Abstract
TEM- and SHV-type extended-spectrum β-lactamases (ESBLs) are the most common ESBLs found in the United States and are prevalent throughout the world. Amino acid substitutions at a number of positions in TEM-1 lead to the ESBL phenotype, although substitutions at residues 104 (E to K), 164 (R to S or H), 238 (G to S), and 240 (E to K) appear to be particularly important in modifying the spectrum of activity of the enzyme. The SHV-1-derived ESBLs are a less diverse collection of enzymes; however, the majority of amino acid substitutions resulting in an ESBL mirror those seen in the TEM-1-derived enzymes. Pyrosequencing by use of the single-nucleotide polymorphism (SNP) protocol was applied to provide sequence data at positions critical for the ESBL phenotype spanning the blaTEM and blaSHV genes. Three novel β-lactamases are described: the ESBLs TEM-155 (Q39K, R164S, E240K) and SHV-105 (I8F, R43S, G156D, G238S, E240K) and a non-ESBL, SHV-48 (V119I). The ceftazidime, ceftriaxone, and aztreonam MICs for an Escherichia coli isolate expressing blaSHV-105 were >128, 128, and >128 μg/ml, respectively. Likewise, the ceftazidime, ceftriaxone, and aztreonam MICs for an E. coli isolate expressing blaTEM-155 were >128, 64, and > 128 μg/ml, respectively. Pyrosequence analysis determined the true identity of the β-lactamase on plasmid R1010 to be SHV-11 rather than SHV-1, as previously reported. Pyrosequencing is a real-time sequencing-by-synthesis approach that was applied to SNP detection for TEM- and SHV-type ESBL identification and represents a robust tool for rapid sequence determination that may have a place in the clinical setting.
The age of penicillin quickly saw the emergence of penicillin resistance in gram-negative pathogens through the expression of hydrolytic enzymes capable of inactivating β-lactams (penicillin, amoxicillin, cefazolin) (4, 40). Two important classes of these resistance determinants are the TEM-type and SHV-type β-lactamases, which were first described in the 1960s (14, 34, 51). With the advent of the oxyimino-cephalosporins (e.g., ceftazidime and cefotaxime) and aztreonam in the 1980s, extended-spectrum β-lactamases (ESBLs) emerged with the first SHV-derived enzyme, SHV-2, which appeared in Germany (24, 25), and the first TEM-derived enzyme, TEM-3, which appeared in France (53). Interestingly, it has been reported that the TEM-12 ESBL may actually have appeared several years earlier than TEM-3 (40). By using the classification scheme of Bush et al. (9), ESBLs are placed in functional group 2be and are defined as β-lactamases that are capable of hydrolyzing oxyimino-cephalosporins (in addition to earlier cephalosporins) and aztreonam and that are inhibited by clavulanic acid and other β-lactamase inhibitors (4, 9, 40). Although CTX-M-type ESBLs are currently the most prevalent extended-spectrum enzymes reported worldwide (10, 30), TEM-type and SHV-type enzymes are still the most prevalent ESBLs in the United States, and worldwide reports of novel TEM-type and SHV-type enzymes are commonplace (29, 35, 44, 49, 58). A further concern is the recent report of complex mutant TEM enzymes that maintain an extended spectrum, in addition to reduced susceptibility to β-lactamase inhibitors (11, 49).
TEM-1, which is usually encoded on a plasmid, is now present in a number of members of the family Enterobacteriaceae (Klebsiella pneumoniae, Enterobacter aerogenes, Morganella morganii, Proteus mirabilis, Salmonella spp.) and has spread into additional gram-negative pathogens, including Pseudomonas aeruginosa, Haemophilus influenzae, and Neisseria gonorrhoeae (4). Amino acid substitutions at a number of positions in TEM-1 can lead to the ESBL phenotype, although substitutions at residues 104 (glutamate to lysine), 164 (arginine to serine or histidine), 238 (glycine to serine), and 240 (glutamate to lysine) appear to be particularly important in modifying the spectrum of activity of the enzyme (16, 20, 39, 43). These residues are located in or around the β-lactam binding site, and substitutions result in a remodeling of the active-site cavity, permitting the binding of the bulky expanded-spectrum cephalosporins (20). At least 160 TEM derivatives have been reported, although not all have an expanded spectrum of activity (http://www.lahey.org/Studies/).
