Abstract
The 773-residue ectodomain of the herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) has been resistant to the use of mutagenic strategies because the majority of the induced mutations result in defective proteins. As an alternative strategy for the identification of functionally important regions and novel inhibitors of infection, we prepared a library of overlapping peptides homologous to the ectodomain of gB and screened for the ability of the peptides to block infection. Seven of 138 15-mer peptides inhibited infection by more than 50% at a concentration of 100 μM. Three peptides (gB94, gB122, and gB131) with 50% effective concentrations (EC50s) below 20 μM were selected for further studies. The gB131 peptide (residues 681 to 695 in HSV-1 gB [gB-1]) was a specific entry inhibitor (EC50, ∼12 μM). The gB122 peptide (residues 636 to 650 in gB-1) blocked viral entry (EC50, ∼18 μM), protected cells from infection (EC50, ∼72 μM), and inactivated virions in solution (EC50, ∼138 μM). We were unable to discern the step or steps inhibited by the gB94 peptide, which is homologous to residues 496 to 510 in gB-1. Substitution of a tyrosine in the gB122 peptide (Y640 in full-length gB-1) reduced the antiviral activity eightfold, suggesting that this residue is critical for inhibition. This peptide-based strategy could lead to the identification of functionally important regions of gB or other membrane proteins and identify novel inhibitors of HSV-1 entry.
Herpes simplex virus type 1 (HSV-1) is a significant human pathogen that primarily causes mucocutaneous ulcers. Less common but more serious infections can result in encephalitis, blinding keratitis, or neonatal herpes (78). Several antivirals are approved for the treatment of HSV infections but are ineffective in certain situations and do not eliminate latent infections (20, 56, 65). Thus, new strategies are needed to deal with these infections.
HSV-1 infection is initiated by the binding of glycoprotein C (gC) to cell surface heparan sulfate (69, 71, 72). The HSV-1 gB protein (gB-1) can also bind to heparan sulfate proteoglycan, but the binding is less efficient (37). Following attachment, gD binds to any of four cellular coreceptors, including HVEM (herpesvirus entry mediator), nectin 1, nectin 2, or 3-O-sulfated heparan sulfate, resulting in a conformational change in gD (18, 28, 41, 50, 51). The conformational change in gD is then thought to trigger the formation of the fusion complex, which includes gB and the gH-gL heterodimer. The coexpression of gD, gB, and gH-gL in the same cell results in cell-cell fusion, indicating that these four proteins constitute the minimal fusion apparatus (10, 24, 52, 61, 75). It has been suggested that gH-gL and gB are recruited to gD independently of one another and possibly interact with each other (3). Recent studies show that HSV-1 fusion is mediated through a hemifusion intermediate involving gH-gL but that the complete fusion event requires the action of gB (73). gB-1 interacts with the cell surface (7, 59, 60), and a cell surface protein, PILRα, has been identified as a gB-1 binding entry coreceptor (66).
The role of gB-1 in the entry/fusion process is poorly understood, even though it is the most highly conserved herpesvirus envelope protein (58). Several studies indicate that gB-1 functions as an oligomer. Mature gB-1 sediments at approximately 200 kDa in sucrose gradients, whereas the monomeric form has an apparent molecular mass of 110 kDa (2, 17, 38). Mutant gB-1 can act in a dominant negative manner to reduce viral titers and inhibits the complementation of null mutants by wild-type gB-1 (13), and a temperature-sensitive gB-1 mutant (mutant tsB5) fails to oligomerize at the nonpermissive temperature (15, 34). Recently, the elucidation of the crystal structure of the postfusion form of gB-1 has confirmed that the protein is a trimer (36) and shares a high degree of structural, but not sequence, homology to the vesicular stomatitis virus G protein (64).
gB-1 is 904 amino acids long and has a 773-residue ectodomain, a 22-residue transmembrane domain, and a 109-residue cytoplasmic tail (36, 57). Monomeric gB-1 contains 10 cysteines that form five intramolecular disulfide bonds, some of which appear to be important for proper folding and processing of the protein (36, 42, 53). Each gB-1 monomer contains five domains (36), and recent studies with gB-specific monoclonal antibodies have identified functional regions in the gB-1 ectodomain that are important for infection (6). A heparan sulfate binding site, which allows gB-1 to substitute for gC-1 in attachment, is located between residues 68 to 76 of gB (37, 43). Single amino acid substitutions in domain I of gB-1, which is structurally analogous to the fusion loop in vesicular stomatitis virus G protein, were shown to ablate syncytium formation and cell-cell fusion in transfected receptor-positive cells that transiently expressed gD, gH-gL, and the mutant gB, suggesting that these residues are important for membrane fusion. Thus, this region is a putative fusion loop in gB-1 (35). Several mutations resulting in a hyperfusion phenotype have been mapped to the cytoplasmic tail of gB-1, indicating that this region plays an important role in regulating the fusogenic activity of the protein (4, 21, 24, 25, 46, 63, 77). Mutations leading to reversion of the syncytium-forming phenotype of gB-1 mutants have been mapped to the UL45 envelope protein (33), suggesting that UL45 may interact with gB-1 and regulate fusogenic activity. gB-1 is also associated with lipid rafts in infected cells (8); however, the region in gB-1 that is involved in this association has not been identified.
The use of mutagenic approaches to identify functionally important regions of the gB-1 ectodomain has been difficult due to misfolding of the majority of the mutant proteins (13, 14, 49, 62). Insertions into α helices or β sheets appear to be particularly disruptive (54). Even approaches that target predicted surface-exposed loops result in a high percentage of mutants that are improperly expressed (44). Recently, a random 5-amino-acid-insertion mutagenesis strategy was utilized; and only 27 of 81 mutants were properly folded, processed, and transported to the cell surface (45). The difficulty with the use of mutagenesis strategies to study the function of the gB-1 ectodomains may be due to the fact that trimer formation involves extensive intermolecular contacts over large areas of the protein (36).
Peptide-based strategies have previously been used to study herpesvirus glycoprotein function. Trybala et al. (74) used a series of synthetic peptides to show that two discontinuous regions of HSV-1 gC were involved in binding to heparan sulfate proteoglycans. Several laboratories have shown that peptides homologous to heptad repeat regions from gB-1, HSV-1 gH, bovine herpes virus type 1 gB, and human cytomegalovirus gB inhibit infection, indicating that these regions of the proteins are functionally significant (27, 29, 48, 55). Rationally designed β-peptides that mimic the human cytomegalovirus gB heptad repeat are effective inhibitors of infection (23). More recently, and while the manuscript for this study was being written, peptides homologous to HSV-1 and bovine herpes virus type 1 gB that had a high probability of forming regions of high interfacial hydrophobicity were shown to inhibit viral infection (26). Those studies clearly indicate that peptide-based approaches can be useful in identifying functionally important regions of herpesvirus glycoproteins. A significant feature of these peptide-based studies is that all of them targeted regions of the proteins with predictable structures or motifs, and these approaches might miss unique regions that play important roles in infection.
To determine if a more generalized peptide-based approach could identify functionally important regions of gB, we synthesized a library of overlapping 15-mer peptides encompassing the ectodomain of gB-1 and tested the peptides for their ability to inhibit HSV infection. Seven of the peptides inhibited infection by 50% or more when they were screened at a concentration of 100 μM. Three peptides were selected for further study, and two primarily blocked the entry of HSV-1 into cells. These results validate the use of overlapping peptide libraries (peptide scanning inhibition) for the identification of functionally important regions in membrane glycoproteins.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal bovine serum and 5% calf serum. High-titer stocks of HSV-1 KOS mutant hrR3, which expresses Escherichia coli β-galactosidase from the early ICP6 promoter (30), were prepared (32) and were further purified on sucrose gradients, as described previously (76).
Peptide synthesis and analysis.
