Abstract
As a part of a nationwide study in Spain, 15 clinical isolates of Acinetobacter genomic species 3 (AG3) were analyzed. The main objective of the study was to characterize the ampC genes from these isolates and to determine their involvement in β-lactam resistance in AG3. The 15 AG3 isolates showed different profiles of resistance to ampicillin (range of MICs, 12 to >256 μg/ml). Nucleotide sequencing of the 15 ampC genes yielded 12 new AmpC enzymes (ADC-12 to ADC-23). The 12 AG3 enzymes showed 93.7 to 96.1% amino acid identity with respect to the AmpC enzyme from Acinetobacter baumannii (ADC-1 enzyme). Eight out of fifteen ampC genes were expressed in Escherichia coli cells under the control of a common promoter, and with the exception of one isolate (isolate 65, which showed lower β-lactam MICs), significant differences in overall β-lactam MICs for E. coli cells expressing AG3 ampC genes were not revealed. No significant differences in ampC gene expression in AG3 clinical isolates were revealed by reverse transcription-PCR analysis. A detailed analysis of the 12 AmpC protein sequences revealed that amino acid replacements (in comparison with those of ADC-1) occurred mainly in the same positions, although none were located in important functional domains such as the Ω- loop or conserved β-lactamase motifs. Kinetic experiments performed with three representative AmpC enzymes (ADC-14, -16, and -18) in some cases revealed dramatic changes in Km and kcat values for β-lactams. No ISAba1 was detected upstream of the ampC genes. Our results reveal 12 new ampC genes in AG3. The enzymes showed a moderate degree of variability, and they are tentatively named ADC-12 to ADC-23.
Species belonging to the genus Acinetobacter are widely distributed in nature (2, 3) and are reported to be the cause of ever-increasing numbers of nosocomial infections. Molecular methods based on DNA-DNA hybridization or sequencing of the 16S subunit of the ribosome have been described for up to 33 different groups (31). Groups 1, 2, 3, and 13 are phenotypically similar and traditionally known as the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. Except for those in group 1, these genomic species are important nosocomial pathogens that frequently cause outbreaks of infection in intensive care units and burn units (18, 20, 32). Although mechanisms of antibiotic resistance in A. baumannii have been described (4, 6, 13, 14, 19, 23, 24, 31, 34, 35), there are few descriptions of the mechanisms of resistance in Acinetobacter genomic species 3 (AG3) (1, 10, 25, 29, 35). With regard to β-lactam resistance in AG3, two metalloenzymes, VIM-2 and IMP-4, and a chromosomal cephalosporinase have been described for this species (1, 10, 34, 35).
As a part of a nationwide, multicenter study in Spain, which included analysis of 244 Acinetobacter sp. isolates (226 A. baumannii, 15 AG3, and 3 unidentified isolates), we aimed to determine the molecular basis of β-lactam resistance and specifically ampicillin resistance in 15 AG3 clinical isolates. For this purpose, the ampC genes from all isolates were sequenced and further characterized to assess their activities and specificities toward β-lactams. Overall, 12 new ampC genes were discovered in AG3. Following a classification that is currently under development, the genes were designated ADC-12 to ADC-23.
MATERIALS AND METHODS
Bacterial strains.
In November 2000, all A. baumannii isolates from clinical samples were assembled from 28 hospitals in Spain. A total of 244 isolates of Acinetobacter spp. were collected: 226 A. baumannii, 15 AG3, and 3 unidentified Acinetobacter sp isolates. The 15 AG3 isolates used for further studies were isolates 14, 20, 21, 52, 56, 60, 65, 67, 69, 90, 103, 109, 128, 195, and 243, which were all isolated from different hospitals. Escherichia coli DH5α [F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 λ− thi-1 gyrA96 relA1] and E. coli BL21 [F− ompT hsdSB(rB− mB−) gal dcm] were used for determining antibiotic MICs and for analysis of expression and purification of proteins, respectively.
Bacterial strains were frozen in Brucella glycerol broth (10%) (BBL Microbiology Systems, Cockeysville, MD) and were maintained at −80°C until analysis. Strains of E. coli were grown at 37°C in Luria-Bertani (LB) medium. When necessary, LB medium was supplemented with ampicillin (20 μg/ml) or kanamycin (50 μg/ml) (Sigma-Genosys Ltd., United Kingdom).
Antimicrobial agents and determination of MICs.
Antibiotic susceptibility profiles were determined by Etest according to the manufacturer's instructions (AB Biodisk, Solna, Sweden). The following antibiotics were purchased from Sigma-Aldrich (Madrid, Spain): ampicillin, piperacillin, cephalothin, cefoxitin, cefuroxime, ceftazidime, and cefotaxime. Cefepime was obtained from Sigma-Genosys Ltd. (United Kingdom), imipenem was obtained from Merck Sharp and Dohme (Madrid, Spain), and meropenem was obtained from AstraZeneca (Madrid, Spain).
ARDRA.
The species were identified by amplified ribosomal DNA restriction analysis (ARDRA) (30). AG3 was also identified by sequencing of the 16S rRNA gene with oligonucleotides P1 and P2 (Table 1).
TABLE 1.
Oligonucleotides used in the study
| Oligonucleotide | Descriptionb | Sequencea | Reference(s) or source |
|---|---|---|---|
| P1 | ARDRA F | 5′-TGGCTCAGATTGAACGCTGGCGGC-3′ | 30 |
| P2 | ARDRA R | 5′-TACCTTGTTACGACTTCACCCC-3′ | 30 |
| P3 | REP F | 5′-TTTGCGCCGTCATCAGGC-3′ | 5, 33 |
| P4 | REP R | 5′-ACGTCTTATCAGGCCTAC-3′ | 5, 33 |
| P5 | ampC F | 5′-aaggatccATGCGATTTAAAAAAATTTC-3′ | 6 |
| P6 | ampC R | 5′-aaaagcttAGGATATGTTTGGTTC-3′ | 6 |
| P7 | ampC R | 5′-aaaagcttTTATTTCTTTATTGCATTC-3′ | 6 |
| P8 | ISAba1 F | 5′-aaaggatccCTCTGTACACGACAAATTTCAC | This study |
| P9 | ISAba1 R | 5′-aaagaattcCTCTGTACAGCATAAAAATAGAT | This study |
| P10 | amp Int R | 5′-GCCGACTTGATAGAA-3′ | This study |
| P11 | ampC F pGEX (isolate 65) | 5′-aaggatccGGCAATACACCAAAAGACCAAG-3′ | This study |
| P12 | ampC F pGEX (isolate103) | 5′-aaggatccGGTAATACACCAAAAGAGCAA-3′ | This study |
| P13 | ampC F pGEX (isolate 195) | 5′aaggatccGGCAATACACCAAAAGAACAAG-3′ | This study |
| P14 | ampC R pGEX | 5′-aagaattcTCTTTTTTATGTTTAGCTACGG-3′ | This study |
| P15 | RT-PCR ampC F | 5′-AAGTTTTAACTTTTTTCAAAG-3′ | This study |
| P16 | RT-PCR ampC R | 5′-AATTACTGTCTAATAAAGTTT-3′ | This study |
| P17 | RT-PCR control gyrA F | 5′-AATCTGCCCGTGTCGTT-3′ | GenBank accession no. AY204699 |
| P18 | RT-PCR control gyrA R | 5′-GCCATACCTACGGCGA-3′ | GenBank accession no. AY204699 |
Lowercase letters represent restriction sites and tail nucleotides.
