harvested and washed twice with 0.15 M isotonic phosphate buffer (pH 7.2), and the cell suspensions were standardized to an optical density at 520 nm of 0.8.
Enzymes.
Bovine pancreatic DNase I (Sigma Chemical Co., St. Louis, MO) with a specific activity of 2,200 Kunitz units/mg, RNase A (Sigma), and proteinase K (Sigma) were used.
Antibiotics.
The antibiotics tested were ampicillin (ICN), cefotaxime (ICN), rifampin (rifampicin; Sigma), levofloxacin (Sigma), and azithromycin (Pfizer).
Planktonic antimicrobial susceptibility testing.
MIC tests were performed in cation-adjusted Mueller-Hinton and Haemophilus test medium broths according to the guidelines of the Clinical and Laboratory Standards Institute for broth microdilution susceptibility testing (11, 12).
Biofilm formation assay.
An inoculum was prepared by using a 24-h broth culture. The inoculum, which contained 7.53 ± 0.22 log10 CFU/ml, was added to the wells of 96-well plates (200 μl/well), 35-mm petri dishes (2 ml), and coverslips that were placed in glass tubes (2 ml) (all from Sarstedt, Germany); and the plates, dishes, and coverslips were incubated at 37°C for 24 h.
Biofilm CFU assay.
Biofilms were grown in 96-well plates for 24 h at 37°C. Liquid medium with bacteria was aspirated from the wells, which were then washed with isotonic phosphate buffer (0.15 M, pH 7.2). The biofilms were scraped thoroughly, with particular attention given to the well edges. The contents of the wells were again aspirated and then placed in 1.0 ml of isotonic phosphate buffer (0.15 M, pH 7.2), and the total number of CFU was determined by the serial dilution method and plating on the appropriate media.
Microscopy.
For visualization of the biofilms by light microscopy, bacteria were grown on glass coverslips for 24 h at 37°C. Established biofilms were treated with various concentrations of DNase I and were incubated for an additional 24 h at 37°C. The coverslips were washed with a continuous spray of isotonic phosphate buffer (0.15 M, pH 7.2) and glued to a slide. The slides were air dried for 20 to 30 min and were fixed by gentle heating and allowed to cool. Then, 0.1% crystal violet (gentian violet in isopropanol-methanol-1× phosphate-buffered saline [PBS; 0.01 M sodium phosphate, pH 7.2, 0.15 M NaCl] [1:1:18]) was placed on each glass slide, and the slides were incubated for 15 min at room temperature. The slides were then washed thoroughly with 1× PBS until the PBS ran clear. The slides were mounted with immersion oil for microscopy (Sigma) and were examined with an Axiolab microscope (Carl Zeiss, Oberkochen, Germany) at a magnification of ×100. Images were acquired with a PowerShot G3 digital camera (Canon, Tokyo, Japan) attached to the microscope.
Quantitative determination of biofilm formation.
Quantitative determination of biofilm formation was performed by the spectrophotometric method, which measures the total biofilm biomass, including bacterial cells and EPS (43). The liquid medium with the bacteria was aspirated from the wells, and the wells were washed with isotonic phosphate buffer (0.15 M, pH 7.2) without disturbing the adherent film. The biofilms were stained with 200 μl of 0.1% crystal violet and incubated at room temperature for 30 min, and excess stain was removed by three gentle washes with sterile distilled water. After the biofilms had dried, the crystal violet was solubilized by adding 200 μl of ethanol-acetone (80:20, wt/wt), and the extent of biofilm formation was determined by measuring the absorbance of the stained adherent film at 570 nm with a microplate reader (Stat-Fax-2100).
Effect of antibiotics and DNase I on established biofilms.
Biofilm formation was carried out in a 96-well plate, as described above for the biofilm formation assay. After 24 h of incubation at 37°C, the supernatant from each well was gently aspirated with a micropipette. Each well was washed three times with isotonic phosphate buffer (0.15 M, pH 7.2) under aseptic conditions to eliminate the unbound bacteria, without disturbing the adherent film; and 200 μl of the particular antibiotic, DNase I, or antibiotic together with a DNase I dilution in the appropriate medium was added. The plates were incubated at 37°C for 24 h. After exposure to antibiotic, DNase I, or antibiotic together with DNase I for 24 h, the solutions were discarded and the wells were filled (200 μl) with isotonic phosphate buffer (0.15 M, pH 7.2). The numbers of CFU were counted or quantitative determination of biofilm formation was carried out.
Isolation of extracellular DNA.