All peptides were synthesized at the University of Wisconsin—Madison Biotechnology Center. Overlapping gB peptides were synthesized by using SynPhase lanterns (rink amide, 8 μM; Mimotopes Pty. Ltd., Australia), according to the manufacturer's instructions. The peptide concentrations were determined by dividing the weight of the crude product by the volume of water used to dissolve the peptide. Note that for the high-throughput initial screen, potential differences in the solubilities of the peptides were not accounted for. The large-scale synthesis and the purification (>95% purity) of the individual peptides were carried out as we described previously (11, 12). The concentrations of the large-scale batches were determined by measuring the absorbance at 215 nm and 225 nm (68).
MTS assay.
Peptide cytotoxicity was determined by a commercially available assay (Celltiter 96 AQqueous One Solution cell proliferation assay reagent; Promega, Madison, WI), as described previously (1). Briefly, Vero cells (1.5 × 104 cells/well) were seeded in a 96-well plate, and the plate was incubated for 24 h at 37°C. A total of 20 μl of medium containing the desired concentration of peptides was added to the cells. Control cells received medium only. After incubation of the cells in the presence of peptide overnight at 37°C, 20 μl of the 96 AQqueous One Solution cell proliferation assay reagent was added to each well. The plate was then incubated for 2 h at 37°C, and the absorbance at 490 nm was determined with a 96-well plate reader (Biotek Instruments, Inc., Winooski, VT).
Antiviral assays.
Vero cells (1 × 104 cells/well) seeded in 96-well plates and incubated for 4 days at 37°C were infected with hrR3 at a multiplicity of infection of 1. Carbonate-free DMEM supplemented with 5% calf serum and 5% fetal bovine serum (S+ medium) was used for all assays. At 6 h postinfection, the β-galactosidase activity was measured by enzyme-linked immunosorbent assay (ELISA) (12). After the cells were lysed with 0.5% Nonidet P-40 and 2 mM MgCl2 in phosphate-buffered saline (PBS; 50 μl/well) for 5 min and the addition of 10 mM chlorophenol red-β-d-galactopyranoside (50 μl/well; Roche Applied Science, Indianapolis, IN) to each well, the absorbance was measured in triplicate at 570 nm in a 96-well plate reader (Biotek Instruments, Inc.).
(i) Comprehensive antiviral assay.
Various concentrations of the peptides were mixed with virus (1 × 106 PFU/ml) in 70 μl of S+ medium and with Vero cells in 96-well plates. Following incubation for 1 h at 37°C, the virus-peptide mixture was added to the treated cells. At 6 h postinfection, β-galactosidase activity was measured as an indicator of infection, as described above.
(ii) Virus inactivation assay.
Virus (1 × 108 PFU/ml) was treated with various concentrations of peptide in 50 μl of S+ medium for 1 h at 37°C. The treated virus was then diluted 200-fold and used to infect cells (40 μl/well) for 1 h at 37°C. The diluted virus-peptide mixtures were replaced with S+ medium, and the β-galactosidase activity was measured 6 h later.
(iii) Cell protection assay.
Vero cells were treated with various concentrations of peptide (50 μl/well) at 37°C. After 1 h, the cells were washed with S+ medium and infected with 40 μl/well of hrR3 (1 × 106 PFU/ml) at 37°C for 1 h. The medium containing virus was replaced with fresh S+ medium, and the β-galactosidase activity was measured 6 h later.
(iv) Entry assay.
Vero cells were fed with S+ medium and incubated at 4°C for 30 min. The medium was removed, and hrR3 (40 μl/well, 5 × 105 PFU/ml) was allowed to attach to the cells at 4°C. After 1 h, the cells were rinsed with S+ medium. Various concentrations of peptide were then added (40 μl/well), and the cultures were incubated at 4°C for 15 min before the incubation temperature was shifted to 37°C. After 1 h, the cells were washed with 1× PBS, and any remaining extracellular virus was inactivated by treatment with citrate buffer (pH 3.0) for 1 min. The infected cells were rinsed again with 1× PBS and fed with S+ medium, and the β-galactosidase activity was measured 6 h later.
Attachment assay.
The hrR3 virus (4 × 105 PFU/well) was allowed to bind to Vero cells in the presence of various concentrations of peptide at 4°C for 1 h. After free virus was rinsed off with serum-free carbonate-free DMEM (S− medium), the cells were fixed with 4% paraformaldehyde in S− medium and blocked for 1 h with 3% bovine serum albumin in S− medium. Absorbed virus was detected with polyclonal rabbit anti-HSV-1 immunoglobulin G (IgG; 1:100 dilution; Dako, Carpinteria, CA) and alkaline phosphatase (AP)-conjugated goat anti-rabbit IgG (1:2,000 dilution; Sigma-Aldrich, St. Louis, MO). After the plates were washed with S− medium, 100 μl of AP substrate (0.2 M diethanolamine, 0.5 mM MgCl2, 1.1 mM l-homoarginine, and 1.7 mM p-nitrophenylphosphate in Tris-buffered saline, pH 9.6) was added to each well, and the absorbance (405 nm) was read in a 96-well plate reader (Biotek Instruments, Inc.) at 1-min intervals for 20 min. The amount of binding was determined from the slope of the initial rate for triplicate samples, and the signal from mock-infected cultures was subtracted from the data.
VP16 transport to the nucleus and ICP0 expression.
Vero cells (8 × 106 cells per well) were seeded into six-well plates. On the following day, the cells were cooled to 4°C, hrR3 was added at a multiplicity of infection of 1, and the cultures were incubated for 1 h to allow attachment. Various concentrations of peptide were then added, and the cultures were incubated for an additional 15 min at 4°C. The cultures were then shifted to 37°C for 1 h, rinsed with citrate buffer, and then incubated for either 4 h (VP16) or 5 h (ICP0). For VP16 translocation, nuclear extracts were isolated by using a NucBuster protein extraction kit (catalog no. 71183-3; EMD Biosciences, Madison, WI), according to the manufacturer's instructions.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting.
All samples were boiled for 5 min in Laemmli sample buffer containing 0.05% β-mercaptoethanol (Bio-Rad, Hercules, CA) and subjected to electrophoresis in 12% sodium dodecyl sulfate-polyacrylamide gels. The proteins were then transferred to nitrocellulose membranes, which were subsequently blocked in 5% (wt/vol) milk powder in washing buffer (0.3% [vol/vol] Tween 20, 100 mM maleic acid, 150 mM NaCl, pH 7.5) for 1 h at room temperature. The membrane was incubated with primary antibody overnight at 4°C. After the membrane was washed, it was incubated with secondary antibody for 1 h at room temperature. The immunoblots were developed with the AP substrate 5-bromo-4 chloro-3-indolylphosphate-nitroblue tetrazolium (FAST; Sigma-Aldrich). The primary antibodies used were polyclonal goat anti-VP16 antibody (1:200 dilution; catalog no. sc-34070; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and mouse monoclonal anti-ICP0 antibody (1:5,000 dilution; catalog no. H1A027-100; Virusys Corporation, Sykesville, MD). The secondary antibodies were AP-conjugated rabbit anti-goat IgG and AP-conjugated goat anti-mouse IgG (1:2,000 dilution; catalog nos. A7650 and A3562, respectively; Sigma-Aldrich).
RESULTS
Screening of the gB homologous peptide library.