F, forward oligonucleotide; R, reverse oligonucleotide.
REP-PCR.
Repetitive extragenic palindromic sequence (REP)-based PCR (REP-PCR) was used to evaluate the possible clonal relationship between the different isolates of AG3 used in the study. The REP-PCR sequence allows amplification of the localized regions between the REP zones. The primers used are described in Table 1 (primers P3 and P4). The amplification reaction was carried out as previously described (5). We consider that two isolates were epidemiologically unrelated when two or more different bands were detected in them (5, 33).
Cellular extract preparation and IEF.
β-Lactamases were obtained by sonication of cultures of all isolates of AG3 grown overnight at 37°C in LB medium and centrifugation at 14,000 rpm (MiniSpin microcentrifuge; Eppendorf, Hamburg, Germany) for 10 min (22). The pI values were determined as previously reported with commercial isoelectric focusing (IEF) gels (pH 3.5 to 9.5; Pharmacia LKB, Piscataway, NJ) by using a PhastSystem electrophoresis system (Pharmacia). Sonicated extracts of microorganisms expressing β-lactamases of known pIs were used as controls.
Cloning experiments and recombinant plasmids.
Total DNA was extracted from 15 AG3 clinical isolates with the MasterPure DNA purification kit (Epicentre, Madison, WI) according to the manufacturer's instructions. The ampC genes from the 15 AG3 clinical isolates described above were cloned by PCR by use of oligonucleotides P5 and P6 (Table 1) for isolates 21, 52, 60, 67, 90, 128, and 243. For the remaining AG3 clinical isolates, oligonucleotides P5 and P7 were used. The Expand High Fidelity PCR system (Roche Diagnostics, Indianapolis, IN) was used for the amplification procedure under the following experimental conditions: denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1.5 min for a total of 28 cycles; an initial cycle of 2 min at 94°C; and a final cycle of 10 min at 72°C. The amplicons were purified with a High Pure PCR product purification kit (Roche Diagnostics, GmbH, Mannheim, Germany). In 8 out of 15 isolates (isolates 14, 20, 56, 65, 69, 109, and 195), and for expression and MIC studies, the amplified fragment was cloned into BamHI and HindIII sites in pBGS18 (harboring a kanamycin resistance marker) under the control of the strong promoter of the CTX-M-14 β-lactamase gene (positions 1501 to 1740 [GenBank accession number AF252622]) (13). With the remaining seven AG3 isolates, the amplified fragment was directly cloned into pBGS18 at the same restriction sites. In both cases, ligation was carried out with a Rapid DNA ligation kit (Roche Diagnostics, Indianapolis, IN). The DNA was electroporated in E. coli DH5α cells, and the clones were selected on LB plates with 20 μg/ml of ampicillin and 50 μg/ml of kanamycin. Plasmids from selected transformants were purified and examined to check the accuracy of the cloning procedure. Two clones from each gene transformation were selected, and nucleotide sequencing was carried out. Sequencing of nucleotides was performed by use of the Taq DyeDeoxiTerminator cycle sequencing kit before analysis using an automatic DNA sequencer (377 Abi-Prism; Perkin-Elmer). Each gene was sequenced on both strands. The ClustalW program (http://infobiogen.fr) was used to align the multiple protein sequences (28).
Detection of ISAba1 in AG3 isolates.
ISAba1-like sequences were previously identified immediately upstream of the blaAmpC gene in ceftazidime-resistant A. baumannii isolates, where the strong promoter of the ISAba1 insertion increased the expression of the blaAmpC gene (16). To find out whether the ISAba1 element was present in AG3, a PCR assay was performed with primer pairs for this element and the ampC gene from AG3 (primer pair P8/P9 was used to detect ISAba1, and primer pair P8/P10 was used to detect ISAba1 upstream of the ampC gene).
β-Lactamase purification and kinetic experiments.
Three representative AmpC enzymes from isolates 65, 103, and 195 were purified for kinetic experiments. For this, the ampC genes were cloned into vector pGEX-6P-1, which allows the production of a fusion protein from glutathione S-transferase (GST) and the AmpC enzyme. The primer pairs used for PCR amplification and cloning into pGEX-6P-1 were P11/P14 for isolate 65, P12/P14 for isolate 103, and P13/P14 for isolate 195 (Table 1). β-Lactamase was purified to homogeneity by use of the GST gene fusion system (Amersham Pharmacia Biotech, Europe, GmbH) according to the manufacturer's instructions. The mature purified proteins lacking the GST fusion protein appeared on sodium dodecyl sulfate-polyacrylamide gels as a band of 43 kDa (≥95% purity). Kinetic experiments were performed at 25°C using a Nicolete Evolution 300 spectrophotometer (Thermo Electron Corporation, Waltham, MA) with quartz cuvettes of optical path lengths of 1 and 0.2 cm. The tests were each repeated three times with phosphate-buffered saline (PBS) with 20 mg/liter bovine serum albumin. The kinetic parameters kcat, Km, and kcat/Km were studied for the antibiotics ampicillin, cephalothin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, aztreonam, and imipenem. The Km values were calculated as Ki values in competitive assays with Centa (Calbiochem, Merck, Darmstadt, Germany) as the substrate, as previously described for putative poor substrates such as imipenem and meropenem. The Vmax was calculated by considering an antibiotic concentration six times the Km and by use of the Michaelis-Menten equation, as previously described (26). Studies of the 50% inhibitory concentration (IC50) were conducted by incubating the purified proteins (1 μg/ml) for 10 min in the presence of inhibitors of class A β-lactamases (clavulanic acid and sulbactam).
Semiquantitative RT-PCR.