Biofilm formation was carried out in petri dishes, as described above for the biofilm formation assay, for 24 h at 37°C. The supernatant was aspirated, and the biofilms were gently washed three times with isotonic phosphate buffer (0.15 M, pH 7.2) without disturbing the adherent film. Biofilms were scraped from the petri dishes in the presence of 0.5 ml of isotonic phosphate buffer (0.15 M, pH 7.2), and the biofilm matrix was separated from the bacteria by centrifugation at 5,000 × g for 10 min (5415 C centrifuge; Eppendorf Geratgebau GmbH, Engelsdorf, Germany). Extracellular DNA was extracted from the biofilm matrix with phenol-chloroform and was precipitated with 100% cold ethanol, as described previously (38).
Electrophoresis.
The size of the extracted DNA was ascertained by electrophoresis on a 1.0% agarose gel (ICN) in the presence of 0.5 μg/ml of ethidium bromide, which was used for DNA staining, and the DNA was visualized under UV light (UV Transilluminator 2000; Bio-Rad).
Digestion with DNase, RNase, and proteinase K.
The nucleic acids that were extracted with phenol-chloroform were then resuspended in 20 μl of sterile distilled water. Twenty microliters of the purified nucleic acids was treated with either DNase I (final concentration, 100 U/ml), RNase A (final concentration, 20 μg/ml), or proteinase K (final concentration, 0.5 mg/ml) (all from Sigma), according to the manufacturer's protocol. The products from the untreated and the treated samples were analyzed by 0.7% agarose gel electrophoresis and ethidium bromide staining.
Preparation of DNA digestion products.
Enzyme-digested DNA was obtained by adding 100 μg/ml of DNase I to standard chicken embryo DNA (ICN), and following incubation for 24 h at 37°C, the enzyme was inactivated by heating in a water bath for 15 min at 100°C.
Effect of DNA digestion products on biofilm formation.
Different concentrations (20, 50, and 100 μg/ml) of the digested DNA in 200 μl of the appropriate medium were simultaneously inoculated together with the 24-h bacterial broth culture in a 96-well plate as described above for the biofilm formation assay, and the plate was incubated at 37°C for 24 h.
Statistical analysis.
All statistic analyses were performed by using the statistics package Statistica for Windows (version 5.0). A P value of <0.05 was considered statistically significant.
RESULTS
Confirmation of the presence of extracellular DNA in biofilm matrix.
Extracellular polymeric molecules with molecular sizes of about 30 kb were found in the matrices of all bacterial biofilms tested after phenol-chloroform extraction. To evaluate the nature of these molecules, they were stained with ethidium bromide, which is known to bind specifically to double-stranded nucleic acids (33). Moreover, enzymatic treatment of 20 μl purified extracellular polymeric molecules with DNase I, RNase A, or proteinase K with subsequent analysis by 0.7% agarose gel electrophoresis indicated that only DNase I treatment resulted in the complete removal of the 30-kb DNA band. The results indicate that the extracellular matrix of bacterial biofilms formed by unrelated gram-positive and gram-negative bacteria contains detectable DNA fragments of about 30 kb.
Antimicrobial susceptibility testing of planktonic suspensions.
We studied the in vitro susceptibilities of bacterial strains in planktonic suspensions to antimicrobial agents, including representatives of classes of antibiotics with different mechanisms of action; and the results are shown in Table 1.
TABLE 1.
In vitro susceptibilities of bacterial strains to ampicillin, cefotaxime, azithromycin, levofloxacin, and rifampin
| Strain | MIC (mg/liter)a
|
||||
|---|---|---|---|---|---|
| Ampicillin | Cefotaxime | Azithromycin | Levofloxacin | Rifampin | |
| A. baumannii | 50.0 | 1.5 | NDb | 8.0 | 8.0 |
| H. influenzae | 0.5 | 0.16 | 1.0 | 0.07 | 0.8 |
| K. pneumoniae | 64.0 | 0.06 | ND | 0.15 | 16 |
| E. coli | 16.0 | 1.5 | ND | 0.035 | 8.0 |
| P. aeruginosa | 128.0 | 64.0 | ND | 0.6 | 16.0 |
| S. aureus | 0.025 | 3.8 | 0.3 | 0.3 | 0.25 |
| S. pyogenes | 0.25 | 0.03 | 0.25 | 0.5 | 0.25 |
The results are representative of those from three to four separate experiments.
ND, not done.
Effect of DNase I on 24-h-old biofilms.
The susceptibility of established biofilms to different concentrations of DNase was studied by microscopy, spectrophotometric analysis, and determination of the numbers of CFU. The results are shown in Table 2.
TABLE 2.