SHV-type ESBLs, having evolved from the SHV-1 enzyme common to K. pneumoniae, are found on both the chromosome and plasmids and have migrated into Citrobacter spp., Escherichia coli, and P. aeruginosa, among others (4). The SHV-1-derived ESBLs are a less diverse collection of enzymes than the TEM family; the majority of SHV-1-derived ESBLs carry the substitution of serine for glycine at position 238. In addition, substitutions at residues 156 (glycine to aspartic acid) and 179 (aspartic acid to glycine, asparagine, or alanine) can lead to the ESBL phenotype, and the substitution at residue 238 is often paired with a second mutation at position 240, lysine for glutamic acid, in SHV-type ESBLs (16, 21, 39). More than 100 SHV derivatives have been reported; however, not all of the SHV derivatives described are ESBLs (http://www.lahey.org/Studies/).
Pyrosequencing is a methodology that combines standard PCR and sequencing by synthesis to rapidly determine the sequence of an intended target in as little as 3 h (15). This methodology has previously been applied to the detection of bacterial resistance genes (19, 36, 52, 57); most recently, this technology was used to define CTX-M-type ESBLs (37, 38). The present analysis utilized the single-nucleotide polymorphism (SNP) protocol for pyrosequence analysis of short stretches of sequence spanning the blaTEM and blaSHV genes and focused on the codons for residues critical to the ESBL phenotype in TEM-type and SHV-type enzymes. A distinct benefit of this approach is that the SNP protocol allows the deconvolution of mixed sequences in bacteria encoding multiple copies of β-lactamase genes, such as a non-ESBL (SHV-1) and an ESBL (SHV-12).
(This study was presented in part previously at the 46th Interscience Conference on Antimicrobial Agents and Chemotherapy [23].)
MATERIALS AND METHODS
Bacterial strains.
Bacterial isolates with well-characterized blaTEM or blaSHV derivatives were used as controls to optimize the study design and reagents. The isolates, along with the encoded TEM- or SHV-type β-lactamase, and the relevant amino acid substitutions that result in the ESBL phenotype are indicated in Table 1. All the isolates were from the Wyeth Research (Pearl River, NY) Strain Collection, including several clinical isolates that were obtained during recent clinical trials.
TABLE 1.
Bacterial strains used in study
| Strain | Organism | blaTEM or blaSHV derivative | Amino acid substitution(s)a | Source or reference |
|---|---|---|---|---|
| GC 1472 (R6K) | E. coli | TEM-1 | NAb | Wyeth Strain Collection |
| GC 1494 (CF102) | E. coli | TEM-3 | Q-39-K, E-104-K, G-238-S | Wyeth Strain Collection |
| GC 1500 | E. coli | TEM-4 | L-21-F, E-104K, G-238-S, T-265-M | Wyeth Strain Collection |
| GC 1505 | E. coli | TEM-7 | Q-39-K, R-164-S | Wyeth Strain Collection |
| GC 1511 (SC15042) | E. coli | TEM-10 | R-164-S, E-240-K | Wyeth Strain Collection |
| GC 2336 | E. coli | TEM-28 | R-164-H, E-240-K | 6 |
| GAR 4649 | K. pneumoniae | TEM-29 | R-164-H | Wyeth Clinical Studies |
| GC 2400 (BA156) | E. coli | TEM-43 | E-240-K, R-164-H, M-182-T | 60 |
| GAR 2830 | E. coli | TEM-1 | NA | Wyeth Clinical Studies |
| GAR 3593 | P. mirabilis | TEM-1/TEM-2 | Q-39-K | Wyeth Clinical Studies |
| GAR 10941 | P. mirabilis | TEM-155 | Q-39-K, R-164-S, E-240-K | Wyeth Clinical Studies |