To detect ampC gene expression, reverse transcriptase PCR (RT-PCR) was carried out with 15 AG3 clinical isolates as well as with an Acinetobacter sp. isolate with a high level of ampC gene expression (as a positive control). Total RNA was extracted from cultures grown overnight in LB medium at 37°C with the Trizol Max bacterial RNA isolator kit (Invitrogen, Carlsbad, CA), and the RNA was then treated with DNase (Sigma-Genosys Ltd., United Kingdom). The Qiagen OneStep RT-PCR kit was used for RT-PCR analysis with a 200-ng sample of total RNA. The primers used for gene amplification were designed to hybridize in highly conserved fragments in all sequences of ampC genes (P15 and P16 in Table 1), which amplified an internal product of 470 bp. As an internal control for the RT-PCR, the gyrA gene from AG3 was used as a template with oligonucleotides P17 and P18 (Table 1), which amplified the 344 bp of this gene. The conditions of the RT-PCR were as follows: an initial cycle of reverse transcription at 50°C for 30 min, followed by amplification of the DNA with a initial cycle of 15 min at 95°C, 23 cycles at 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min and a final cycle of 10 min at 72°C. Aliquots were removed during the amplification process at cycles 14, 18, and 22 (exponential phase of amplification). The bands were revealed in agarose gels, as described above. The intensity of ampC gene bands was compared with that of gyrA gene bands.
Western blot analysis.
Western blot analysis was used to detect and assess AmpC expression in the AG3 isolates with polyclonal antibodies raised against ADC-7 (19). Bacterial extracts were obtained as described above for pI analysis and were loaded onto sodium dodecyl sulfate-polyacrylamide gels (12%) in a minigel apparatus (Bio-Rad, Hercules, CA). The proteins were transferred onto Immobilon-P membranes (Millipore, Billerica, MA). The membranes were blocked with 5% (wt/vol) blocking agent (skim milk) in PBS-Tween. After the membranes were washed with PBS-Tween, they were incubated with a mixture of the polyclonal rabbit anti-ADC-7 antiserum diluted 1:1,500. The membranes were then washed and incubated with secondary conjugated rabbit antiserum diluted 1:2,000 (ECL Western blotting reagent; Amersham Pharmacia Biotech, United Kingdom). All incubations were done for 1 h at 25°C. The Western blot was revealed by overlaying the membranes with luminol, a substrate of the horseradish peroxidase enzyme, ligated to secondary antibody.
OMP purification.
Purification and analysis of outer membrane proteins (OMPs) were performed as previously described (13, 14) with bacterial AG3 isolates 20 and 103 (with clear ampicillin resistance) and isolates 21, 56, and 128 (ampicillin susceptible).
Determination of antibiotic MICs in the presence of cloxacillin and the efflux pump inhibitors PAN and CCCP.
To assess the putative role of an efflux pump mechanism involved in β-lactam resistance in AG3, antibiotic MICs in the presence and absence of chemical efflux pump inhibitors were determined. Thus, MICs were determined in the presence of 150 μg/ml of cloxacillin as an inhibitor of cephalosporinase activity (7) and of efflux pump inhibitors, such as carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma) at 25 μM (5.1 μg/ml) (15) and Phe-Arg β-naphthylamide dihydrochloride (PAN; Sigma) at 25 μg/ml (8), alone and with antibiotics.
Theoretical modeling of the three-dimensional structure of AmpC enzymes.
The theoretical three-dimensional structure of the ADC-12 enzyme was obtained by theoretical homology modeling with different computer software packages: BLAST (to align sequences), Deep View (to obtain the theoretical structure from previously aligned genes), and UCSF Chimera (protein model edition). For this, the ADC-12 sequence was compared with the previously published structure of CMY-10 (Protein Data Bank accession number 1ZKJ) (21), which showed the highest amino acid identity (45%). We used the Deep View program in combination with the Swiss-model server to generate a homology-based model of the enzyme. As part of the default pipeline of the ProModII modeling program (implemented in Swiss-model), a final step of structure “dumping” was performed, resulting in an unrefined, fast energy minimization process, mainly with the purpose of avoiding atomic clashes. No further minimization or molecular dynamic equilibration was executed.
Nucleotide sequence accession numbers.
The nucleotide sequences of the ADC-type enzymes have been submitted to the GenBank database under accession numbers AM283529 (ADC-12), AM283528 (ADC-13), AM283527 (ADC-14), AM283526 (ADC-15), AM283525 (ADC-16), AM283524 (ADC-17), AM283523 (ADC-18), AM283522 (ADC-19), AM283521 (ADC-20), AM283520 (ADC-21), AM283519 (ADC-22), and AM283518 (ADC-23).
RESULTS
IEF analysis and antimicrobial susceptibility pattern.
IEF was performed with sonicated extracts obtained from 15 AG3 isolates. A single pI of ca. 9 was detected in all strains, probably corresponding to a chromosomal cephalosporinase. The antibiotic MICs determined by Etest for the 15 AG3 clinical isolates are shown in Table 2. High MICs of cephalothin and cefoxitin and a moderate degree of resistance to cefuroxime and cefotaxime were observed with all AG3 clinical isolates. Although most of the AG3 isolates were resistant to ampicillin (as deduced from the Clinical and Laboratory Standards Institute breakpoints for the Enterobacteriaceae determined for ampicillin) (12), a wide range of MICs was obtained (12 to >256 μg/ml). With two of the isolates, isolates 20 and 103, high MICs of ampicillin were obtained (256 and >256 μg/ml, respectively). Interestingly, meropenem MICs were 6 and 3 μg/ml for the same two isolates, respectively. The MICs of meropenem for the remaining AG3 isolates were within the range of 0.19 to 1 μg/ml.
TABLE 2.
β-Lactam susceptibility profile of AG3 isolates included in the study
| β-Lactam | MIC (μg/ml) for AG3 isolate:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 20 | 21 | 52 | 56 | 60 | 65 | 67 | 69 | 90 | 103 | 109 | 128 | 195 | 243 | |
| Ampicillin | 32 | 256 | 24 | 48 | 24 | 32 | 32 | 48 | 32 | 12 | >256 | 48 | 24 | 32 | 12 |
| Piperacillin | 16 | 24 | 24 | 12 | 12 | 24 | 16 | 16 | 16 | 12 | 24 | 16 | 12 | 24 | 32 |
| Cephalothin | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| Cefoxitin | >256 | >256 | >256 | >256 | 64 | >256 | 128 | >256 | >256 | 128 | 192 | >256 | 128 | >256 | 48 |
| Cefuroxime | 32 | 48 | 48 | 32 | 16 | 64 | 24 | 48 | 32 | 16 | 48 | 32 | 32 | 64 | 32 |
| Ceftazidime | 3 | 4 | 6 | 3 | 1.5 | 3 | 4 | 3 | 4 | 6 | 4 | 3 | 3 | 4 | 4 |
| Cefotaxime | 24 | 16 | 24 | 12 | 4 | 24 | 12 | 16 | 16 | 12 | 16 | 16 | 12 | 24 | 16 |
| Cefepime | 3 | 4 | 12 | 2 | 2 | 4 | 3 | 4 | 3 | 16 | 3 | 2 | 2 | 6 | 3 |
| Imipenem | 0.25 | 1 | 0.38 | 0.25 | 0.19 | 0.25 | 0.19 | 0.19 | 0.25 | 0.38 | 0.75 | 0.19 | 0.12 | 0.19 | 0.19 |
| Meropenem | 0.50 | 6 | 1 | 0.50 | 0.38 | 0.75 | 0.50 | 0.50 | 0.25 | 0.75 | 3 | 0.38 | 0.19 | 1 | 0.25 |
REP-PCR analysis.
