Abstract
Natural products are leads for new antibiotics as a result of their structural complexity and diversity. We have isolated a series of structurally related polyketide-derived natural products from Streptomyces venezuelae ISP5230. The most active of these jadomycin analogues showed good activity against a variety of staphylococci, including methicillin-resistant Staphylococcus aureus.
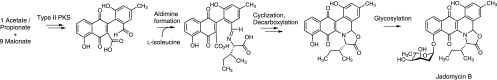
Natural products, or natural-product derivatives, comprise the majority of clinically utilized therapeutics (11). A significant proportion of these are polyketides produced by plants, bacteria, and fungi by a type II polyketide synthase enzyme complex (4). Access to structural variants of polyketide synthase (PKS)-derived natural products may expedite the discovery of improved therapeutics. However, despite significant advances in understanding the genetics and mechanism of action of nature's biosynthetic machinery, significant effort is still required to generate relevant structural diversity through combinatorial biosynthesis (9, 10, 16, 17, 19). Under specific nutrient and stress conditions, the soil microbe Streptomyces venezuelae ISP5230 produces jadomycin B (3), a type II PKS-derived natural product. The skeleton of this glycosylated natural product is unique, as it contains a five-membered oxazolone ring (Fig. 1). The formation of the oxazolone ring arises through the reaction of the amino acid present in the culture medium with a biosynthetic aldehyde precursor (14, 15). The pathway generates a reactive aldimine that undergoes a series of transformations to furnish the cyclized product (14), as outlined in Fig. 1. We have explored the utility of this unique biosynthetic reaction to produce jadomycin B derivatives where the amino acid within the minimal medium used for secondary metabolite production is incorporated into the oxazolone ring (5, 6). We have recently reported the isolation of a series of jadomycin B analogues using aromatic, polar, and aliphatic α-l-amino acids and α-d-amino acids (2). We have shown that there is no racemization of the amino acid incorporated into the natural product, providing supporting evidence that the amino acid incorporation into the jadomycin scaffold occurs through a nonenzymatic process. Herein we present data regarding the effect of several isolated jadomycin B analogues on a series of pathogenic microorganisms.
FIG. 1.
Jadomycin B biosynthetic precursors.
All microbial strains were obtained from the American Type Culture Collection, except for Staphylococcus epidermidis C621 and methicillin-resistant Staphylococcus aureus (MRSA) C623, which were originally isolated by A. Chow at the University of British Columbia. Preliminary MICs were determined as described previously (12) by measuring culture growth using the broth microdilution method of Amsterdam (1) as modified by Wu and Hancock (18). Stock solutions of the jadomycins were made by dissolving in 100% dimethyl sulfoxide (DMSO) and diluted to give a concentration of 2.56 mg ml−1 of antibiotic and 5% DMSO. Fifty-microliter aliquots of a diluted working stock in the first well, having a final concentration of 0.5% DMSO, were serially diluted twofold with broth in 96-well polypropylene microtiter plates (Costar; Corning Incorporated, Corning, NY). A control of the same concentration of DMSO was included without test compounds in the plate. The bacterial strains were grown overnight to the mid-logarithmic phase in Mueller-Hinton (MH) broth and diluted 100-fold into fresh broth. After a 4-h incubation, the cells were diluted 10−6 into fresh broth. An aliquot (50 μl) of a given indicator bacterial strain was added to each well, and the plate was incubated overnight at the appropriate temperature. Inhibition was defined as growth less than or equal to one-half of the growth observed in the control wells to which no jadomycin analogue was added. However, for all organisms, complete inhibition (no growth) was achieved at the lowest inhibitory concentration. Growth was assessed visually. Three replicates were performed for each indicator bacterial strain, and no replicates differed by more than 1 dilution. Subsequently, the MICs were determined by the broth microdilution method for selected bacterial strains using the Clinical and Laboratory Standards Institute (CLSI) method (8). The cationic adjusted MH broth used provided a final concentration of 5 × 105 CFU ml−1 or 5 × 104 CFU/well of the test bacterial strain. The jadomycin analogue and vancomycin working stock solutions were made by diluting them in the cationic adjusted MH broth to 256 μg ml−1, as per the protocol. The jadomycins were isolated as previously reported (2) using our improved isolation methodology (7).