Effect of DNase I on 24-h-old biofilm biomass
| Strain | Optical density at 570 nm of biofilm biomass after treatment with DNase I at the following concn (μg/ml)a:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0 (control) | 0.5 | 1.0 | 5.0 | 10.0 | 20.0 | 100.0 | 1,000.0 | |
| A. baumannii | 1.157 | 0.789 | 0.738 | 0.645 | 0.534 | 0.486 | 0.456 | 0.345 |
| H. influenzae | 1.132 | 0.766 | 0.728 | 0.608 | 0.534 | 0.467 | 0.428 | 0.389 |
| K. pneumoniae | 1.039 | 0.721 | 0.673 | 0.586 | 0.517 | 0.453 | 0.407 | 0.321 |
| E. coli | 1.067 | 0.734 | 0.674 | 0.590 | 0.495 | 0.457 | 0.412 | 0.341 |
| P. aeruginosa | 1.373 | 0.939 | 0.899 | 0.777 | 0.664 | 0.586 | 0.533 | 0.445 |
| S. aureus | 1.509 | 1.092 | 1.035 | 0.928 | 0.790 | 0.724 | 0.652 | 0.524 |
| S. pyogenes | 1.458 | 1.048 | 0.981 | 0.888 | 0.736 | 0.665 | 0.609 | 0.479 |
The results are representative of those from three to four independent experiments.
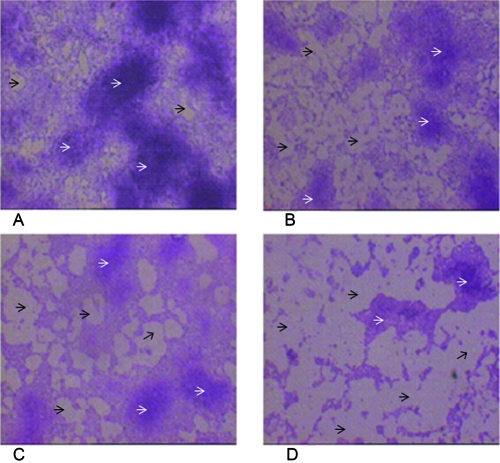
Microscopy of biofilms treated with different concentrations of DNase I revealed changes in the morphologies of the bacterial communities (Fig. 1). Biofilms that were not treated with DNase I formed a confluent biofilm (Fig. 1A). A microscopic study of the biofilms treated with different concentrations of DNase I (Fig. 1B to D) displayed regions of various sizes that were practically free of cells. Microcolonies were not confluent, and their sizes and numbers were decreased in biofilms treated with DNase I compared with their sizes and numbers in the control that was not treated with DNase I. These changes were common for all strains tested, as the original appearance was not restored during the subsequent 120 h of biofilm growth.
FIG. 1.
Photomicrographs illustrating the effects of different DNase I concentrations on established E. coli ATCC 25922 biofilm formation. Biofilms were formed on the coverslips within 24 h at 37°C. Established biofilms were treated with DNase I at 0.5 μg/ml, 1.0 μg/ml, or 5.0 μg/ml for 24 h at 37°C; stained with 0.1% crystal violet; and photographed at ×100 magnification. The black arrows denote the regions that are free of cells; microcolonies are indicated by white arrows. Bars, 50 μm. (A) Control (untreated); (B to D) treatment with DNase I at 0.5 μg/ml, 1.0 μg/ml, and 5.0 μg/ml, respectively, for 24 h.
Established 24-h-old biofilms were treated with various concentrations of DNase I for 24 h at 37°C, and then the biomass was subsequently determined spectrophotometrically. Thereafter, the biomass of the biofilm was determined and compared with the biomass of the biofilm not treated with DNase I.
The results of four independent experiments, summarized in Table 2, indicate that DNase I has a consistent effect on established biofilms of various bacteria. This dose-ranging study revealed a DNase concentration-dependent reduction in biomass. DNase I at a concentration 5.0 μg/ml decreased the biofilm biomass by approximately 40% for all strains tested but did not change the numbers of CFU (Table 3). Thus, we used DNase I at a concentration of 5.0 μg/ml in the subsequent experiments.
TABLE 3.