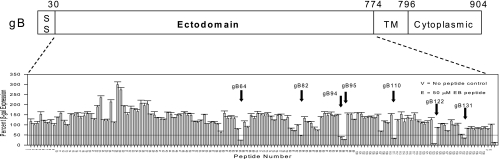
To test the feasibility of using a more general peptide strategy for identifying functionally important regions of membrane glycoproteins and potential inhibitors, a library of 15-mer peptides extending from residues 30 to 730 of the gB-1 ectodomain and overlapping each other by 10 residues was synthesized. The library was initially screened for peptides with inhibitory activity by using a high-throughput comprehensive antiviral assay in which both virus and cells were exposed to the peptides prior to infection and the peptides remained present throughout the assay. Infectivity was measured by determination of the β-galactosidase activity expressed as a reporter gene in the hrR3 virus. The criterion for designating a peptide positive was the inhibition of β-galactosidase activity by 50% or more at a presumptive concentration of 100 μM. Figure 1 shows the locations of the peptides in the library and their antiviral activities in the initial screen. Of the 138 peptides in the library, 7, all of whose sequences clustered in the carboxy-terminal half of the ectodomain, inhibited infection by 50% or more (Fig. 1). The seven peptides corresponded to residues 346 to 360 (gB64), 436 to 450 (gB82), 496 to 510 (gB94), 501 to 515 (gB95), 576 to 590 (gB110), 636 to 650 (gB122), and 681 to 695 (gB131) (Fig. 2 and Table 1). A peptide known to inhibit HSV-1 entry, the EB peptide (12), was also included as a positive control in this assay. Eight peptides clustering in the amino-terminal half of the gB-1 ectodomain appeared to enhance infection by more than twofold. These peptides were not explored further.
FIG. 1.
Identification of gB-1 peptides that inhibit HSV-1 infection. A library of 138 overlapping peptides spanning residues 30 to 730 (ectodomain) of gB-1 were synthesized and tested at a concentration of 100 μM in a comprehensive antiviral assay for the ability to inhibit β-galactosidase (β-gal) expression by the hrR3 virus. The top line shows a schematic representation of the gB-1 protein, and the activities of the peptides are shown at the bottom. Arrows denote the seven peptides that were positive in the assay. V, virus-only control; E, inhibition by the EB peptide as a positive control (12); SS, signal sequence; TM, transmembrane domain.
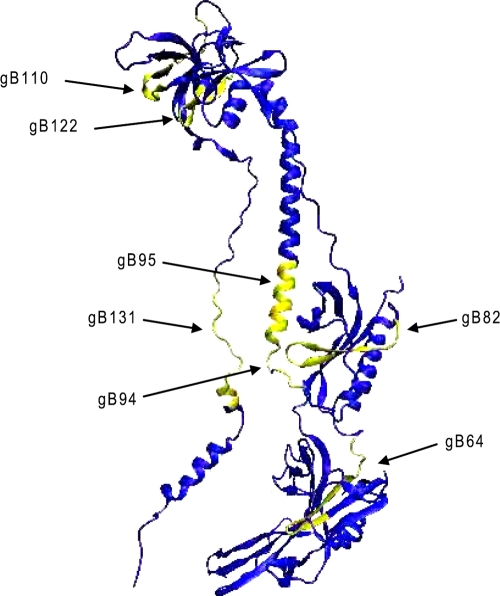
FIG. 2.
Schematic representation of the locations of the seven inhibitory peptides on the gB-1 monomer. The structural coordinates were obtained from http://www.rcsb.or/pdb/home/home/do. The gB-1 structure is designated 2GUM in the PDB and was visualized by use of the visual molecular dynamics program (version 1.8.4) at http://www.ks.uiuc.edu/Research/vmd (36, 40).
TABLE 1.
Sequences of the seven inhibitory peptides, locations in gB-1, and EC50sa
| Peptideb | Sequence | gB location (amino acid residues) | Domainc | EC50 (μM)d |
|---|---|---|---|---|
| gB64 | TTPKFTVAWDWVPKR | 346-360 | I | 85 |
| gB82 | VGQPQYYLANGGFLI | 436-450 | II | NDe |
| gB94 | KTTSSIEFARLQFTY | 496-510 | III | 6.5 |
| gB95 | IEFARLQFTYNHIQR | 501-515 | III | ND |
| gB110 | VAADNVIVQNSMRIS | 576-590 | IV | ND |
| gB122 | GHRRYFTFGGGYVYF | 636-650 | IV | 15 |
| gB131 | HEVVPLEVYTRHEIK | 681-695 | V | 18 |
At least one peptide maps to each of the five subdomains of gB-1.
The peptides are designated sequentially starting at residue 30 of mature gB-1 and extending to the start site of the transmembrane segment.
The domain nomenclature is that of Heldwein et al. (36).
Comprehensive assay.
ND, not determined due to poor solubility.
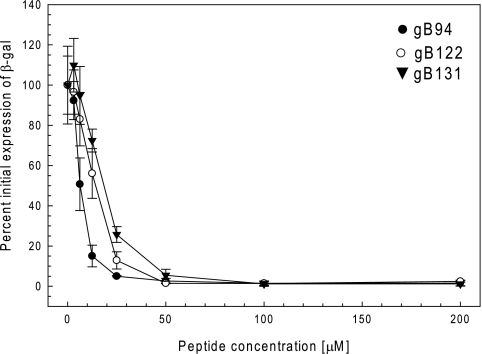
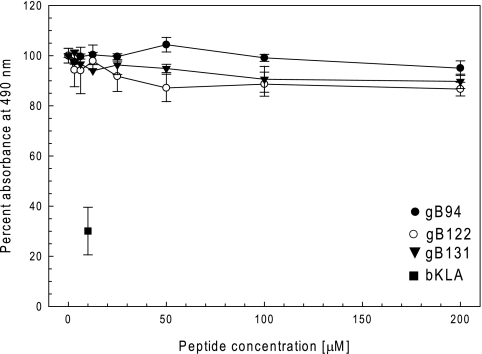
To further confirm that the active peptides inhibited infection, each peptide was resynthesized on a larger scale and with a higher purity. Resynthesized peptides gB82, gB95, and gB110 had reduced solubility in aqueous solution compared to the solubilities of the other peptides and were not studied further. The remaining peptides were tested in the comprehensive assay, and the EC50s were determined. The results confirmed that gB94, gB122, and gB131 inhibited viral infection with EC50s of less than 20 μM (6.5, 15, and 18 μM, respectively; Fig. 3 and Table 2). Peptide gB64 had an EC50 of 85 μM and was not considered further. The locations of the inhibitory peptides on the monomeric gB-1 crystal structure are shown in Fig. 2. None of the selected inhibitory peptides (gB94, gB122, and gB131) were cytotoxic at a concentration of 200 μM by an MTS assay (Fig. 4).
FIG. 3.
Antiviral activities of gB94, gB122, and gB131 in the comprehensive assay. The peptides were newly synthesized and purified, and their antiviral activities were confirmed in the comprehensive assay, as described in Materials and Methods. Briefly, both cells and virus were incubated with various concentrations of peptides for 1 h at 37°C. Following incubation, the peptides on the cells were replaced with virus-peptide mixtures and the cultures were incubated further. Six hours later, the β-galactosidase (β-gal) activity was determined as a measure of infectivity. The graph is a representative result from at least two independent experiments. Each point and error bar represents the mean ± standard deviation of triplicate samples.
TABLE 2.
EC50s of gB peptides in various antiviral assays
| Peptide | EC50 (μM) in the following assaysa:
|
||||
|---|---|---|---|---|---|
| Comprehensive | Entry | Cell protection | Virus inactivation | Attachment | |
| gB94 | 6.5 ± 1.3 | >200 | 165.0 ± 71.9 | 125 ± 11.4 | >200 |
| gB122 | 15.0 ± 2.2 | 17.6 ± 2.6 | 71.5 ± 17.4 | 118 ± 11.6 | >200 |
| gB131 | 18.7 ± 2.70 | 12.2 ± 6.56 | >200 | >200 | >200 |
The EC50s are the means ± standard deviations determined from at least two independent experiments.
FIG. 4.
The gB94, gB122, and gB131 peptides are not cytotoxic for Vero cells. Vero cell monolayers in a 96-well plate were treated with various concentrations of the peptides overnight. The next day, 20 μl of Celltiter 96 AQqueous One Solution cell proliferation assay reagent (Promega) was added to each well, and 2 h later, the absorbance at 490 nm was read in a 96-well plate reader. The bKLA peptide is a cationic amphipathic peptide that is toxic to cells (1, 11) and was used at 10 μM as the positive control. Each point and error bar represents the mean ± standard deviation of triplicate samples. The peptide concentration denoted zero is the no-peptide control, whose activity was set equal to 100%.