The inhibitory activities of the jadomycins evaluated by two methods are listed in Tables 1 and 2, while the jadomycins used in this study are shown in Fig. 2. In our initial screen for antimicrobial activity, the jadomycins were evaluated against several S. aureus, S. epidermidis, Pseudomonas aeruginosa, Enterococcus faecalis, and Bacillus subtilis strains. Greater activity was observed against gram-positive microorganisms than gram-negative ones. The erythromycin control had comparable or better activity against many of the cultures except MRSA. The compounds were then evaluated in more detail against three of the more active strains, C623 (MRSA), S. aureus 305, and S. epidermidis C960, using reference methods from the CLSI. Vancomycin was added as a control since it is used clinically against MRSA infections. The CLSI reference method differs from our initial screening in that higher CFU are used in the cultures to determine the MICs; consequently, the MICs obtained were two or three times greater than those obtained in our initial screen. Nevertheless, the same structure activity trends were observed between the jadomycins and across the different strains.
TABLE 1.
Screening jadomycins against several gram-positive and gram-negative bacterial strains
| Jadomycin analogue | MIC (μg ml−1) for indicated straina
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus C622 (ATCC 25923) | S. aureus C623 (MRSA) | S. aureus 305 | S. aureus Becker CP8 (ATCC 49525) | S. aureus Becker Lyc12 CP336 (ATCC 55804) | S. epidermidis C960 (ATCC 14990) | S. epidermidis C621 (clinical isolate) | P. aeruginosa H187 | P. aeruginosa H188 | E. faecalis C625 (ATCC 29212) | B. subtilis C971 (ATCC 6633) | |
| B | 2 | <1 | <1 | 2 | 4 | <1 | <1 | >128 | >128 | 32 | 4 |
| L | <1 | <1 | 8 | 8 | 16 | <1 | 4 | >128 | >128 | >128 | 16 |
| F | 1 | 1 | 4 | 8 | 16 | <1 | 2 | >128 | >128 | >128 | 16 |
| Y | 8 | 8 | 4 | 4 | 16 | <1 | 2 | >128 | >128 | >128 | 8 |
| DM | 2 | 4 | 32 | 16 | 64 | <1 | 2 | >128 | >128 | >128 | 16 |
| T | 8 | 8 | 4 | 4 | 16 | <1 | 2 | >128 | >128 | >128 | 8 |
| DT | 16 | 16 | 32 | 32 | 64 | 2 | 8 | >128 | >128 | >128 | 64 |
| S | n/t | n/t | 8 | 8 | 32 | 1 | 4 | >128 | >128 | >128 | 16 |
| DS | 8 | 8 | 4 | 8 | 16 | 1 | 4 | >128 | >128 | >128 | 16 |
| G | 64 | 32 | 64 | 32 | 64 | 2 | 8 | >128 | >128 | >128 | 64 |
| N | 32 | 16 | 8 | 16 | 32 | <1 | 8 | >128 | >128 | >128 | 32 |
| Erythromycin | <1 | >128 | <0.25 | <0.25 | <0.25 | <0.125 | <0.125 | >2 | >2 | 1 | <0.125 |
n/t, not tested.
TABLE 2.
MICs of jadomycin B analogues evaluated using CLSI reference methods
| Jadomycin analogue | MIC (μg ml−1) for indicated strain
|
||
|---|---|---|---|
| S. aureus C623 (MRSA) | S. aureus 305 | S. epidermidis C960 (ATCC 14990) | |
| B | 8 | 4 | 0.5 |
| L | 8 | 8 | 0.5 |
| F | 8 | 4 | 0.5 |
| DM | 16 | 32 | 2 |
| T | 32 | 64 | 1 |
| DT | 32 | 32 | 4 |
| S | 32 | 32 | 0.5 |
| DS | 16 | 32 | 2 |
| G | >128 | >128 | 32 |
| N | 128 | >128 | 16 |
| Vancomycin | 0.5 | 0.5 | 1 |
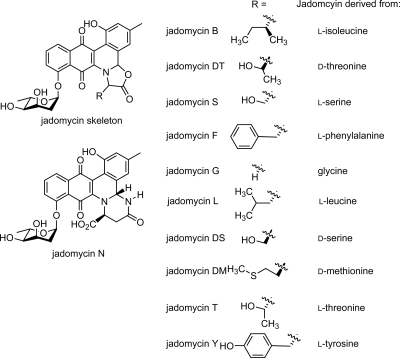
FIG. 2.