A different DNA band pattern was obtained for each AG3 isolate by REP-PCR, and the isolates were therefore assumed to be genetically unrelated (data not shown).
Amino acid sequence diversity of AmpCs of AG3.
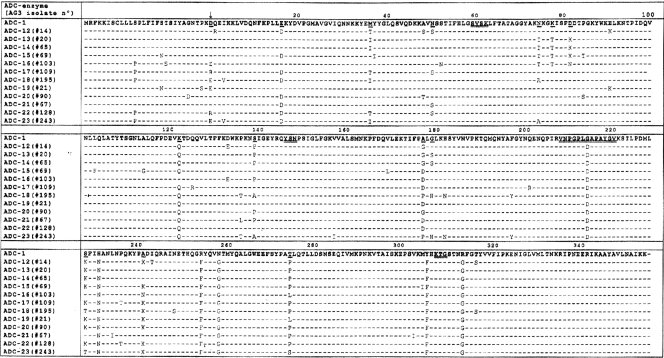
The ampC genes from AG3 clinical isolates were amplified and sequenced as described in Materials and Methods. The 15 ampC genes were composed of 1,152 nucleotides, which encode an open reading frame of 384 amino acids. The ampC genes of isolates 52, 56, 60, and 195 were identical; thus, there was a total of 12 different AmpC-encoding gene sequences (Fig. 1). These sequences differ from those of the previously reported ADC-type genes, and following the recently developed uniform numerical system for this family of AmpC β-lactamases, we tentatively named them ADC-12 to ADC-23. To explain whether or not differences in antibiotic MICs (mainly ampicillin) were due to differences in the amino acid compositions of AmpC enzymes, a detailed examination of the amino acid sequences of the AmpC enzymes was carried out and compared with that of AmpC from A. baumannii or ADC-1 (Fig. 1). Overall, the genes showed 93.7 to 96.1% identity with ADC-1. Although a moderate degree of genetic variability was observed, the pattern does not appear to follow a random profile, as some positions are more likely to be replaced than others. Indeed, some amino acid positions were replaced in at least four of the ADC-type enzymes analyzed with more than one residue (Fig. 1) and are shown in Fig. 3. Graphical analysis revealed no changes in the relationships with important domains or catalytic regions of the AmpC enzyme. Analysis of amino acid sequences of ADC-13 and ADC-16 (isolated from AG3 isolates 20 and 103, for which the highest ampicillin MICs were obtained) did not reveal any significant differences (in amino acid composition or position) with respect to the remaining ADC-type enzymes.
FIG. 1.
Amino acid sequence alignment among the 12 AmpC β-lactamases from AG3 (ADC-12 to -23) with respect to that of A. baumannii (ADC-1). Amino acid differences are indicated. The typical β-lactamase domains (SVSK, YSN, and KTG) and the Ω-loop are double underlined. The amino acid replacements present in at least four or more proteins (relative to that of ADC-1) with more than one residue are underlined. The vertical arrow indicates the position of the +1 amino acid (after cleavage of signal peptide). ADC-1, AmpC from A. baumannii (see reference 6).
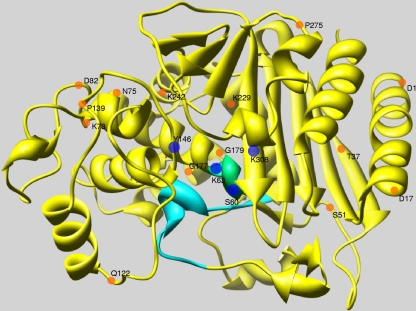
FIG. 3.
Theoretical model of the crystallographic structure of the ADC-12 AmpC β-lactamase. Amino acids belonging to the main β-lactamase domains (SVSK, YSN, and KTG) are represented in the figure as blue circles (60-63SXXK, Y146, and K308). The SVSK domain is also shown in green. The Ω-loop is shown in turquoise. Residues at positions 1, 17, 37, 51, 75, 78, 82, 122, 139, 177, 229, 242, and 275 (Fig. 1), which were replaced in four or more ADC-type enzymes (relative to ADC-1), are represented by orange circles (starting at position +1 of the mature protein with D). The diagram was drawn with the USCF Chimera software package.
Cloning and expression of ampC genes in the E. coli host.
To confirm whether or not amino acid changes have any effect on the phenotype of ampicillin or β-lactam resistance, we cloned and expressed several ampC genes in an E. coli host. For this, ampC genes from clinical isolates 20 and 103 (ampicillin MICs of 256 and >256 μg/ml) and those from clinical isolates 14, 56, 65, 69, 109, and 195 (ampicillin MIC ranges of 24 to 48 μg/ml) were cloned into pBGS18 under the control of a common external CTX-M-14 gene promoter and were then transformed into a common E. coli DH5α host, and the β-lactam MICs were determined (Table 3). The MICs of different β-lactams, including ampicillin and meropenem, were similar for all E. coli transformants except the transformant that expresses the β-lactamase AmpC of AG3 isolate 65 (ADC-14), for which the MICs were slightly lower.
TABLE 3.