Peptides gB94, gB122, and gB131 have different modes of action.
To determine the step in infection inhibited by gB94, gB122, and gB131, we varied the time during infection when the peptide was added. Initially, to test the inhibition of attachment, the virions were mixed with peptides and added to cells, and the amount of bound virus was determined by a cell-based ELISA. To determine if the peptides were virucidal, virions were incubated with various concentrations of peptides for an hour at 37°C and then serially diluted, and the titers were determined. To determine if pretreatment of the cells was protective, the cells were incubated with the peptides for 1 h at 37°C and rinsed prior to infection. The ability of the peptides to block entry was tested by adding virions to the cells at 4°C, followed by the addition of the peptides. The cultures were incubated at 4°C for an additional 15 min, at which time they were shifted to 37°C and infection was monitored. For all these assays except the attachment assay, infectivity was determined by measuring the β-galactosidase activity at 6 h postinfection. The means of the EC50s determined in these assays are summarized in Table 2.
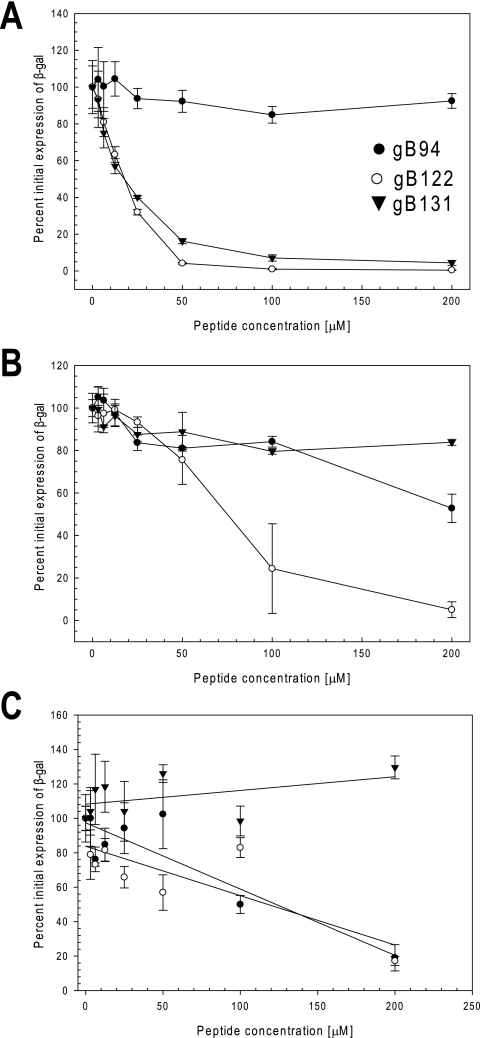
(i) Entry-blocking activities of peptides gB94, gB122, and gB131.
As shown in Fig. 5A, the gB94 peptide had no effect in the entry assay. In contrast, both gB122 and gB131 clearly inhibited viral infection at the entry step in a dose-dependent manner with EC50s of 18 and 12 μM, respectively. These values were comparable to the EC50s of 15 and 18 μM for gB122 and gB131, respectively, in the comprehensive assay. To rule out the possibility that peptides gB94, gB122, and gB131 blocked virus binding to cells, the ability of the peptides to inhibit attachment was tested by the cell-based ELISA. As summarized in Table 2, the peptides at concentrations as high as 200 μM did not interfere with attachment (EC50s > 200 μM). These results indicate that the predominant antiviral mechanism of peptides gB122 and gB131 is at the viral entry step, whereas peptide gB94 does not appear to block viral entry.
FIG. 5.
Antiviral activities of the gB94, gB122, and gB131 peptides. (A) To test for entry inhibition, virus was attached to cells at 4°C, peptide was added, and the cultures were shifted to 37°C. (B) To test for cell protection, Vero cells were incubated with various concentrations of peptide for 1 h at 37°C, rinsed, and then infected with hrR3. (C) To test for virucidal activity, peptide was incubated with virus for 1 h at 37°C and serially diluted, and then the titers were determined on Vero cells. For all studies, β-galactosidase (β-gal) activity was determined at 6 h postinfection as a measure of infectivity. The graph is a representative result from at least two independent experiments. Each point and error bar represents the mean ± standard deviation of triplicate samples.
(ii) Effects of gB94, gB122, and gB131 on cellular protection.
Figure 5B shows the antiviral activities of peptides gB94, gB122, and gB131 when Vero cell monolayers were pretreated with one of the peptides and rinsed prior to infection. Pretreatment of the cells with the gB94 peptide inhibited infection by 45% at the highest concentration tested (200 μM). When the cells were pretreated with gB122, protection was seen with an EC50 of 70 μM. These results suggest that peptides gB94 and gB122 had some effect on the cells, but this could not account for the majority of the antiviral activity seen in the comprehensive assay. The gB131 peptide was inactive when the cells were pretreated with the peptide prior to infection.
(iii) Effects of gB94, gB122, and gB131 on inactivation of virions in solution.
Having shown that gB94, gB122, and gB131 vary in their abilities to block viral entry and confer cellular resistance to infection, we next tested them for their virucidal activities. When virus was incubated with the gB94 peptide prior to infection, the EC50 was approximately 125 μM (Fig. 5C). This was significantly higher than that observed in the comprehensive assay, suggesting that the gB94 peptide can irreversibly inactivate virions in solution only at high concentrations. When virus was pretreated with the gB122 peptide, the EC50 was 118 μM (Fig. 5C), indicating that the antiviral activity of gB122 is not primarily due to virucidal activity. Pretreatment of virus with the gB131 peptide had no inhibitory effect at concentrations as high as 200 μM (Fig. 5C). In summary, Table 2 and Fig. 3 to 5 demonstrate that gB94, gB122, and gB131 all inhibited HSV-1 infection but did so by distinct mechanisms.
Peptide gB122 blocks postentry events, as measured by nuclear VP16 translocation and ICP0 expression.
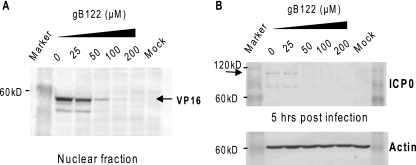
A comparison of previously identified functionally important regions in gB with the locations of the seven inhibitory peptides indicated that only one of the peptides, gB122, mapped to a putative functionally important region (38, 42). A comparison of the gB122 sequence in all herpesvirus gB proteins indicated that this region is highly conserved among the alphaherpesvirus subfamily (Table 3). The gB122 peptide also partly overlaps a peptide identified as having a propensity for high interfacial hydrophobicity (26). Because of these features, we selected the gB122 peptide for further studies. To confirm that gB122 acted at an early step in infection, the translocation of the VP16 tegument protein to the nucleus and expression of the ICP0 immediate early protein in the presence of gB122 were determined. The translocation of VP16 to the nucleus has previously been used as an indicator of entry (16, 67). In the absence of gB122, VP16 was detected in the nucleus by 4 h postinfection (Fig. 6A). There was a dose-dependent decrease in the amount of nuclear VP16 in the presence of gB122, and at concentrations of 100 μM or higher, VP16 was not detected in the nuclear fraction. The ICP0 protein is one of five HSV-1 immediate early proteins that are expressed before any other viral proteins are expressed. Similar to the VP16 translocation, there was a dose-dependent reduction in ICP0 expression in the presence of peptide, and no expression was detected at peptide concentrations of 50 μM or higher (Fig. 6B). Both of these results are consistent with a mechanism by which gB122 acts to block infection at the entry step.