Structures of jadomycin B analogues used in this study.
The least active jadomycins were jadomycin G and jadomycin N against all strains evaluated. These two jadomycins are structurally distinct from the other natural products evaluated in this study, since jadomycin G (2) does not contain a substituent extending from the oxazolone ring, while for jadomycin N, the oxazolone ring is replaced by a unique six-membered ring containing two nitrogen atoms (2), presumably formed by the cyclization of the primary amide side chain of l-asparagine rather than by ring closure through the carboxylate functionality, as is usual for other jadomycin B analogues. The most active compounds against C623 (a clinical MRSA strain) were jadomycins B, L, and F. In terms of structure, two of these analogues have aliphatic side chains and one has a phenyl ring extending from the oxazolone ring. Due to a lack of material, jadomycin Y, the derivative with a hydroxyphenyl side chain, was not evaluated using the CLSI protocol; however, from the initial data, it was observed to be less effective as an antibiotic than jadomycin F, indicating that the additional hydroxyl functionality present in jadomycin Y was detrimental to the antimicrobial activity. Jadomycins B, L, and F were more active by a factor of 2 or 3 than the remaining analogues derived from serine, threonine, or methionine when evaluated against C623. The isolated jadomycins from the incorporation of l- and d-threonine and l- and d-serine provided an opportunity to observe the effects of altering the stereochemistry of the amino acid in the oxazolone ring upon the antibacterial activity. We have previously demonstrated that the chiral center in each amino acid is not inverted or racemized during the biosynthesis of the jadomycin analogues (2). The MIC data obtained using the CLSI method against the two S. aureus strains differed by only a factor of 2 between jadomycins S and DS and jadomycins T and DT, likely indicating that there was no significant difference due to the change in stereochemistry. Against S. epidermidis C960, jadomycins S and T were more active than their diastereomeric counterparts by a factor of 4.
This study represents the first investigation into the antimicrobial effects of a series of jadomycins. It demonstrates that generating structural analogues of PKS-derived natural products provides a significant range of biological activity. Analysis of our data indicates that the structure and substitution around the oxazolone ring confer differing antimicrobial activities on the jadomycin analogues and provides further impetus to explore the unique biosynthetic machinery involved in jadomycin production, a trend consistent with our initial study investigating the anticancer effects of a series of jadomycin analogues (2). However, in our initial evaluation of several jadomycin analogues against a human breast cancer and a melanoma cell line (T-47D and MDA-MB-435, respectively [13]), we observed a different rank order of structure activity to that observed in the antimicrobial activities. In the study using the breast cancer cell line, it was observed that (i) the more active compounds were those with hydroxylmethylene side chains, (ii) the least active compounds were those with aromatic side chains, and (iii) the stereochemistry at the α-carbon atom of the amino acid did not significantly alter anticancer activity. In the antimicrobial study reported herein, jadomycins B and L, both compounds containing aliphatic side chains extending from the oxazolone ring α-carbon, and then jadomycin F, an analogue with a benzyl group connected to the oxazolone α-carbon, were observed to have the most significant activity against MRSA (see Fig. 2 for structures). These contrasting rank orders of biological activity are most likely due to the different susceptibilities of the cell lines and bacteria toward the compounds, potentially due to different mechanisms of action or different affinities for their biological target(s). Studies elucidating the mode of action, cellular toxicity, and specificity of the jadomycins are currently under way.
Acknowledgments
This work was supported by the Atlantic chapter of the Canadian Breast Cancer Foundation, Dalhousie University Pharmacy Endowment Fund, the Canadian Institutes of Health Research, Rx&D, and the National Research Council of Canada Institute for Marine Biosciences.
Footnotes
Published ahead of print on 15 December 2008.
REFERENCES
- 1.Amsterdam, D. 1996. Susceptibility testing for antimicrobials in liquid media, p. 52-111. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, MD.