Bactericidal activities of various antibiotics, DNase I, or each antibiotic together with DNase I against bacteria in 24-h-old biofilms
| Treatment (n = 3) | Bacterial count (log10 CFU/well)a
|
||||||
|---|---|---|---|---|---|---|---|
| A. baumannii | H. influenzae | K. pneumoniae | E. coli | P. aeruginosa | S. aureus | S. pyogenes | |
| Control | 8.32 ± 0.30 | 8.46 ± 0.65 | 8.67 ± 1.30 | 8.92 ± 0.55 | 8.71 ± 0.65 | 9.83 ± 0.51 | 8.32 ± 1.58 |
| DNase I | 8.34 ± 0.20 | 8.41 ± 0.25 | 8.60 ± 1.11 | 8.37 ± 0.47 | 8.58 ± 0.90* | 9.80 ± 0.45* | 8.24 ± 1.10 |
| Ampicillin | 8.29 ± 0.15 | 8.44 ± 0.30 | 8.33 ± 1.00 | 8.55 ± 0.77* | 8.62 ± 1.00 | 9.51 ± 0.40* | 8.16 ± 0.77 |
| Ampicillin + DNase I | 7.30 ± 0.11** | 7.34 ± 0.41** | 7.37 ± 0.50** | 7.27 ± 0.28** | 7.48 ± 1.34** | 8.60 ± 1.09** | 7.18 ± 0.81 |
| Cefotaxime | 8.32 ± 0.26 | 8.45 ± 0.43 | 8.54 ± 0.97 | 8.66 ± 0.81 | 8.68 ± 0.40 | 9.67 ± 0.36* | 8.20 ± 0.25 |
| Cefotaxime + DNase I | 7.26 ± 0.10** | 7.38 ± 0.20** | 7.53 ± 0.77** | 7.51 ± 0.97** | 7.38 ± 0.72** | 8.20 ± 0.55** | 6.70 ± 1.00** |
| Levofloxacin | 8.26 ± 0.15 | 8.25 ± 0.65 | 8.62 ± 0.87 | 8.46 ± 0.76* | 8.69 ± 1.15 | 7.39 ± 0.35* | 6.39 ± 0.40 |
| Levofloxacin + DNase I | 6.76 ± 1.52** | 6.78 ± 0.77** | 7.28 ± 0.62** | 6.75 ± 1.36** | 6.60 ± 0.52** | 6.35 ± 0.64** | 5.31 ± 0.65** |
| Rifampin | 8.15 ± 0.41 | 8.14 ± 0.55* | 8.50 ± 0.75 | 8.58 ± 0.73* | 8.56 ± 0.65 | 7.50 ± 0.41* | 6.44 ± 0.55 |
| Rifampin + DNase I | 6.68 ± 1.05** | 6.79 ± 1.68 | 7.18 ± 0.68** | 7.19 ± 0.61** | 6.38 ± 1.25** | 6.60 ± 1.07** | 5.65 ± 1.93** |
| Azithromycin | ND*** | 8.14 ± 0.89 | ND | ND | ND | 9.75 ± 1.06 | 8.24 ± 0.68 |
| Azithromycin + DNase I | ND | 7.19 ± 0.36 | ND | ND | ND | 9.19 ± 0.87** | 8.10 ± 0.43** |
Viable cells (log10 CFU/well) were measured after 24 h of growth in the presence of antibiotic at 50 times the MIC or DNase I (5 μg/ml), or both, as described in Materials and Methods. The results are the means ± standard errors of the means from three independent experiments, with viable counts done in triplicate. *, significantly different from the corresponding value for the control (P < 0.05); all other values are not significantly different; **, significantly different from the corresponding value for the antibiotic-treated biofilms (P < 0.05); all other values are not significantly different; ND, not done.
Effects of DNA digestion products on biofilm biomass.
The toxicity of the decay products formed during the digestion of extracellular DNA might be another cause of the observed changes in the amounts and the characteristics of the biofilm. To exclude such an effect, we have added to an E. coli biofilm a DNA digest obtained in vitro by the digestion of DNA in the presence of DNase I. The properties of the biofilms formed in the presence of the DNA digest at concentrations ranging from 20 to 100 μg/ml of DNase I remained unchanged.
Effects of antibiotics and combined use of DNase I and antibiotics on 24-h-old biofilms.