TABLE 3.
gB protein sequences among herpesviruses
| Herpesvirus | Peptide sequencea
|
||
|---|---|---|---|
| gB94 | gB122 | gB131 | |
| HSV-1 KOS | KTTSSIEFARLQFTY | GHRRYFTFGGGYVYF | HEFVPLEVYTRHEIK |
| HSV-1 strain F | KTTSSIEFARLQFTY | GHRRYFTFGGGYVYF | HEFVPLEVYTRHEIK |
| HSV-1 strain 17 | KTTSSIEFARLQFTY | GHRRYFTFGGGYVYF | HEFVPLEVYTRHEIK |
| HSV-1 Patton | KTTSSIEFARLQFTY | GHRRYFTFGGGYVYF | HEFVPLEVYTRHEIK |
| HSV-1 Angelotti Path | KTTSSIEFARLQFTY | GHRRYFTFGGGYVYF | HEFVPLEVYTRHEIK |
| HSV-1 HF | KTTSSIEFARLQFTY | GHRRYFTFGGGYVYF | HEFVPLEVYTRHEIK |
| HSV-1 HSZP | KTTSSIEFARLQFTY | GHRRYFTFGGGYVYF | HEFVPLEVYTRHEIK |
| HSV-1 MTD-1730 | KTTSSIEFARLQFTY | GHRRYFTFGGGYVYF | HEFVPLEVYTRHEIK |
| HSV-2 333 | KTTSSIEFARLQFTY | GHRRYFIFGGGYVYF | HEFVPLEVYTRHEIK |
| HSV-2 Burr | KTTSSIEFARLQFTY | GHRRYFIFGGGYVYF | HEFVPLEVYTRHEIK |
| HSV-2 BBKC | KTTSSIEFARLQFTY | GHRRYFIFGGGYVYF | HEFVPLEVYTRHEIK |
| HSV-2 HG52 | KTTSSIEFARLQFTY | GHRGYFIFGGGYVYF | HEFVPLEVYTRHEIK |
| Chimp alpha-1 herpesvirus | KTTSSIEFARLQFTY | GHRRYFIFGGGYVYF | HEFVPLEVYTRHEIK |
| SA8 B264 | KTTSSVEFARLQFTYa | GHRRYFTFGAGYVYF | HEFVPLEVYTRQEIK |
| Cercopithecine herpesvirus 1 E2490 | KTTSSVEFARLQFTY | GHRRYFTFGAGYVYF | HEFVPLEVYTRQEIK |
| Pseudorabies virus Ea | -TTGSAEFARLQFTY | NHRRYFKLGGGYVYY | REFLPLEVYTREELA |
| Varicella-zoster virus Varivax | TTTSSVEFAMLQFTY | NHKRYFLFGHHYVYY | REFMPLQVYTRDELR |
| Epstein-Barr virus B95-8 | GTLNNPATVQIQFAY | TSQYYFQSGNEIHVY | IDFASLELYSRDEQR |
| Cytomegalovirus Towne | ESVHNLVYAQLQFTY | PSLKIFIAGNSAYEY | TDFRVLELYSQKELR |
| Human herpesvirus 6 U1102 | KSRHDILYVQLQYLY | PSTKIFLSGNHAHVY | ADFKVLDLYSPDELS |
Boldface amino acids represent changes from the original gB peptide sequence.
FIG. 6.
The gB122 peptide inhibits VP16 translocation to the nucleus and ICP0 expression. (A) VP16 translocation. Virus was allowed to attach to cells at 4°C for 1 h. Various concentrations of gB122 were then added, and 15 min later the cells were transferred to 37°C. One hour later, the cultures were treated with pH 3.0 citrate buffer, rinsed, and refed with fresh medium. At 4 h of citrate treatment, the cells were harvested, nuclear extracts were prepared and electrophoresed, and nuclear VP16 was detected by immunoblotting. (B) ICP0 expression. Cells infected as described for panel A were harvested at 5 h postinfection, and the ICP0 in total cell extracts was detected by immunoblotting with ICP0-specific antiserum. The blot was stripped and reprobed with actin-specific antibody to determine if all lanes were equally loaded.
Residues critical for antiviral activity of gB122.
To identify the residues in gB122 critical for the antiviral activity, a series of altered peptides were synthesized, and their EC50s were determined. The results are summarized in Table 4. Deletion of the first glycine residue [gB122(2-15)] increased the EC50 only about twofold (P < 0.05). The gB122 peptide contains two hydrophobic amino acid blocks that are separated by a 3-residue glycine linker. Deletion of 3 residues from the N-terminal end of gB122 (gB122Δ1-3) resulted in a 2.5-fold increase in the EC50, whereas the removal of 6 residues at the N-terminal end (gB122Δ1-6), which included 2 hydrophobic residues, resulted in a greater reduction in antiviral activity (EC50 > 100 μM). Deletion of 3 residues (hydrophobic residues V, Y, and F) from the C-terminal end of gB122 (gB122Δ13-15) resulted in a sixfold increase in the EC50. The removal of 6 residues at the C terminus (gB122Δ10-15) resulted in the complete loss of antiviral activity. When they are considered together, these results suggest that deletions extending into either hydrophobic region were detrimental to the activity of gB122.
TABLE 4.
Sequence and EC50s of structural variants of peptide gB122 in the comprehensive assay
| Peptide | Sequence | EC50 (μM)a |
|---|---|---|
| gB122 | GHRRYFTFGGGYVYF | 15.0 ± 2.2 |
| gB122(2-15)b | --------------- | 31.2 ± 5.1 |
| gB122R4A | ---A----------- | 26.4 ± 13.5 |
| gB122Y5A | ----A---------- | 119.2 ± 26.6 |
| gB122F6A | -----A--------- | 67.6 ± 8.1 |
| gB122F8A | -------A------- | 37.5 ± 4.2 |
| gB122G10L | ---------L----- | 25.1 ± 4.9 |
| gB122Y14A | -------------A- | 58.0 ± 5.2 |
| gB122F15A | --------------A | 48.6 ± 6.2 |
| gB122Δ1-3 | ------------ | 34.9 ± 1.8 |
| gB122Δ1-6 | --------- | >100 |
| gB122Δ13-15 | ------------ | 91.4 ± 9.8 |
| gB122Δ10-15 | --------- | >100 |
The EC50s are the means ± standard deviations determined from at least two independent experiments. The gB122Δ1-6 and gB122Δ10-15 peptides had no effect on HSV-1 infection at the highest concentration tested (100 μM).
The numbering system refers to the position of the residue by counting the first glycine as 1 and does not refer to the residue in the full length gB-1 protein. The glycine at position 1 in the peptide corresponds to residue 636 in full-length gB-1. Note that except for gB122, a glycine residue at the first position is not present in the peptides.
Replacement of the central residue of the glycine triplet with leucine (gB122G10L) and an alanine substitution for the second arginine (gB122R4A) increased the EC50 (25 μM) about 1.7-fold, suggesting that the glycine and the arginine in this region are not critical for the antiviral activity. Single alanine replacements of most of the amino acids in the two hydrophobic regions (gB122F6A, gB122F8A, gB122Y14A, and gB122F15A) reduced the antiviral activity approximately 2.5- to 5-fold. Of the single-residue substitutions, the greatest decrease in the antiviral activity (eightfold) resulted from replacement of the tyrosine in the first group of hydrophobic residues (gB122Y5A), suggesting that this residue is important for the antiviral activity of gB122. This corresponds to tyrosine 640 (Y640) in the full-length gB-1 protein.
DISCUSSION
Currently, gB-1 is thought to act downstream of gD and in concert with gH-gL as part of the fusion complex; however, little is known about the specific role of gB in the entry process. To date, only a few functionally important residues in gB have been identified (2, 4, 5, 21, 24, 25, 27, 38, 39, 43, 63). The gB-1 ectodomain has been difficult to study by the use of mutagenesis strategies because a high percentage of the mutations severely disrupt the structure and processing of the protein and therefore affect its function (13, 45, 54). The targeting of exposed loop regions for mutagenesis also results in a high percentage of altered proteins (44). The fact that the ectodomain is intolerant of mutation is consistent with the extensive protein-protein contacts revealed in the crystal structure of the gB-1 trimer (36).