- 2.Borissow, C. N., C. L. Graham, R. T. Syvitski, T. R. Reid, J. Blay, and D. L. Jakeman. 2007. Stereochemical integrity of oxazolone ring-containing jadomycins. Chembiochem 8:1198-1203. [DOI] [PubMed] [Google Scholar]
- 3.Doull, J. L., S. W. Ayer, A. K. Singh, and P. Thibault. 1993. Production of a novel polyketide antibiotic, jadomycin B, by Streptomyces venezuelae following heat shock. J. Antibiot. 46:869-871. [DOI] [PubMed] [Google Scholar]
- 4.Hertweck, C., A. Luzhetskyy, Y. Rebets, and A. Bechthold. 2007. Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 24:162-190. [DOI] [PubMed] [Google Scholar]
- 5.Jakeman, D. L., S. Farrell, W. Young, R. J. Doucet, and S. C. Timmons. 2005. Novel jadomycins: incorporation of non-natural and natural amino acids. Bioorg. Med. Chem. Lett. 15:1447-1449. [DOI] [PubMed] [Google Scholar]
- 6.Jakeman, D. L., C. L. Graham, and T. R. Reid. 2005. Novel and expanded jadomycins incorporating non-proteogenic amino acids. Bioorg. Med. Chem. Lett. 15:5280-5283. [DOI] [PubMed] [Google Scholar]
- 7.Jakeman, D. L., C. L. Graham, W. Young, and L. C. Vining. 2006. Culture conditions improving the production of jadomycin B. J. Ind. Microbiol. Biotechnol. 33:767-772. [DOI] [PubMed] [Google Scholar]
- 8.Jorgensen, J. H., and J. D. Turnidge. 2003. Susceptibility test methods: dilution and disk diffusion methods, p. 1108-1127. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, DC.
- 9.Kittendorf, J. D., and D. H. Sherman. 2006. Developing tools for engineering hybrid polyketide synthetic pathways. Curr. Opin. Biotechnol. 17:597-605. [DOI] [PubMed] [Google Scholar]
- 10.Menzella, H. G., and C. D. Reeves. 2007. Combinatorial biosynthesis for drug development. Curr. Opin. Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 11.Newman, D. J., and G. M. Cragg. 2007. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70:461-477. [DOI] [PubMed] [Google Scholar]
- 12.Patrzykat, A., J. W. Gallant, J. K. Seo, J. Pytyck, and S. E. Douglas. 2003. Novel antimicrobial peptides derived from flatfish genes. Antimicrob. Agents Chemother. 47:2464-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rae, J. M., C. J. Creighton, J. M. Meck, B. R. Haddad, and M. D. Johnson. 2007. MDA-MB-435 cells are derived from M14 melanoma cells—a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res. Treat. 104:13-19. [DOI] [PubMed] [Google Scholar]
- 14.Rix, U., C. C. Wang, Y. H. Chen, F. M. Lipata, L. L. R. Rix, L. M. Greenwell, L. C. Vining, K. Q. Yang, and J. Rohr. 2005. The oxidative ring cleavage in jadomycin biosynthesis: a multistep oxygenation cascade in a biosynthetic black box. Chembiochem 6:838-845. [DOI] [PubMed] [Google Scholar]
- 15.Rix, U., J. Zheng, L. L. Remsing Rix, L. Greenwell, K. Yang, and J. Rohr. 2004. The dynamic structure of jadomycin B and the amino acid incorporation step of its biosynthesis. J. Am. Chem. Soc. 126:4496-4497. [DOI] [PubMed] [Google Scholar]
- 16.Salas, J. A., and C. Mendez. 2007. Engineering the glycosylation of natural products in actinomycetes. Trends Microbiol. 15:219-232. [DOI] [PubMed] [Google Scholar]
- 17.Weissman, K. J., and P. F. Leadlay. 2005. Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Microbiol. 3:925-936. [DOI] [PubMed] [Google Scholar]
- 18.Wu, M., and R. E. Hancock. 1999. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J. Biol. Chem. 274:29-35. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, M. Q., S. Gaisser, M. Nur-E-Alam, L. S. Sheehan, W. A. Vousden, N. Gaitatzis, G. Peck, N. J. Coates, S. J. Moss, M. Radzom, T. A. Foster, R. M. Sheridan, M. A. Gregory, S. M. Roe, C. Prodromou, L. Pearl, S. M. Boyd, B. Wilkinson, and C. J. Martin. 2008. Optimizing natural products by biosynthetic engineering: discovery of nonquinone Hsp90 inhibitors. J. Med. Chem. 51:5494-5497. [DOI] [PubMed] [Google Scholar]