| GAR 10711 | K. pneumoniae | SHV-1 | NA | Wyeth Clinical Studies |
| GAR 11829 | K. pneumoniae | SHV-2 | G-238-S | Wyeth Clinical Studies |
| GC 2871 (PAB-C16) | E. coli | SHV-4 | R-205-L, G-238-S, E-240-K | 7 |
| GAR 9191 | K. pneumoniae | SHV-5 | G-238-S, E-240-K | Wyeth Clinical Studies |
| GC 2904 (PAB-C41) | E. coli | SHV-6 | D-179-A | 7 |
| GC 2337 | E. coli | SHV-7 | I-8-F, R-43-S, G-238-S, E-240-K | 8 |
| GC 5901 | E. coli | SHV-8 | D-179-N | 47 (gift of F. Tenover) |
| GAR 5951 | K. pneumoniae | SHV-11 | L-35-Q | Wyeth Clinical Studies |
| GC 1709 (R1010)c | E. coli | SHV-11 | L-35-Q | 50 (Wyeth Strain Collection) |
| GC 6326 (KL-1) | E. coli | SHV-12 | L-35-Q, G-238-S, E-240-K | Wyeth Strain Collection (gift of H. Hachler) |
| GAR 12767 | K. pneumoniae | SHV-27 | G-156-D | Wyeth Clinical Studies |
| GC 7991 (A1220) | K. pneumoniae | SHV-1, SHV-5, SHV-105 | I-8-F, R-43-S, G-156-D, G-238-S, E-240-K | Wyeth Strain Collection (gift of L. Saiman) |
| GC 7644 (KP1691) | K. pneumoniae | SHV-1, SHV-5 | G-238-S, E-240-K | Wyeth Strain Collection (gift of C. Urban) |
| GAR 2332 | K. pneumoniae | SHV-11, SHV-12 | L-35-Q, G-238-S, E-240-K | Wyeth Clinical Studies |
| GAR 11218 | K. pneumoniae | SHV-1, SHV-5, SHV-11, SHV-12 | L-35-Q, G-238-S, E-240-K | Wyeth Clinical Studies |
| GAR 8110 | K. pneumoniae | SHV-1, SHV-5, SHV-45 | G-156-D, G-238-S, E-240-K | Wyeth Clinical Studies |
| ATCC 25922 | E. coli | None | NA | www.atcc.org |
| GC 8130d | E. coli DH5α | SHV-1 | NA | This study |
| GC 7668d | E. coli DH5α | SHV-48 | I-119-V | This study |
| GC 8128d | E. coli DH5α | SHV-105 | I-8-F, R-43-S, G-156-D, G-238-S, E-240-K | This study |
| GC 8133d | E. coli DH5α | TEM-155 | Q-39-K, R-164-S, E-240-K | This study |
Amino acid numbering according to Ambler et al. (1).
NA, not applicable.
Strain originally reported to carry SHV-1.
Respective β-lactamase genes shotgun cloned into pCLL2300.
Antibiotics.
Standard powders of all drugs were used. Piperacillin and tazobactam powder were obtained from Wyeth Research. Cefepime, ceftriaxone, and imipenem were obtained from the U.S. Pharmacopeia, Rockville, MD. Amoxicillin, ampicillin, cefoxitin, cefotaxime, and ceftazidime were obtained from Sigma-Aldrich, St. Louis, MO. Clavulanic acid was obtained from DSM Anti-Infectives, Heerlen, The Netherlands; and aztreonam was obtained from the Squibb Institute for Medical Research, Princeton, NJ.
Susceptibility testing.
The in vitro activities of the antibiotics were determined according to standard practice and the recommendations of the CLSI (12). Microtiter plates containing serial dilutions of each antimicrobial agent were inoculated with each organism to yield the appropriate density (105 CFU/ml) in a 100-μl final volume. The plates were incubated for 18 to 22 h at 35°C in ambient air. For all isolates, the MIC was defined as the lowest concentration of antimicrobial agent that completely inhibited the growth of the organism, as detected by the unaided eye. To identify strains that expressed ESBLs, isolates with ceftazidime MICs of ≥2 μg/ml were subject to a confirmatory test by using the Etest ESBL test strips (AB Biodisk, Solna, Sweden) with both ceftazidime and cefotaxime with and without clavulanic acid, according to the manufacturer's instructions.
Pyrosequencing.