In this study, we have described an alternative approach for identifying functionally important regions of gB-1 and other membrane-bound glycoproteins on the basis of the findings of previous studies that showed that peptides that match protein-protein interaction interfaces have inhibitory activity (9, 19, 22, 31, 47, 70, 74). The approach involved the synthesis of a library of overlapping 15-mer peptides that encompassed the entire ectodomain of gB-1 extending from residues 30 to 730. The library of 138 peptides was then screened for antiviral activity. Seven of the 138 peptides tested positive for reducing infection by 50% or more at a presumed concentration of 100 μM in the initial high-throughput screen. When we retested highly purified peptides, three of the seven peptides (gB94, gB122, gB131) had EC50s less than 20 μM in our comprehensive assay and were selected for further studies. These peptides represent novel inhibitors of HSV-1 infection and validate the strategy (peptide scanning inhibition) for the identification of antiviral drug targets. Evidence that the regions of gB-1 corresponding to the active peptides are functionally important comes from the finding that a linker insertion at T690 in the gB region corresponding to gB131 altered the fusogenic activity (45), and a monoclonal antibody that recognizes gB residues 640 to 670 (gB122) neutralized virus and blocked the binding of gB to cells (6). The gB122 peptide also partly overlaps a region of gB and has properties predictive of a membrane-partitioning domain (26). Further studies of the regions of gB-1 corresponding to gB122 and gB131 are clearly warranted.
Dose-response studies showed that peptides gB94, gB122, and gB131 inhibited infection with EC50s of less than 20 μM when the peptides were continuously present over the entire assay period. However, when the peptides were introduced at various times during infection, the three peptides displayed different antiviral activity profiles, suggesting that they possess different inhibitory mechanisms. The gB131 peptide was clearly shown to be an entry blocker, since the EC50s obtained in the comprehensive and entry assays were essentially identical and it was inactive in the other assays. The gB122 peptide primarily blocked viral infection at a postattachment entry step, but it also had other activities, as infection was inhibited when either cells or the virus was exposed to the peptide prior to infection. Although the gB94 peptide efficiently inhibited viral infection in a dose-dependent manner with an EC50 of 6.5 μM in the comprehensive assay, it did not significantly block infection when the peptide was added to either cells or the virus before infection. Moreover, the gB94 peptide showed no antiviral activity in the entry assay. It is possible that gB94 is active intracellularly, which will require additional studies. Currently, it is unclear at which stage in HSV-1 infection gB94 is acting.
Additional studies were focused on the gB122 peptide for several reasons. First, it was the only peptide displaying at least some antiviral activity in all of the assays except the attachment assay. Second, gB122 overlapped a region that had previously been suggested to play a role in the oligomerization of gB, and a linker insertion in this region altered the fusogenic activity (38, 42, 45). The gB122 peptide consists of two 4-residue hydrophobic sequences separated by a 3-residue glycine linker and has high tyrosine and phenylalanine contents. The hydrophobic amino acids may be important for the antiviral activity, since peptides with deletions in these sequences had higher EC50s or lost antiviral activity. With the exception of tyrosine in the fifth position (residue 640 in the gB-1 protein), single alanine substitutions for individual hydrophobic residues raised the EC50s only slightly, suggesting that multiple hydrophobic residues are important for the activity of gB122. The tyrosine at the fifth position in the gB122 peptide is a critical residue for antiviral activity and corresponds to residue 640 (Y640) in the full-length gB-1 protein. Studies to determine if Y640 is important for the function of gB-1 are in progress.
A comparison of the herpesvirus gB protein sequences (Table 3) revealed that the sequences corresponding to gB94 are completely conserved across all HSV-1 and HSV-2 sequences that are available and suggest that gB94 would have broad activity against HSV-1 and -2 strains. The sequences corresponding to gB122 match exactly for the gB-1 sequences that are available but differ by 1 or 2 residues for HSV-2 strains. Thus, gB122 would be predicted to block HSV-1. Further studies will be needed to see if gB122 acts on HSV-2 strains. For gB131, the sequence is highly conserved among the herpes simplex viruses, and this result would be predicted to block HSV-1 and HSV-2 strains. The three corresponding peptide sequences are not highly conserved in pseudorabies and varicella-zoster viruses, but only a single sequence is available for each virus. The corresponding sequences for Epstein-Barr virus, cytomegalovirus Towne, and human herpesvirus 6 are poorly conserved. It is therefore likely that gB94, gB122, and gB131 would most likely be specific for HSVs. Clearly, additional studies of the spectrum of activity of the three peptides are needed.
There are several possible mechanisms by which gB122 and gB131 could be inhibiting HSV-1 entry. These peptides could disrupt the oligomeric structure of gB-1. If this were true, the gB122 and gB131 peptides would be expected to be virucidal. However, our data showed that gB122-treated virions lose their infectivity only at higher concentrations and that gB131 was not virucidal. It is also possible that gB122 or gB131 could either prematurely trigger or inhibit a conformational change in the gB-1 molecule required for entry. Finally, gB122 or gB131 may be acting by blocking a protein-protein interaction. In particular, these peptides might inhibit gB binding to PILRα, gD, or gH-gL. The region of gB corresponding to gB122 is predicted to have a high propensity for interfacial hydrophobicity (26) and could function as a binding site for gD, gH, or a cellular coreceptor such as PICRα (66).
According to the crystal structure (36), the region of gB corresponding to gB122 forms a shallow pocket on the side of the globular head opposite the viral envelope and would be in a position to interact with another protein. Tyrosine 640, which appears to be important for the activity of the gB122 peptide, lies at the center of the shallow pocket where the side chain would be available for binding. Thus, future studies focusing on this region may provide important information on the function of gB. Our observation that preincubation of the cells with gB122 was not inhibitory except at high peptide concentrations leads us to favor the possibility that gB122 is not acting through cellular effects. The gB131 peptide lies near the envelope proximal end of the stalk portion of gB (36), and the crystal structure reveals that residues corresponding to gB131 are in a position to form interactions between the monomers stabilizing the trimer (36). It is not likely that gB131 is acting by destabilizing the trimer, since gB131 was not virucidal; however, the gB131 peptide could be interfering with a conformational change involving interactions between residues in this region or by blocking a binding interaction.
The gB peptides identified by peptide scanning inhibition could be useful in several ways. Further testing and development could lead to their use directly as antiviral agents. The fact that peptides generally enter cells poorly suggests that the peptides would likely be most useful for topical applications, perhaps as a microbicide. Alternatively, or in addition, molecular modeling and drug design efforts focusing on the corresponding regions of gB could lead to the identification of small molecular inhibitors. Finally, these peptides may also be useful as reagents in the screening of small-molecule libraries for inhibitors of the function of gB.
Acknowledgments
We thank Donna Peters, Stacey Schultz-Cherry, Hermann Bultmann, Sharon Altmann, Gilbert Jose, and Jeremy Teuton for comments on the manuscript.
These studies were supported by grants from the NIH (PO1-AI52049), an NEI Core Grant for Vision Research (P30-016665), a Research to Prevent Blindness Senior Scientist award to C.R.B., and an unrestricted grant from Research to Prevent Blindness, Inc., to the Department of Ophthalmology and Visual Sciences.
Footnotes
Published ahead of print on 22 December 2008.