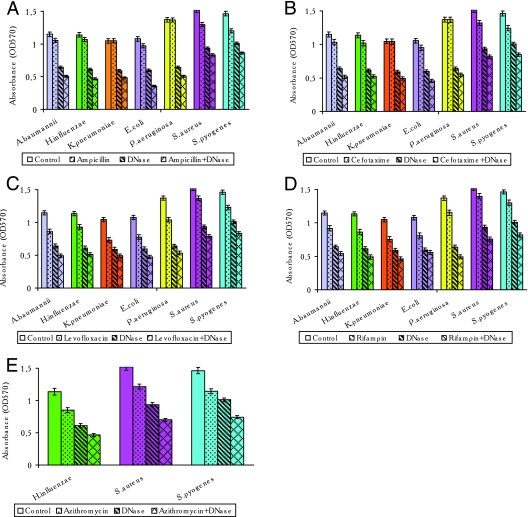
Different concentrations of antibiotics used alone or together with DNase I were added to 24-h-old biofilms, and the mixtures were incubated for 24 h at 37°C. Antibiotics at concentrations 1 to 10 times the MIC did not change either the biomass of the 24-h-old biofilms or the numbers of CFU of the bacterial strains evaluated in this study. Addition of antibiotics at concentrations of 50 times the MIC to the 24-h-old established biofilms decreased the biofilm biomass by 0.0 to 30.0% (Fig. 2A to E). We therefore used the antibiotics at concentrations 50 times their MICs to evaluate their effects on established biofilms. β-Lactams decreased the biomasses of the E. coli, A. baumannii, and H. influenzae biofilms only slightly (6 to 10%) but did not change the characteristics of the P. aeruginosa and K. pneumoniae biofilms (Fig. 2A and B). Under the same conditions, when β-lactams and DNase I at 5.0 μg/ml were added together to the 24-h-old microbial communities, the biomasses of the E. coli, A. baumannii, H. influenzae, P. aeruginosa, and K. pneumoniae biofilms were reduced by 53.0% to 67.0%. Gram-positive bacteria were much more susceptible to ampicillin and cefotaxime, which reduced the biomasses of the S. aureus and S. pyogenes biofilms by 10 to 15%. The combined use of ß-lactams and DNase I at 5.0 μg/ml caused reductions in the biomasses of these gram-positive bacterial biofilms of 41.0 to 46.0%. Levofloxacin and rifampin caused 10 to 30% decreases in the biomasses of 24-h-old biofilms of both gram-negative and gram-positive bacteria. When DNase I was used together with levofloxacin or rifampin, the biofilm biomasses in microbial communities formed by gram-positive and gram-negative bacteria were reduced by 43 to 64% (Fig. 2C and D). Azithromycin was tested only against gram-positive bacteria and H. influenzae, and the biomasses were reduced by 20 to 25%. The combination of azithromycin and DNase at 5.0 μg/ml reduced the biomass of the H. influenzae biofilm by 59% and those of gram-positive bacterial biofilms by 49 to 54% (Fig. 2E).
FIG. 2.
Quantitative biofilm analysis of the effect of the combined use of DNase I and various antibiotics on 24-h-old biofilms. Preformed 24-h-old biofilms were treated with a particular antibiotic (50 times the MIC), DNase I (5 μg/ml), or the antibiotic (50 times the MIC) plus DNase I (5 μg/ml) in the appropriate medium and were incubated at 37°C for 24 h. The bacteria adhering to the microtiter plates were stained, and the absorbance (the optical density at 570 nm [OD570]) was measured as described in Materials and Methods. Each bar represents the average of three or four experiments, with error bars representing the standard errors of the means. A P value of <0.05 was considered statistically significant. (A) Treatment with ampicillin, DNase I, or ampicillin plus DNase I; (B) treatment with cefotaxime, DNase I, or cefotaxime plus DNase I; (C) treatment with levofloxacin, DNase I, or levofloxacin plus DNase I; (D) treatment with rifampin, DNase I, or rifampin plus DNase I; (E) treatment with azithromycin, DNase I, or azithromycin plus DNase I.
The numbers of CFU were changed by different antibiotics, although the changes in this parameter was less variable than the changes in the biofilm biomass. β-Lactams did not statistically significantly reduce the numbers of CFU of the gram-positive and gram-negative bacteria tested, although combinations of β-lactams with DNase I at 5.0 μg/ml caused 10- to 15-fold decreases in viable bacterial counts (Table 3).
Levofloxacin and rifampin each reduced the numbers of CFU of S. aureus and S pyogenes in biofilms by on the order of 102 but did not affect the viability of gram-negative bacteria. The combination of levofloxacin or rifampin with DNase I reduced the numbers of CFU in the E. coli, A. baumannii, H. influenzae, P. aeruginosa, and K. pneumoniae biofilms by up to 102. DNase I in combination with levofloxacin or rifampin had activity against gram-positive bacteria 10 times greater than that of each antibiotic alone.
Azithromycin by itself did not reduce the numbers of CFU of gram-positive bacteria but decreased the viability of H. influenzae by four to eight times.
Azithromycin plus DNase I at 5.0 μg/ml decreased the viability of gram-positive bacterial biofilms by 2- to 4-fold and that of H. influenzae biofilms by 20-fold.
Thus, when they were added together, antibiotics and DNase I significantly reduced the biofilm biomasses of all bacteria studied. The numbers of CFU were also more significantly reduced by the use of the antibiotics in combination with DNase I than by the use of the antibiotics alone.
DISCUSSION
The use of biofilms for the development of new principles for the enhancement of the clinical effectiveness of antibiotics is a subject of intensive investigation (8, 34, 42, 53). We have evaluated extracellular DNA of about 30 kb that appeared to be a universal component of the matrices of all biofilms formed by the bacteria tested. The presence of extracellular DNA in human and animal plasma, ocean sediments, and bacterial biofilms indicates that it is an important component of the microbiome and pangenome (4, 16, 47, 56).