MICs of β-lactam antibiotics tested with E. coli DH5α carrying the ampC genes under the control of a common promotera
| β-Lactam | MIC (μg/ml) for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| E. coli DH5α |
E. coli carrying ampC from AG3 isolate:
|
||||||||
| 14 | 20 | 56 | 65 | 69 | 103 | 109 | 195 | ||
| Ampicillin | 3 | 128 | 96 | 96 | 32 | 96 | 128 | 64 | 96 |
| Piperacillin | 0.38 | 4 | 4 | 3 | 4 | 3 | 4 | 4 | 4 |
| Cephalothin | 3 | >256 | >256 | >256 | 192 | >256 | >256 | >256 | >256 |
| Cefoxitin | 2 | 32 | 32 | 16 | 3 | 16 | 32 | 24 | 24 |
| Cefuroxime | 1.5 | 64 | 96 | 48 | 4 | 32 | 64 | 32 | 48 |
| Ceftazidime | 0.19 | 0.38 | 0.75 | 0.25 | 0.25 | 0.19 | 0.50 | 0.19 | 0.50 |
| Cefotaxime | 0.064 | 0.75 | 0.125 | 0.38 | 025 | 0.38 | 0.75 | 0.38 | 0.75 |
| Cefepime | 0.016 | 0.023 | 0.19 | 0.023 | 0.032 | 0.023 | 0.032 | 0.023 | 0.032 |
| Imipenem | 0.125 | 0.125 | 0.023 | 0.19 | 0.19 | 0.19 | 0.125 | 0.12 | 0.25 |
| Meropenem | 0.008 | 0.023 | 0.023 | 0.023 | 0.023 | 0.023 | 0.023 | 0.032 | 0.032 |
Identical MICs were obtained with three different transformants.
Kinetic experiments.
To confirm that differences in AmpC amino acid sequence are related to differences in the catalytic efficiency of AmpC enzymes toward β-lactams, three representative AmpC enzymes were chosen for further biochemical experiments. Those of AG3 isolates 65 (ADC-14), 103 (ADC-16), and 195 (ADC-18) were purified to homogeneity, and the kinetic parameters Km, kcat, and kcat/Km were determined (Table 4). The three AmpC enzymes showed almost identical catalytic efficiencies (kcat/Km) toward ampicillin and cephalothin, although ADC-14 showed important differences regarding the Km and kcat values, thus revealing differences in the biochemical behavior. Indeed, the kcat for ampicillin of ADC-14 was between 13 and 8 times lower than the corresponding values for ADC-18 and -16, respectively, and also 25 and 24 times lower for cephalothin, respectively. Moreover, ADC-14 showed a lower catalytic efficiency for cefoxitin than ADC-18 and ADC-16 (2.1 and 1.9 times, respectively), cefuroxime (3.4 and 4.4 times, respectively), and cefotaxime (6.4 and 4.4 times, respectively), which are consistent with MICs obtained with E. coli harboring ampC genes (Table 3). With regard to imipenem, the three enzymes had similar Km values, although the kcat/Km values for ADC-18 were almost seven times higher. No hydrolysis was detected with aztreonam.
TABLE 4.
Kinetic experiments performed with AmpC enzymes from the indicated AG3 isolatesa
| Drug | AmpC from isolate (ADC type)b:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 65 (ADC-14)
|
103 (ADC-16)
|
195 (ADC-18)
|
|||||||
| Mean Km (μM) (SD) | Mean kcat (s−1) (SD) | kcat/Km (s−1 mM−1) | Mean Km (μM) (SD) | Mean kcat (s−1) (SD) | kcat/Km (s−1 mM−1) | Mean Km (μM) (SD) | Mean kcat (s−1) (SD) | kcat/Km (s−1 mM−1) | |
| Ampicillin | 0.12 (±0.04) | 0.30 (±0.08) | 2,447.15 | 1.03 (±0.4) | 2.6 (±0.66) | 2,518.91 | 1.5 (±0.47) | 4.05 (±1.06) | 2,700.94 |
| Cephalothin | 6.73 (±2.3) | 56.52 (±22.38) | 8,398.16 | 139.24 (±16.72) | 1,380.80 (± 490.13) | 9,916.65 | 148.27 (±13.23) | 1,418.36 (±318.51) | 9,566.33 |
| Cefoxitin | 0.02 (±0.01) | 0.02 (±0.004) | 941.18 | 0.18 (±0.04) | 0.33 (±0.02) | 1,875.71 | 0.13 (±0.03) | 0.27 (±0.004) | 1,992.54 |
| Cefuroxime | 0.02 (±0.01) | 0.01 (±0.002) | 529.41 | 0.27 (±0.03) | 0.62 (±0.20) | 2,342.1 | 0.27 (±0.04) | 0.48 (±0.17) | 1,791.05 |
| Cefotaxime | 0.08 (±0.02) | 0.004 (±0.002) | 48.19 | 0.32 (±0.12) | 0.07 (±0.01) | 211.84 | 0.2 (±0.01) | 0.06 (±0.004) | 306.93 |
| Ceftazidime | 26.09 (±4.49) | 0.01 (±0.001) | 0.38 | 110.34 (±6.44) | 0.02 (±0.003) | 0.22 | 84.02 (±21.12) | 0.02 (±0.006) | 0.25 |
| Aztreonam | 6.01 (±1.26) | ND | NC | 3.85 (±0.49) | ND | NC | 4.54 (±0.41) | ND | NC |
| Imipenem | 2.29 (±0.60) | 0.003 (±0.001) | 1.31 | 2.56 (±0.73) | 0.004 (±0.0005) | 1.56 | 2.51 (±0.86) | 0.02 (±0.003) | 9.57 |
The IC50 values of clavulanate were 235.52 μM (± 01.71), 1,462.48 μM (±111.07), and 1,928.02 μM (±357.02) for isolates 65, 103, and 195, respectively; the IC50 values of sulbactam were 1.12 μM (±0.29), 7.75 μM (±1.20), and 11.65 μM (±2.03) for isolates 65, 103, and 195, respectively.
ND, not done; NC, not calculated.
Regarding the inhibition studies, IC50s showed a typical class C profile, with high clavulanic acid IC50s. However, there was a moderate degree of sulbactam inhibition, and the IC50s for clavulanic acid and sulbactam with ADC-14 were lower, which indicates that differences in amino acid sequence (primary structure of the enzyme) are related to differences in the catalytic properties of the ADC-type enzymes.
Expression of the ampC gene in AG3.
The next step was to assess whether or not differences in β-lactam MICs in AG3 clinical isolates may be due to differences in ampC gene expression. For this, Western blot analysis and RT-PCR were carried out.
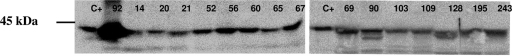
Western blot analysis of the sonicated AG3 extracts with polyclonal rabbit antiserum against ADC-7 enzyme revealed a protein band in each isolate, which corresponded to the expected molecular mass (43 kDa) of the Acinetobacter sp. AmpC and which migrated at the same level of ADC-7 (Fig. 2). Furthermore, the presence of the previously sequenced ISAba1 (involved in high-level blaAmpC expression in A. baumannii) (16) in the genome of the 15 AG3 isolates under study was discounted by the results of a series of PCR-based experiments (data not shown).
FIG. 2.
Western blotting with anti-ADC-7 antibody and with protein extracts (10 μg) obtained from the 15 different AG3 isolates indicated above the gel. Purified ADC-7 enzyme (15 ng) (a gift from R. A. Bonomo) (C+) and protein extracts of an Acinetobacter sp. isolate overexpressing the AmpC enzyme (isolate 92) were included as positive controls.