REFERENCES
- 1.Akkarawongsa, R., A. E. Cullinan, A. Zinkel, J. Clarin, and C. R. Brandt. 2006. Corneal toxicity of cell-penetrating peptides that inhibit herpes simplex virus entry. J. Ocul. Pharmacol. Ther. 22:279-289. [DOI] [PubMed] [Google Scholar]
- 2.Ali, M. A. 1990. Oligomerization of herpes simplex virus glycoprotein B occurs in the endoplasmic reticulum and a 102 amino acid cytosolic domain is dispensable for dimer assembly. Virology 178:588-592. [DOI] [PubMed] [Google Scholar]
- 3.Avitabile, E., C. Forghieri, and G. Campadelli-Fiume. 2007. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J. Virol. 81:11532-11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baghian, A., L. Huang, S. Newman, S. Jayachandra, and K. G. Kousoulas. 1993. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J. Virol. 67:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beitia Ortiz de Zarate, I., K. Kaelin, and F. Rozenberg. 2004. Effects of mutations in the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B on intracellular transport and infectivity. J. Virol. 78:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender, F. C., M. Samanta, E. E. Heldwein, M. P. de Leon, E. Bilman, H. Lou, J. C. Whitbeck, R. J. Eisenberg, and G. H. Cohen. 2007. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J. Virol. 81:3827-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender, F. C., J. C. Whitbeck, H. Lou, G. H. Cohen, and R. J. Eisenberg. 2005. Herpes simplex virus glycoprotein B binds to cell surfaces independently of heparan sulfate and blocks virus entry. J. Virol. 79:11588-11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender, F. C., J. C. Whitbeck, M. Ponce de Leon, H. Lou, R. J. Eisenberg, and G. H. Cohen. 2003. Specific association of glycoprotein B with lipid rafts during herpes simplex virus entry. J. Virol. 77:9542-9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandt, C. R., B. Spencer, P. Imesch, M. Garneau, and R. Deziel. 1996. Evaluation of a peptidomimetic ribonucleotide reductase inhibitor with a murine model of herpes simplex virus type 1 ocular disease. Antimicrob. Agents Chemother. 40:1078-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browne, H., B. Bruun, and T. Minson. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419-1422. [DOI] [PubMed] [Google Scholar]
- 11.Bultmann, H., and C. R. Brandt. 2002. Peptides containing membrane-transiting motifs inhibit virus entry. J. Biol. Chem. 277:36018-36023. [DOI] [PubMed] [Google Scholar]
- 12.Bultmann, H., J. S. Busse, and C. R. Brandt. 2001. Modified FGF4 signal peptide inhibits entry of herpes simplex virus type 1. J. Virol. 75:2634-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai, W. Z., S. Person, C. DebRoy, and B. H. Gu. 1988. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1. An analysis of linker insertion mutants. J. Mol. Biol. 201:575-588. [DOI] [PubMed] [Google Scholar]
- 14.Cai, W. Z., S. Person, S. C. Warner, J. H. Zhou, and N. A. DeLuca. 1987. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J. Virol. 61:714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapsal, J. M., and L. Pereira. 1988. Characterization of epitopes on native and denatured forms of herpes simplex virus glycoprotein B. Virology 164:427-434. [DOI] [PubMed] [Google Scholar]
- 16.Cheshenko, N., B. Del Rosario, C. Woda, D. Marcellino, L. M. Satlin, and B. C. Herold. 2003. Herpes simplex virus triggers activation of calcium-signaling pathways. J. Cell Biol. 163:283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claesson-Welsh, L., and P. G. Spear. 1986. Oligomerization of herpes simplex virus glycoprotein B. J. Virol. 60:803-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cocchi, F., D. Fusco, L. Menotti, T. Gianni, R. J. Eisenberg, G. H. Cohen, and G. Campadelli-Fiume. 2004. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc. Natl. Acad. Sci. USA 101:7445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen, E. A., P. Gaudreau, P. Brazeau, and Y. Langelier. 1986. Specific inhibition of herpesvirus ribonucleotide reductase by a nonapeptide derived from the carboxy terminus of subunit 2. Nature 321:441-443. [DOI] [PubMed] [Google Scholar]
- 20.Corey, L., A. Wald, R. Patel, S. L. Sacks, S. K. Tyring, T. Warren, J. M. Douglas, Jr., J. Paavonen, R. A. Morrow, K. R. Beutner, L. S. Stratchounsky, G. Mertz, O. N. Keene, H. A. Watson, D. Tait, and M. Vargas-Cortes. 2004. Once-daily valacyclovir to reduce the risk of transmission of genital herpes. N. Engl. J. Med. 350:11-20. [DOI] [PubMed] [Google Scholar]
- 21.Diakidi-Kosta, A., G. Michailidou, G. Kontogounis, A. Sivropoulou, and M. Arsenakis. 2003. A single amino acid substitution in the cytoplasmic tail of the glycoprotein B of herpes simplex virus 1 affects both syncytium formation and binding to intracellular heparan sulfate. Virus Res. 93:99-108. [DOI] [PubMed] [Google Scholar]
- 22.Dutia, B. M., M. C. Frame, J. H. Subak-Sharpe, W. N. Clark, and H. S. Marsden. 1986. Specific inhibition of herpesvirus ribonucleotide reductase by synthetic peptides. Nature 321:439-441. [DOI] [PubMed] [Google Scholar]
- 23.English, E. P., R. S. Chumanov, S. H. Gellman, and T. Compton. 2006. Rational development of beta-peptide inhibitors of human cytomegalovirus entry. J. Biol. Chem. 281:2661-2667. [DOI] [PubMed] [Google Scholar]
- 24.Foster, T. P., J. M. Melancon, and K. G. Kousoulas. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18-29. [DOI] [PubMed] [Google Scholar]
- 25.Gage, P. J., M. Levine, and J. C. Glorioso. 1993. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J. Virol. 67:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galdiero, S., M. Vitiello, M. D'Isanto, A. Falanga, M. Cantisani, H. Browne, C. Pedone, and M. Galdiero. 2008. The identification and characterization of fusogenic domains in herpes virus glycoprotein B molecules. Chembiochem 9:758-767. [DOI] [PubMed] [Google Scholar]
- 27.Galdiero, S., M. Vitiello, M. D'Isanto, A. Falanga, C. Collins, K. Raieta, C. Pedone, H. Browne, and M. Galdiero. 2006. Analysis of synthetic peptides from heptad-repeat domains of herpes simplex virus type 1 glycoproteins H and B. J. Gen. Virol. 87:1085-1097. [DOI] [PubMed] [Google Scholar]
- 28.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 29.Gianni, T., A. Piccoli, C. Bertucci, and G. Campadelli-Fiume. 2006. Heptad repeat 2 in herpes simplex virus 1 gH interacts with heptad repeat 1 and is critical for virus entry and fusion. J. Virol. 80:2216-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein, D. J., and S. K. Weller. 1988. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZ insertion mutant. J. Virol. 62:196-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottlieb, J., A. I. Marcy, D. M. Coen, and M. D. Challberg. 1990. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J. Virol. 64:5976-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grau, D. R., R. J. Visalli, and C. R. Brandt. 1989. Herpes simplex virus stromal keratitis is not titer-dependent and does not correlate with neurovirulence. Investig. Ophthalmol. Vis. Sci. 30:2474-2480. [PubMed] [Google Scholar]
- 33.Haanes, E. J., C. M. Nelson, C. L. Soule, and J. L. Goodman. 1994. The UL45 gene product is required for herpes simplex virus type 1 glycoprotein B-induced fusion. J. Virol. 68:5825-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haffey, M. L., and P. G. Spear. 1980. Alterations in glycoprotein gB specified by mutants and their partial revertants in herpes simplex virus type 1 and relationship to other mutant phenotypes. J. Virol. 35:114-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hannah, B. P., E. E. Heldwein, F. C. Bender, G. H. Cohen, and R. J. Eisenberg. 2007. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J. Virol. 81:4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 37.Herold, B. C., R. J. Visalli, N. Susmarski, C. R. Brandt, and P. G. Spear. 1994. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulphate and glycoprotein B. J. Gen. Virol. 75(Pt 6):1211-1222. [DOI] [PubMed] [Google Scholar]