The fact that DNase I reduced the established biofilms suggests that extracellular DNA is essential for the maintenance of these bacterial communities. There are several proposals concerning the role of extracellular DNA in biofilm function. The possible roles of extracellular DNA as a polymeric substance (37, 46), a nutrient (20), and a gene transporter (25) have been considered. Our data suggest that the alteration of the biofilm biomass in the presence of DNase I is broadly observed in the biofilms of different unrelated gram-positive and gram-negative bacteria. We suppose that this effect is a result of extracellular DNA destruction, as the DNase that we have used does not penetrate bacteria (36) and its effect is realized outside the cells. We have demonstrated that this effect, which was observed on 24-h-old biofilms formed by diverse bacteria, is constant and concentration dependent and results in 28% to 70% reductions in the community biomass with DNase I concentrations varying from 0.5 to 1,000 μg/ml. The overall morphology of the biofilms, as revealed by microscopy, is modified in the presence of DNase I, although the numbers of CFU remained unchanged. The biomass of biofilms is defined by the bacteria and the extracellular matrix (8). The bacteria within biofilms can be viable (as determined from CFU counts), viable but nonculturable, or dead (39, 41, 62). The discrepancy between the relatively constant number of viable bacteria and the reduction of the biofilm biomass in the presence of DNase I or antibiotics measured spectrophotometrically may result from the decreased amount of the extracellular matrix and the decreased number of viable but nonculturable or dead bacteria. Thus, the number of CFU remains constant, even though the total biofilm biomass is reduced. Because the number of CFU after treatment of the biofilm with DNase I remains unchanged, spectrophotometric measurement of the amount of crystal violet incorporated into a biofilm allows estimation of the amount of the extracellular matrix together with the amount of viable but nonculturable and dead bacteria or the strength of biofilm adhesion to a surface. A decrease in the biofilm biomass by addition of DNase I to 24-h-old biofilms may be realized because of the decreased amount of the extracellular matrix or the formation of a more weakly bound biofilm. Thus, spectrophotometric analysis of biofilm biomass allowed us to compare the characteristics of biofilms treated or not treated with DNase I.
The possible toxic effect of DNase I or DNA digestion decay products as an explanation for the alteration of the biofilm biomass cannot be completely ruled out. The addition of a DNA digest suggested that neither DNase I itself nor the decay products formed as a result of extracellular DNA digestion possess a toxic effect on the bacteria or the biofilms.
It is known that the bacteria within biofilms are much less susceptible to antibiotics because of the poor penetration of antimicrobials through the surface film that covers the microbial community and because of inactivation of the antimicrobials by the extracellular matrix (8, 9).
The effects of DNase I and various antibiotics when they were used separately and together on the established biofilms of various gram-positive and gram-negative bacteria were noted. The addition of antibiotics at concentrations 50 times their MICs decreased the numbers of CFU in the biofilms of some species by 101 to 104 but had no notable effects on others. At the same time, the use of a combination of antibiotics at concentrations 50 times their MICs with DNase I at 5.0 μg/ml (which had no effect on the numbers of CFU of 24-h-old biofilms when it was used alone) decreased the numbers of CFU 2- to 15-fold more than the use of the antibiotics alone did. The use of a combination of antibiotics and DNase I resulted in a significant decrease in the established biofilm biomass compared to the reduction of biomass achieved when each antibiotic or DNase I was used alone. It is not clear what role extracellular DNA plays in biofilm formation and maintenance. Our data indicate that the destruction of extracellular DNA by DNase I leads to a decrease in the extracellular matrix/EPS, and as a result, antibacterial agents act more effectively to reduce the biofilm biomass and the numbers of CFU.
Acknowledgments
We thank Steve Lerner and Michael Cynamon for helpful comments during preparation of the manuscript and editing our English.
Footnotes
Published ahead of print on 8 December 2008.