A band was observed at 470 bp by RT-PCR, which corresponded to the ampC gene in all 15 strains. A band of 344 bp of the gyrA gene (as an internal control) was observed in all AG3 isolates. The ratios of ampC/gyrA among AG3 isolates 20, 21, 28, 65, 103 and 243 and one Acinetobacter sp. isolate overexpressing the AmpC enzyme (isolate 92) were as follows: 1.9, 1.8, 1.7, 1.7, 1.6, 1.7, and 4.2, respectively. Therefore, no differences in the amounts of mRNA in the ampC genes of AG3 clinical isolates 20 and 103 or the remaining AG3 clinical isolates (with lower ampicillin MICs) were observed by semiquantitative RT-PCR (with the exception of positive control isolate 92). Overall, AG3 clinical strains 20 and 103 (with high ampicillin MICs) did not show higher ampC gene expression than the remaining AG3 isolates.
OMP analysis.
To assess whether or not differences in OMP expression were associated with differences in susceptibility to β-lactams, OMP profiles were obtained from isolates 20 and 103 as well as from representative ampicillin-susceptible isolates 21, 56, and 128. No differences in OMPs at molecular masses of 22, 29, 33 to 36, 40, and 45 kDa were visualized among the bacterial isolates (data not shown), thereby ruling out the involvement of OMPs in ampicillin or β-lactam resistance in AG3 isolates.
Effect of cloxacillin and efflux pumps inhibitors on MICs.
As no differences in either OMP or AmpC expression among AG3 isolates or catalytic efficiency against ampicillin were observed among AmpCs, we proposed that other mechanisms, such as an efflux pump, may also be operating in some AG3 clinical isolates and that this may account for differences in β-lactam MICs. The MICs of ampicillin, cephalothin, and meropenem were therefore determined in the presence of cloxacillin, CCCP, and PAN (Table 5). The antibiotic MICs for all AG3 clinical isolates were moderately lower in the presence of cloxacillin, although the effect was most apparent with ampicillin (between two and eight times lower). In the presence of the efflux pump inhibitors PAN or CCCP, the antibiotic MICs were slightly lower with some isolates, although the effect was most evident with AG3 isolate 103, for which the ampicillin MICs were at least 10.6 and 8 times lower with PAN and CCCP, respectively. The meropenem MICs were four and three times lower with the latter isolate, respectively. Cloxacillin and CCCP also exerted a synergistic effect on cephalothin MICs (>256 μg/ml without inhibitors compared with 12 to 96 μg/ml when both are added), which suggests the presence of an efflux operating at a constitutively low level in AG3. MICs of cloxacillin, PAN, and CCCP alone for AG3 isolates were higher than the concentration used in combination with antibiotics at 150 μg/ml, 25 μg/ml, and 25 μM (5.1 μg/ml), respectively. Therefore, MICs were the final effect of the antibiotic-inhibitor combination rather than of the inhibitor by itself (Table 5).
TABLE 5.
MICs for AG3 isolates in the presence of the indicated inhibitors
| AG3 isolate | MIC (μg/ml)a
|
MIC of CCCP (μM)b | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative control in the presence of:
|
Cloxacillin in the presence of:
|
PAN in the presence of:
|
CCCP in the presence of:
|
Cloxacillin + PAN in the presence of:
|
Cloxacillin + CCCP in the presence of:
|
Cloxacillinb | PANb | ||||||||||||||
| AMP | CE | MEM | AMP | CE | MEM | AMP | CE | MEM | AMP | CE | MEM | AMP | CE | MEM | AMP | CE | MEM | ||||
| 14 | 32 | >256 | 0.5 | 4 | 128 | 0.25 | 32 | >256 | 0.25 | 24 | >256 | 0.25 | 16 | 128 | 0.38 | 6 | 24 | 0.25 | 2,048 | >400 | 400 |
| 20 | 256 | >256 | 6 | 128 | >256 | 3 | 96 | >256 | 3 | 192 | >256 | 4 | 128 | >256 | 3 | 64 | 24 | 1.5 | 1,024 | >400 | 400 |
| 21 | 24 | >256 | 1 | 8 | >256 | 0.5 | 12 | >256 | 0.47 | 12 | >256 | 0.5 | 4 | 192 | 0.64 | 6 | 48 | 0.25 | 512 | >400 | >400 |
| 52 | 48 | >256 | 0.5 | 8 | >256 | 0.19 | 48 | >256 | 0.38 | 16 | >256 | 0.38 | 8 | >256 | 0.5 | 6 | 12 | 0.094 | 1,024 | 400 | >400 |
| 56 | 24 | 192 | 0.38 | 6 | >256 | 0.38 | 16 | >256 | 0.094 | 24 | 96 | 0.25 | 8 | >256 | 0.38 | 6 | 16 | 0.125 | 512 | >400 | >400 |
| 60 | 32 | >256 | 0.75 | 16 | >256 | 0.75 | 32 | >256 | 0.19 | 32 | >256 | 0.5 | 12 | >256 | 0.75 | 8 | 28 | 0.25 | 1,024 | 400 | >400 |
| 65 | 32 | >256 | 0.5 | 16 | 128 | 0.19 | 32 | >256 | 0.25 | 16 | 128 | 0.25 | 16 | 128 | 0.38 | 12 | 24 | 0.19 | 1,024 | >400 | >400 |
| 67 | 48 | >256 | 0.5 | 8 | 128 | 0.25 | 48 | >256 | 0.25 | 32 | 192 | 0.25 | 8 | 128 | 0.25 | 8 | 24 | 0.19 | 1,875 | 400 | >400 |
| 69 | 32 | >256 | 0.25 | 8 | >256 | 0.19 | 32 | >256 | 0.25 | 24 | >256 | 0.25 | 8 | >256 | 0.38 | 6 | 24 | 0.19 | >2,048 | 400 | >400 |
| 90 | 12 | >256 | 0.75 | 6 | 192 | 1 | 12 | >256 | 0.38 | 8 | >256 | 1.5 | 6 | 64 | 0.25 | 2 | 24 | 0.38 | 512 | >400 | >400 |
| 103 | >256 | >256 | 3 | 64 | 192 | 1.5 | 48 | >256 | 0.75 | 64 | >256 | 1 | 12 | 192 | 0.75 | 16 | 24 | 1 | 1,024 | 400 | >400 |
| 109 | 48 | >256 | 0.38 | 6 | 128 | 0.25 | 48 | >256 | 0.25 | 32 | >256 | 0.25 | 8 | 128 | 0.38 | 4 | 24 | 0.19 | 2,048 | 400 | >400 |
| 128 | 24 | >256 | 0.19 | 8 | 48 | 0.25 | 24 | >256 | 0.125 | 24 | >256 | 0.19 | 8 | 48 | 0.19 | 4 | 32 | 0.094 | 2,048 | 400 | >400 |
| 195 | 32 | >256 | 1 | 16 | >256 | 1 | 48 | >256 | 1 | 32 | >256 | 0.5 | 16 | >256 | 1 | 16 | 96 | 0.38 | 1,024 | 400 | >400 |
| 243 | 12 | >256 | 0.25 | 4 | 192 | 0.125 | 12 | >256 | 0.19 | 8 | >256 | 0.25 | 4 | 192 | 0.19 | 2 | 12 | 0.094 | 2,048 | >400 | 400 |
AMP, ampicillin; CE, cephalothin; MEM, meropenem; negative control, no inhibitor added.