- 38.Highlander, S. L., W. F. Goins, S. Person, T. C. Holland, M. Levine, and J. C. Glorioso. 1991. Oligomer formation of the gB glycoprotein of herpes simplex virus type 1. J. Virol. 65:4275-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huff, V., W. Cai, J. C. Glorioso, and M. Levine. 1988. The carboxy-terminal 41 amino acids of herpes simplex virus type 1 glycoprotein B are not essential for production of infectious virus particles. J. Virol. 62:4403-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:27-28, 33-38. [DOI] [PubMed] [Google Scholar]
- 41.Krummenacher, C., V. M. Supekar, J. C. Whitbeck, E. Lazear, S. A. Connolly, R. J. Eisenberg, G. H. Cohen, D. C. Wiley, and A. Carfi. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laquerre, S., D. B. Anderson, R. Argnani, and J. C. Glorioso. 1998. Herpes simplex virus type 1 glycoprotein B requires a cysteine residue at position 633 for folding, processing, and incorporation into mature infectious virus particles. J. Virol. 72:4940-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laquerre, S., R. Argnani, D. B. Anderson, S. Zucchini, R. Manservigi, and J. C. Glorioso. 1998. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J. Virol. 72:6119-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, W., T. J. Minova-Foster, D. D. Norton, and M. I. Muggeridge. 2006. Identification of functional domains in herpes simplex virus 2 glycoprotein B. J. Virol. 80:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, E., and P. G. Spear. 2007. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc. Natl. Acad. Sci. USA 104:13140-13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lingen, M., T. Seck, K. Weise, and D. Falke. 1995. Single amino acid substitutions in the glycoprotein B carboxy terminus influence the fusion from without property of herpes simplex virus type 1. J. Gen. Virol. 76(Pt 7):1843-1849. [DOI] [PubMed] [Google Scholar]
- 47.Liuzzi, M., R. Deziel, N. Moss, P. Beaulieu, A. M. Bonneau, C. Bousquet, J. G. Chafouleas, M. Garneau, J. Jaramillo, R. L. Krogsrud, et al. 1994. A potent peptidomimetic inhibitor of HSV ribonucleotide reductase with antiviral activity in vivo. Nature 372:695-698. [DOI] [PubMed] [Google Scholar]
- 48.Lopper, M., and T. Compton. 2004. Coiled-coil domains in glycoproteins B and H are involved in human cytomegalovirus membrane fusion. J. Virol. 78:8333-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marlin, S. D., S. L. Highlander, T. C. Holland, M. Levine, and J. C. Glorioso. 1986. Antigenic variation (mar mutations) in herpes simplex virus glycoprotein B can induce temperature-dependent alterations in gB processing and virus production. J. Virol. 59:142-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milne, R. S., S. L. Hanna, A. H. Rux, S. H. Willis, G. H. Cohen, and R. J. Eisenberg. 2003. Function of herpes simplex virus type 1 gD mutants with different receptor-binding affinities in virus entry and fusion. J. Virol. 77:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 52.Muggeridge, M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 81:2017-2027. [DOI] [PubMed] [Google Scholar]
- 53.Norais, N., D. Tang, S. Kaur, S. H. Chamberlain, F. R. Masiarz, R. L. Burke, and F. Marcus. 1996. Disulfide bonds of herpes simplex virus type 2 glycoprotein gB. J. Virol. 70:7379-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norton, D. D., D. S. Dwyer, and M. I. Muggeridge. 1998. Use of a neural network secondary structure prediction to define targets for mutagenesis of herpes simplex virus glycoprotein B. Virus Res. 55:37-48. [DOI] [PubMed] [Google Scholar]
- 55.Okazaki, K., and H. Kida. 2004. A synthetic peptide from a heptad repeat region of herpesvirus glycoprotein B inhibits virus replication. J. Gen. Virol. 85:2131-2137. [DOI] [PubMed] [Google Scholar]
- 56.Patel, R. 2004. Antiviral agents for the prevention of the sexual transmission of herpes simplex in discordant couples. Curr. Opin. Infect. Dis. 17:45-48. [DOI] [PubMed] [Google Scholar]
- 57.Pellett, P. E., K. G. Kousoulas, L. Pereira, and B. Roizman. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pereira, L. 1994. Function of glycoprotein B homologues of the family herpesviridae. Infect. Agents Dis. 3:9-28. [PubMed] [Google Scholar]
- 59.Perez, A., Q. X. Li, P. Perez-Romero, G. Delassus, S. R. Lopez, S. Sutter, N. McLaren, and A. O. Fuller. 2005. A new class of receptor for herpes simplex virus has heptad repeat motifs that are common to membrane fusion proteins. J. Virol. 79:7419-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez-Romero, P., and A. O. Fuller. 2005. The C terminus of the B5 receptor for herpes simplex virus contains a functional region important for infection. J. Virol. 79:7431-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 62.Qadri, I., C. Gimeno, D. Navarro, and L. Pereira. 1991. Mutations in conformation-dependent domains of herpes simplex virus 1 glycoprotein B affect the antigenic properties, dimerization, and transport of the molecule. Virology 180:135-152. [DOI] [PubMed] [Google Scholar]
- 63.Rasile, L., K. Ghosh, K. Raviprakash, and H. P. Ghosh. 1993. Effects of deletions in the carboxy-terminal hydrophobic region of herpes simplex virus glycoprotein gB on intracellular transport and membrane anchoring. J. Virol. 67:4856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roche, S., S. Bressanelli, F. A. Rey, and Y. Gaudin. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187-191. [DOI] [PubMed] [Google Scholar]
- 65.Sacks, S. L., P. D. Griffiths, L. Corey, C. Cohen, A. Cunningham, G. M. Dusheiko, S. Self, S. Spruance, L. R. Stanberry, A. Wald, and R. J. Whitley. 2004. HSV-2 transmission. Antivir. Res. 63:S27-S35. [DOI] [PubMed] [Google Scholar]
- 66.Satoh, T., J. Arii, T. Suenaga, J. Wang, A. Kogure, J. Uehori, N. Arase, I. Shiratori, S. Tanaka, Y. Kawaguchi, P. G. Spear, L. L. Lanier, and H. Arase. 2008. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwartz, J. A., E. K. Lium, and S. J. Silverstein. 2001. Herpes simplex virus type 1 entry is inhibited by the cobalt chelate complex CTC-96. J. Virol. 75:4117-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Segel, I. H. 1976. Biochemical calculations, 2nd ed. John Wiley & Sons, Inc., New York, NY.
- 69.Shukla, D., and P. G. Spear. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Investig. 108:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simmen, K. A., A. Newell, M. Robinson, J. S. Mills, G. Canning, R. Handa, K. Parkes, N. Borkakoti, and R. Jupp. 1997. Protein interactions in the herpes simplex virus type 1 VP16-induced complex: VP16 peptide inhibition and mutational analysis of host cell factor requirements. J. Virol. 71:3886-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spear, P. G. 1993. Entry of alphaherpesviruses into cells. Semin. Virol. 4:167-180. [Google Scholar]
- 72.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 73.Subramanian, R. P., and R. J. Geraghty. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. USA 104:2903-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trybala, E., T. Bergstrom, B. Svennerholm, S. Jeansson, J. C. Glorioso, and S. Olofsson. 1994. Localization of a functional site on herpes simplex virus type 1 glycoprotein C involved in binding to cell surface heparan sulphate. J. Gen. Virol. 75(Pt 4):743-752. [DOI] [PubMed] [Google Scholar]
- 75.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Visalli, R. J., and C. R. Brandt. 1993. The HSV-1 UL45 18 kDa gene product is a true late protein and a component of the virion. Virus Res. 29:167-178. [DOI] [PubMed] [Google Scholar]
- 77.Walev, I., M. Lingen, M. Lazzaro, K. Weise, and D. Falke. 1994. Cyclosporin A resistance of herpes simplex virus-induced “fusion from within” as a phenotypical marker of mutations in the Syn 3 locus of the glycoprotein B gene. Virus Genes 8:83-86. [DOI] [PubMed] [Google Scholar]
- 78.Whitley, R. J. 1996. Herpes simplex viruses, p. 2297-2342. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, vol. 2, 3rd ed. Lippincott-Raven, Philadelphia, PA. [Google Scholar]