The primers used for both PCR and sequencing analysis of SNPs at defined positions in the blaTEM and blaSHV genes of interest are presented in Tables 2 and 3, respectively. DNA was prepared by using a DNeasy tissue kit, according to the manufacturer's instructions (Qiagen Sciences, Valencia, CA). Pyrosequencing was carried out according to the manufacturer's instructions (Biotage AB, Uppsala, Sweden). PCR primers were designed to amplify the blaTEM and blaSHV genes in two pieces each (5′ half, 3′ half), such that the fragments were approximately 500 bp in length, which is optimal for pyrosequencing analysis (Tables 2 and 3). To permit SNP analysis of both the coding and the noncoding strands, two sets of primers were designed for each amplification, with one of the primers from each set biotinylated at the 5′ end to permit purification of the single-stranded templates through streptavidin affinity. Pyrosequencing reactions were carried out in 96-well plates. Strand separation and purification of the single-stranded template were facilitated by the use of the Vacuum Prep Tool (Biotage AB). The purified single-stranded template was then added to the sequencing mixture containing annealing buffer and sequencing primers (0.3 to 0.5 μM), incubated at 90°C for 2 min, and allowed to cool to room temperature. Pyrosequencing was carried out in a PyroMark ID instrument (Biotage AB), and the data were analyzed by using sequence analysis software from the manufacturer.
TABLE 2.
Amplification and pyrosequencing primers for the blaTEM gene used in the study
| Primer | Position of sequence start or size of amplicon | Sequence (5′-3′)a | Comment |
|---|---|---|---|
| TEM_F1_bio (TEM_F3) | 518 bp | 5′-AGACAATAACCCTGGTAAATGC | PCR primer forward set, 5′ half of the gene, 5′ biotin labeled on forward primer (TEM_F1_bio) |
| TEM_R1 (TEM_R3_bio) | 5′-CAAGGCGAGTTACATGATCC | PCR primer reverse set, 5′ biotin on reverse primer (TEM_R3_bio) | |
| TEM_F2_bio (TEM_F4) | 460 bp | 5′-ACTTACTTCTGACAACGATCGG | PCR primer forward set, 3′ half of gene, 5′ biotin labeled on forward primer (TEM_F2_bio) |
| TEM_R2 (TEM_R4_bio) | 5′-TTACCAATGCTTAATCAGTGAGG | PCR primer reverse set, 5′ biotin on reverse primer (TEM_R4_bio) | |
| TEM_dwn_280 | 828, noncoding | 5′-CAGTGAGGCACCTATCTC | Sequencing primer for use with forward set PCR products |
| TEM_dwn_165n | 492, noncoding | 5′-GGCTTCATTCAGCTCCGG | Sequencing primer for use with forward set PCR products |
| TEM_dwn_104 | 310, noncoding | 5′-GATGCTTTTCTGTGACTGGTG | Sequencing primer for use with forward set PCR products |
| TEM_up_265n | 773, coding | 5′-CGTATCGTAGTTATCTACACG | Sequencing primer for use with reverse set PCR products |
| TEM_up_238 | 705, coding | 5′-TTGCTGATAAATCTGGAGCC | Sequencing primer for use with reverse set PCR products |
| TEM_up_182n | 537, coding | 5′-AACGACGAGCGTGACACCACG | Sequencing primer for use with reverse set PCR products |
| TEM_up_69 | 194, coding | 5′-TCGCCCCGAAGAACGTTTTCC | Sequencing primer for use with reverse set PCR products |
| TEM_up_39 | 106, coding | 5′-TGAAAGTAAAAGATGCTGAAG | Sequencing primer for use with reverse set PCR products |
| TEM_up_21 | 55, coding | 5′-TCCCTTTTTTGCGGCATTTTGC | Sequencing primer for use with reverse set PCR products |
The GenBank accession number for the blaTEM-1 sequence is AY394610.
TABLE 3.