REFERENCES
- 1.Allesen-Holm, M., K. Barken, L. Yang, M. Klausen, J. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114-1128. [DOI] [PubMed] [Google Scholar]
- 2.Anderl, J. N., M. J. Franklin, and P. S. Stewart. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44:1818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anker, P., and M. Stroun. 2000. Circulating nucleic acids in plasma or serum. Medicina (Buenos Aires) 60(5 Pt 2):699-702 . [PubMed] [Google Scholar]
- 4.Anker, P., H. Mulcahy, X. Chen, and M. Stroun. 1999. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metast. Rev. 18:65-73. [DOI] [PubMed] [Google Scholar]
- 5.Baker-Austin, C., M. Wright, R. Stepanauskas, and J. V. McArthur. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14:176-182. [DOI] [PubMed] [Google Scholar]
- 6.Böckelmann, U., A. Janke, R. Kuhn, T. R. Neu, J. Wecke, J. R. Lawrence, and U. Szewzyk. 2006. Bacterial extracellular DNA forming a defined network-like structure. FEMS Microbiol. Lett. 262:31-39. [DOI] [PubMed] [Google Scholar]
- 7.Burton, E., P. Gawande, N. Yakandawala, K. Vetri, G. Zhanel, T. Romeo, A. Freisen, and S. Madhyastha. 2006. Antibiofilm of GlmU enzyme inhibitors against catheter-associated uropathogens. Antimicrob. Agents Chemother. 50:1835-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambless, J., S. Hunt, and P. Stewart. 2006. A three-dimensional computer model of four hypothetical mechanisms protecting biofilms from antimicrobials. Appl. Environ. Microbiol. 72:2005-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaw, K. C., M. Manimaran, and F. E. Tay. 2005. Role of silver ions in destabilization of intermolecular adhesion forces measured by atomic force microscopy in Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 49:4853-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241-249. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 1999. Methods for determining bactericidal activity of antimicrobial agents; approved standard, 6th ed. Clinical and Laboratory Standards Institute document M26-A. Clinical and Laboratory Standards Institute, Wayne, PA.
- 12.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed. Clinical and Laboratory Standards Institute document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 13.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 14.Davies, D. 2003. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2:114-122. [DOI] [PubMed] [Google Scholar]
- 15.Dell'Anno, A., and R. Danovaro. 2005. Extracellular DNA plays a key role in deep-sea ecosystem functioning. Science 309:2179. [DOI] [PubMed] [Google Scholar]
- 16.Dell'Anno, A., S. Bompadre, and R. Danovaro. 2002. Quantification, base composition, and fate of extracellular DNA in marine sediments. Limnol. Oceanogr. 47:899-905. [Google Scholar]
- 17.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorward, D., C. Garon, and R. Judd. 1989. Export and intracellular transfer of DNA via membrane Neisseria gonorrhoeae. J. Bacteriol. 171:2499-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Draghi, J., and P. Turner. 2006. DNA secretion and gene-level secretion in bacteria. Microbiology 152:2683-2688. [DOI] [PubMed] [Google Scholar]
- 20.Finkel, S. E., and R. Kolter. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183:6288-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemming, H. C., R. N. Thomas, and D. J. Wozniak. 2007. The EPS matrix: the “house of biofilm cells”. J. Bacteriol. 189:7945-7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fux, C., S. Wilson, and P. Stoodley. 2004. Detachment characteristics and oxacillin resistance of Staphylococcus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 186:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34-40. [DOI] [PubMed] [Google Scholar]
- 24.Gander, S. 1996. Bacterial biofilms: resistance to antimicrobial agents. J. Antimicrob. Chemother. 37:1047-1050. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton, H. L., and J. P. Dillard. 2006. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59:376-385. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton, H. L., N. M. Domínguez, K. J. Schwartz, K. T. Hackett, and J. D. Dillard. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704-1721. [DOI] [PubMed] [Google Scholar]
- 27.Harrison, J., H. Ceri, N. Roper, E. Badry, K. Sproule, and R. Turner. 2005. Persister cells mediate tolerance to metal oxyanions in Escherichia coli. Microbiology 151:3181-3195. [DOI] [PubMed] [Google Scholar]
- 28.Hoyle, B., and J. Costerton. 1991. Bacterial resistance to antibiotics: the role of biofilms. Prog. Drug Res. 37:91-105. [DOI] [PubMed] [Google Scholar]
- 29.Hunt, S. M., M. A. Hamilton, and P. S. Stewart. 2005. A 3D model of antimicrobial action on biofilms. Water Sci. Technol. 52:143-148. [Google Scholar]
- 30.Izano, E. A., M. A. Amarante, W. B. Kher, and J. B. Kaplan. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keren, I., N. Kaldalu, A. Spoering, Y. Wang, and K. Lewis. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13-18. [DOI] [PubMed] [Google Scholar]