Only inhibitor added.
Theoretical modeling of the ADC enzymes.
The theoretical overall folding three-dimensional structure of the ADC-12 enzyme is shown in Fig. 3. β-Lactamase domains (blue circles, with domain I in green) and the Ω-loop (turquoise) are shown in Fig. 3, as are the amino acid replacements (orange circles) in the ADC-type enzymes (relative to ADC-1) (Fig. 1). Amino acid replacements are randomly located along the protein sequence regardless of the phenotype of conferred ampicillin or β-lactam resistance, and none of them appear to affect the Ω-loop or any of β-lactamase domains (Fig. 3).
DISCUSSION
During the course of a multicenter study carried out in 2000, 244 Acinetobacter sp. isolates were analyzed. Fifteen of the isolates were identified as belonging to AG3 (6.1%); these microorganisms may represent an emergent genomic species of Acinetobacter (11, 29). Very few reports have been made regarding the role of β-lactamases in β-lactam resistance mechanisms in AG3 (1, 10, 35). In the present study, the range of ampicillin MICs for AG3 isolates was 12 to >256 μg/ml, with two isolates, isolates 20 and 103, showing high MICs (256 and >256 μg/ml, respectively).
A possible involvement of AmpC hyperexpression in clinical isolates was ruled out by RT-PCR analysis. Moreover, determination of MICs in the presence of cloxacillin revealed a slight decrease in β-lactam MICs. Therefore, although constitutively present, the role of AmpC in β-lactam resistance in AG3 appears to be moderate (which is consistent with the absence of ISAba1 upstream of the ampC gene); thus, the treatment of infections caused by AG3 isolates with β-lactams should be avoided, as it cannot be ruled out that in the near future, an ISAba1-like element may be inserted in the upstream region of the ampC gene, thus hyperexpressing the protein.
The sequencing of nucleotides and the deduced amino acid sequence of the 13 ampC genes reported here showed some degree of genetic variability. Although 4 out of 15 AG3 clinical isolates harbored an identical ampC gene (for which any epidemiological relationship was clearly discounted), the AmpC enzymes under study revealed a set of amino acid replacements, which, for unknown reasons, are located in specific positions in the amino acid sequence (Fig. 1). A steric view of these mutations in the overall fold structure of the modeled ADC-12 β-lactamase (Fig. 3) revealed that these replacements are located far from the active site of the enzyme. However, although there were no significant differences in MICs among different ADC-type enzymes, the biochemical analysis of three representative AmpC enzymes (ADC-14, -16, and -18) (one isolated from a highly ampicillin-resistant AG3 isolate and the other two isolated from ampicillin-susceptible AG3 strains) did reveal significant differences in some of the measured kinetic parameters (Table 4). This supports the idea that whereas some amino acid changes may be neutral, others are associated with dramatic changes in the catalytic efficiency or biochemical parameters of the AmpC enzymes (see Km and/or kcat values for ADC-14 and cephalosporins in comparison with ADC-16 and -18).
Other mechanisms such as a loss of porins, efflux pumps, and penicillin binding protein alterations may be involved in β-lactam resistance in Acinetobacter spp. (13, 14, 24). To study the involvement of non-AmpC-related mechanisms in some β-lactam MICs, we studied OMP and efflux pump expression.
Efflux pumps have been described for A. baumannii and AG3 (9, 17, 27) and have been detected in the recently revealed genome of Acinetobacter baylyi (www.genoscope.fr). Thus, experiments were carried out with the chemical inhibitors PAN and CCCP. The ampicillin MICs decreased (by at least 10 times) only for AG3 isolate 103 in the presence of either PAN or CCCP, thus revealing that efflux pumps may operate by pumping out β-lactams in some AG3 strains.
In summary, we report here the identification and analysis of 12 new ampC genes from AG3. We also report further information regarding β-lactam resistance in AG3. A uniform numerical system for the classification of cephalosporinase from Acinetobacter spp. is currently under development, and in accordance with this classification, we tentatively named our 12 new AmpC enzymes ADC-12 to ADC-23.
Acknowledgments
This work was supported by a scholarship (A.B.) from the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC); a research grant from Merck Sharp and Dohme, Spain; the Spanish Network for Research in Infectious Diseases (REIPI) (Instituto de Salud Carlos III, grant RD06/0008/0025), the Consellería de Sanidad, SERGAS PS07/90; and Fondo de Investigaciones Sanitarias (grants PI061368 and PI081613).
We thank R. A. Bonomo for the kind gift of anti-ADC-7 antibody.
Footnotes
Published ahead of print on 24 November 2008.