Amplification and pyrosequencing primers for the blaSHV gene used in the study
| Primer | Position of sequence start or size of amplicon | Sequence (5′-3′)a | Comment |
|---|---|---|---|
| SHV_F1_bio (SHV_F3) | 486 bp | 5′TTTACTCGCCTTTATCGGC | PCR primer forward set, 5′ half of gene, 5′ biotin labeled on forward primer (SHV_F1_bio) |
| SHV_R1 (SHV_R3_bio) | 5′AAAGGCAGTCAATCCTGC | PCR primer reverse set, 5′ biotin on reverse primer (SHV-R3_bio) | |
| SHV_F2_bio (SHV_F4) | 402 bp | 5′CCATTACCATGAGCGATAAC | PCR primer forward set, 3′ half of gene, 5′ biotin labeled on forward primer (SHV_F2_bio) |
| SHV_R2 (SHV_R4_bio) | 5′ACCACAATGCGCTCTGC | PCR primer reverse set, 5′ biotin on reverse primer (SHV_R4_bio) | |
| SHV_dwn_179 | 527, noncoding | 5′ATGCTGGCCGGGGTAGTG | Sequencing primer for use with forward set PCR products |
| SHV_dwn_119 | 352, noncoding | 5′TAATGGCGGCGGCGCAGAG | Sequencing primer for use with forward set PCR products |
| SHV_dwn_43 | 117, noncoding | 5′ATCCATTTCTATCATGCCTAC | Sequencing primer for use with forward set PCR products |
| SHV_up_238n1 | 699, coding | 5′CGCCGATAAGACCGGAGC | Sequencing primer for use with reverse set PCR products |
| SHV_up_205n | 597, coding | 5′CGTCTGAGCGCCCGTTCG | Sequencing primer for use with reverse set PCR products |
| SHV_up_156n | 451, coding | 5′GACTGCCTTTTTGCGCCAG | Sequencing primer for use with reverse set PCR products |
| SHV_up_35 | 84, coding | 5′CCGCAGCCGCTTGAGC | Sequencing primer for use with reverse set PCR products |
| SHV_up_8 | 6, coding | 5′GGATGTATTGTGGTTATGCG | Sequencing primer for use with reverse set PCR products |
The GenBank accession number for the blaSHV-1 sequences is AF124984.
Standard sequencing.
PCR amplicons were sequenced by a standard methodology with an ABI Prism BigDye Terminator cycle sequencing ready reaction kit mix (version 3.1; Applied Biosystems, Foster City, CA). The amplification reactions were performed on a model PTC-225 cycler (Bio-Rad, Hercules, CA) for 40 cycles, and the products were separated, following denaturation, by electrophoresis on an ABI 3730 genetic analyzer (Applied Biosystems), as recommended by the manufacturer.
Nucleotide sequence accession numbers.
The complete sequences of the novel TEM-type and SHV-type β-lactamases have been deposited in the GenBank database under the following accession numbers: for TEM-155, DQ679961; for SHV-48, AY263404; for SHV-105, J194944; and for SHV-11 from resistance plasmid R1010, FJ483937.
RESULTS
Design and validation of approach.
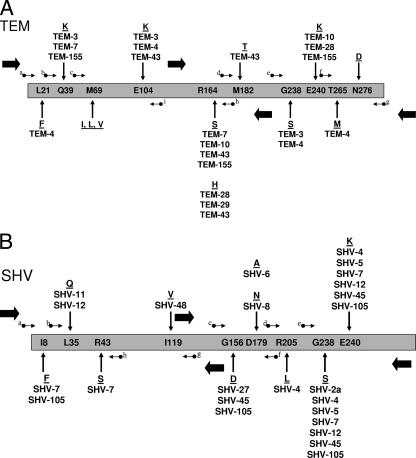
At the time of this study, there were approximately 160 TEM-type and 105 SHV-type β-lactamase derivatives reported to the database designed to track ESBLs (www.lahey.org/studies). Not all of these derivatives have an extended spectrum of activity; however, with the large number of genetic variants, those areas where changes lead to the ESBL phenotype are clearly defined (Fig. 1). In both TEM- and SHV-type ESBLs, a serine substitution for glycine at position 238 is the most common change associated with the alteration of the β-lactam spectrum of activity (4). This substitution is often paired with a lysine substitution for glutamic acid at position 240 in TEM-type ESBLs, a change that is also fairly common in SHV-type enzymes. The other substitutions that appear to be particularly important for the ESBL phenotype in TEM-type enzymes are glutamic acid to lysine at position 104 and arginine to serine or histidine at residue 164 (27, 45). Inhibitor-resistant derivatives of TEM-type enzymes (previously called IRTs) have been described, and the amino acid substitutions that are common to a number of enzymes displaying this phenotype occur at positions 69 and 276 (4, 11). Additionally, there are amino acid substitutions that are seen in many derivatives that are not associated with the ESBL phenotype; nevertheless, for epidemiological purposes, it would be useful to monitor these positions: the leucine at position 21, the glutamine at position 39, the methionine at position 182, and the threonine at position 265. In addition to the substitutions at positions 238 and 240 in SHV-type ESBLs, two other important residues are located at positions 156 (glycine to aspartic acid) and 179 (aspartic acid to alanine, asparagine, or glycine); substitutions at these positions also result in the ESBL phenotype (3, 13, 47). Similarly to the TEM-type enzymes, substitutions in SHV-type proteins that do not result in the ESBL phenotype are often seen: isoleucine to phenylalanine at position 8 and leucine to glutamine at position 35. This data set is well suited for SNP analysis as a tool for the rapid typing of ESBLs (Fig. 1).