- 32.Larsen, T. 2002. Susceptibility of Porphyromonas gingivalis in biofilms to amoxicillin, doxycycline and metronidazole. Oral Microbiol. Immunol. 17:267-271. [DOI] [PubMed] [Google Scholar]
- 33.LePecq, J.-B., and C. Paoletti. 1966. A new fluorometric method for RNA and DNA determination. Ann. Biochem. 17:100-107. [DOI] [PubMed] [Google Scholar]
- 34.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis, K. 2007. Persister cells, dormancy and infectious disease. Nature 5:48-56. [DOI] [PubMed] [Google Scholar]
- 36.Liao, T. H., and J. Salnikow. 1973. Bovine pancreatic deoxyribonuclease A. J. Biol. Chem. 248:1489-1495. [PubMed] [Google Scholar]
- 37.Maier, B., and J. Radler. 2000. DNA on fluid membranes: a model polymer in two dimensions. Macromolecules 33:7185-7194. [Google Scholar]
- 38.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 39.McDougald, D., A. Scott, A. Rice, D. Weichart, and S. Kjelleberg. 1998. Nonculturability: adaptation or debilitation? FEMS Microbiol. Ecol. 25:1-9. [Google Scholar]
- 40.Moscoso, M., E. García, and R. López. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 188:7785-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliver, J. D. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 43:93-100. [PubMed] [Google Scholar]
- 42.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 43.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 44.Paul, J. H., W. H. Jeffrey, and M. F. DeFlaun. 1987. Dynamics of extracellular DNA in the marine environment. Appl. Environ. Microbiol. 53:170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pertl, B., and D. Bianchi. 2001. Fetal DNA in maternal plasma: emerging clinical applications. Obstet. Gynecol. 98:483-490. [DOI] [PubMed] [Google Scholar]
- 46.Rice, K. C., E. E. Mann, J. L. Endres, E. C. Weiss, J. E. Cassat, M. S. Smeltzer, and K. W. Bayles. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 104:8113-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robe, P., R. Nalin, M. Capellanob, T. M. Vogel, and P. Simonetc. 2003. Extraction of DNA from soil. Eur. J. Soil Biol. 39:183-190. [Google Scholar]
- 48.Shingaki, R., Y. Kasahara, T. Inoue, S. Kokeguchi, and K. Fukui. 2003. Chromosome DNA fragmentation and excretion caused by defective prophage gene expression in the early-exponential-phase culture of Bacillus subtilis. Can. J. Microbiol. 49:313-325. [DOI] [PubMed] [Google Scholar]
- 49.Sponza, D. T. 2003. Investigation of extracellular polymer substances (EPS) and physicochemical properties of different activated sludge flocs under steady-state conditions. Enzyme Microb. Technol. 32:375-385. [Google Scholar]
- 50.Steinberger, R. E., and P. A. Holden. 2005. Extracellular DNA in single- and multiple-species unsaturated biofilms. Appl. Environ. Microbiol. 71:5404-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart, P. S. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292:107-113. [DOI] [PubMed] [Google Scholar]
- 52.Stewart, P. S., and J. W. C osterton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 53.Suga, H., and K. Smith. 2003. Molecular mechanisms of bacterial quorum sensing as a new drug target. Curr. Opin. Chem. Biol. 7:586-591. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi, N., K. Ishihara, R. Kimizuka, K. Okuda, and T. Kato. 2006. The effects of tetracycline, minocycline, doxycycline and ofloxacin on Prevotella intermedia biofilm. Oral Microbiol. Immunol. 21:366-371. [DOI] [PubMed] [Google Scholar]
- 55.Tetz, V. V. 1996. The effect of antimicrobial agents and mutagen on bacterial cells in colonies. Med. Microbiol. Lett. 5:426-436. [Google Scholar]
- 56.Tetz, V. V. 2005. The pangenome concept: a unifying view on genetic information. Med. Sci. Monit. 11:24-29. [PubMed] [Google Scholar]
- 57.Tetz, V. V., V. P. Korobov, N. K. Artemenko, L. M. Lemkina, N. V. Panjkova, and G. V. Tetz. 2004. Extracellular phospholipids of isolated bacterial communities. Biofilms 1:149-155. [Google Scholar]
- 58.Tetz, V. V., O. V. Rybalchenko, and G. A. Sakova. 1993. Surface films of Escherichia coli colonies. FEMS Microbiol. Lett. 107:231-240. [DOI] [PubMed] [Google Scholar]
- 59.Tetz, V. V., O. V. Rybalchenko., and G. A. Savkova. 1993. Ultrastructure of surface film of bacterial colonies. J. Microbiol. 139:855-858. [DOI] [PubMed] [Google Scholar]
- 60.Trevors, J. T. 1996. DNA in soil: adsorption, genetic transformation, molecular evolution and genetic microchip. Antonie van Leeuwenhoek 70:1-10. [DOI] [PubMed] [Google Scholar]
- 61.Umetani, N., J. Kim, S. Hiramatsu, H. Reber, O. Hines, O. Bilchik, and D. Hoon. 2006. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: direct quantitative PCR for ALU repeats. Clin. Chem. 52:1062-1069. [DOI] [PubMed] [Google Scholar]
- 62.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 64.Xavier, J. B., and K. R. Foster. 2007. Cooperation and conflict in microbial biofilms. Proc. Natl. Acad. Sci. USA 104:876-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yaron, S., L. Kolling, L. Simon, and K. Matthews. 2000. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 66:4414-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]