REFERENCES
- 1.Beceiro, A., L. Dominguez, A. Ribera, J. Vila, F. Molina, R. Villanueva, J. M. Eiros, and G. Bou. 2004. Molecular characterization of the gene encoding a new AmpC β-lactamase in a clinical strain of Acinetobacter genomic species 3. Antimicrob. Agents Chemother. 48:1374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berlau, J., H. M. Aucken, E. Houang, and T. L. Pitt. 1999. Isolation of Acinetobacter spp. including A. baumannii from vegetables: implications for hospital-acquired infections. J. Hosp. Infect. 3:201-204. [DOI] [PubMed] [Google Scholar]
- 3.Berlau, J., H. M. Aucken, H. Malnick, and T. L. Pitt. 1999. Distribution of Acinetobacter species on skin of healthy humans. Eur. J. Clin. Microbiol. Infect. Dis. 18:179-183. [DOI] [PubMed] [Google Scholar]
- 4.Bonomo, R. A., and D. Szabo. 2006. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43:49-56. [DOI] [PubMed] [Google Scholar]
- 5.Bou, G., G. Cervero, M. A. Dominguez, C. Quereda, and J. Martinez-Beltran. 2000. PCR-based DNA fingerprinting (REP-PCR, AP-PCR) and pulsed-field gel electrophoresis characterization of a nosocomial outbreak caused by imipenem- and meropenem-resistant Acinetobacter baumannii. Clin. Microbiol. Infect. 6:635-643. [DOI] [PubMed] [Google Scholar]
- 6.Bou, G., and J. Martinez-Beltran. 2000. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casin, I., J. Breuil, J. P. Darchis, C. Guelpa, and E. Collatz. 2003. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica Typhimurium isolates in humans. Emerg. Infect. Dis. 11:1455-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chau, S.-L., Y. W. Chu, and T. S. Houang. 2004. Novel resistance-nodulation-cell division efflux system AdeDE in Acinetobacter genomic DNA group 3. Antimicrob. Agents Chemother. 48:4054-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu, Y. W., M. Afzal-Shah, E. T. Houang, M. I. Palepou, D. J. Lyon, N. Woodford, and D. M. Livermore. 2001. IMP-4, a novel metallo-β-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob. Agents Chemother. 45:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu, Y. W., C. M. Leung, E. T. Houang, K. C. Ng, C. B. Leung, H. Y. Leung, and A. F. Cheng. 1999. Skin carriage of acinetobacters in Hong Kong. J. Clin. Microbiol. 37:2962-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S16, vol. 26, no. 1. Clinical and Laboratory Standards Institute, Wayne, PA.
- 13.del Mar Tomás, M., A. Beceiro, A. Pérez, D. Velasco, R. Moure, R. Villanueva, J. Martínez-Beltrán, and G. Bou. 2005. Cloning and functional analysis of the gene encoding the 33- to 36-kilodalton outer membrane protein associated with carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:5172-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Cuenca, F., L. Martinez-Martinez, M. C. Conejo, J. A. Ayala, E. J. Perea, and A. Pascual. 2003. Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 51:565-574. [DOI] [PubMed] [Google Scholar]
- 15.Giovanetti, E., A. Brenciani, R. Burioni, and P. E. Varaldo. 2002. A novel efflux system in inducibly erythromycin-resistant strains of Streptococcus pyogenes. Antimicrob. Agents Chemother. 46:3750-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heritier, C., L. Poirel, and P. Nordmann. 2006. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 12:123-130. [DOI] [PubMed] [Google Scholar]
- 17.Higgins, P. G., H. Wisplinghoff, D. Stefanik, and H. Seifert. 2004. Selection of topoisomerase mutations and overexpression of adeB mRNA transcripts during an outbreak of Acinetobacter baumannii. J. Antimicrob. Chemother. 54:821-823. [DOI] [PubMed] [Google Scholar]
- 18.Horrevorts, A., K. Bergman, L. Kollee, I. Breuker, I. Tjernberg, and L. Dijkshoorn. 1995. Clinical and epidemiological investigations of Acinetobacter genomospecies 3 in a neonatal intensive care unit. J. Clin. Microbiol. 33:1567-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hujer, K. M., N. S. Hamza, A. M. Hujer, F. Perez, M. S. Helfand, C. R. Bethel, J. M. Thomson, V. E. Anderson, M. Barlow, L. B. Rice, F. C. Tenover, and R. A. Bonomo. 2005. Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 β-lactamase: defining a unique family of class C enzymes. Antimicrob. Agents Chemother. 49:2941-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong, S. H., I. K. Bae, K. O. Park, Y. J. An, S. G. Sohn, S. J. Jang, K. H. Sung, K. S. Yang, K. Lee, D. Young, and S. H. Lee. 2006. Outbreaks of imipenem-resistant Acinetobacter baumannii producing carbapenemases in Korea. J. Microbiol. 44:423-431. [PubMed] [Google Scholar]
- 21.Kim, J. Y., H. I. Jung, Y. J. An, J. H. Lee, S. J. Kim, S. H. Jeong, K. J. Lee, P. G. Suh, H. S. Lee, S. H. Lee, and S. S. Cha. 2006. Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase. Mol. Microbiol. 60:907-916. [DOI] [PubMed] [Google Scholar]
- 22.Matthew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 23.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 86:321-331. [DOI] [PubMed] [Google Scholar]
- 24.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 25.Ribera, A., F. Fernández-Cuenca, A. Beceiro, G. Bou, L. Martínez-Martínez, A. Pascual, J. M. Cisneros, J. Rodríguez-Baño, J. Pachón, J. Vila, and the Spanish Group for Nosocomial Infection. 2004. Antimicrobial susceptibility and mechanisms of resistance to quinolones and β-lactams in Acinetobacter genospecies 3. Antimicrob. Agents Chemother. 48:1430-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santillana, E., A. Beceiro, G. Bou, and A. Romero. 2007. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc. Natl. Acad. Sci. USA 104:5354-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su, X.-Z., J. Chen, T. Mizushima, T. Kuroda, and T. Tsuchiya. 2005. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 49:4362-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Traub, W. H., and D. Bauer. 2000. Surveillance of nosocomial cross-infections due to three Acinetobacter genospecies (Acinetobacter baumannii, genospecies 3 and genospecies 13) during a 10-year observation period: serotyping, macrorestriction analysis of genomic DNA and antibiotic susceptibilities. Chemotherapy 46:282-292. [DOI] [PubMed] [Google Scholar]
- 30.Vaneechoutte, M., L. Dijkshoorn, I. Tjernberg, A. Elaichouni, P. de Vos, G. Claeys, and G. Verschraegen. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Looveren, M., H. Goossens, and the ARPAC Steering Group. 2004. Antimicrobial resistance of Acinetobacter spp. in Europe. Clin. Microbiol. Infect. 10:684-704. [DOI] [PubMed] [Google Scholar]
- 32.Vegas, E. Z., B. Nieves, M. Araque, E. Velasco, J. Ruiz, and J. Vila. 2006. Outbreak of infection with Acinetobacter strain RUH 1139 in an intensive care unit. Infect. Control Hosp. Epidemiol. 27:397-403. [DOI] [PubMed] [Google Scholar]
- 33.Vila, J., M. A. Marcos, and M. T. Jiménez de Anta. 1996. A comparative study of different PCR-based DNA fingerprinting techniques for typing of the Acinetobacter calcoaceticus-A. baumannii complex. J. Med. Microbiol. 44:482-489. [DOI] [PubMed] [Google Scholar]
- 34.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 182:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yum, J. H., K. Yi, H. Lee, D. Yong, K. Lee, J. M. Kim, G. M. Rossolini, and Y. Chong. 2002. Molecular characterization of metallo-beta-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: identification of two new integrons carrying the bla(VIM-2) gene cassettes. J. Antimicrob. Chemother. 49:837-840. [DOI] [PubMed] [Google Scholar]