FIG. 1.
Schematic representation of the blaTEM (A) and blaSHV (B) genes. Codons for the amino acid positions listed within the bar representing the TEM-type (A) and SHV-type (B) β-lactamases were subjected to pyrosequence analysis. The amino acid numbering is according to Ambler et al. (1). Small arrows indicate the positions of the primers used to direct pyrosequencing and are for visual reference only: in panel A (TEM), a, TEM_up_21; b, TEM_up_39; c, TEM-up_69; d, TEM_up_182n; e, TEM_up_238; f, TEM_up_265n; g, TEM_dwn_280; h, TEM_dwn_165n; and i, TEM_dwn_104; in panel B (SHV), a, SHV_up_8; b, SHV_up_35; c, SHV_up_156n; d, SHV_up_205n; e, SHV_up_238n1; f, SHV_dwn_179; g, SHV_dwn_119; and h, SHV_dwn_43. The precise locations of the primers are indicated in Tables 2 and 3. The large, filled arrows indicate the positions of the PCR primers used to amplify the blaTEM and blaSHV genes. Substitutions pertinent to the present study found in TEM-type (A) and SHV-type (B) ESBLs are indicated above and below the sequences of TEM-1 and SHV-1. ESBLs often contain more than one amino acid substitution. Inhibitor-resistant TEM (IRT) enzymes were not identified in the present study; therefore, the amino acid substitutions at positions 69 and 276 of the TEM-type sequence were not identified.
In order to validate the utility of using the SNP protocol for the determination of nucleotide substitutions that correlate with the ESBL phenotype, a selection of control isolates with previously characterized TEM-type and SHV-type ESBLs and non-ESBLs were analyzed. The control isolates (seven isolates with TEM-type enzymes and nine isolates with SHV-type enzymes) and the respective encoded β-lactamases, as determined by standard sequencing, are indicated in Table 1; and the primers used for amplification and pyrosequence analysis are presented in Table 2 (for TEM-type enzymes) and Table 3 (for SHV-type enzymes). The amino acid encoded at each of 10 positions that were monitored is shown in Table 4 for the control blaTEM-encoding isolates. Similarly, Table 5 shows the nine amino acid positions that were monitored in blaSHV-encoding control isolates. In all cases, the SNP protocol for pyrosequence analysis confirmed that the expected β-lactamase was present in each of the control isolates (Tables 4 and 5).
TABLE 4.
Pyrosequence analysis of blaTEM enzymes
| Strain | β-Lactamase | Amino acid at positiona:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 21 | 39 | 69 | 104 | 164 | 182 | 238 | 240 | 265 | 276 | ||
| GC 1472 | TEM-1 | L | Q | M | E | R | M | G | E | T | N |
| GC 1494 | TEM-3 | L | K | M | K | R | M | S | E | T | N |
| GC 1500 | TEM-4 | F | Q | M | K | R | M | S | E | M | N |
| GC 1505 | TEM-7 | L | K | M | E | S | M | G | E | T | N |
| GC 1511 | TEM-10 | L | Q | M | E | S | M | G | K | T | N |
| GC 2336 | TEM-28 | L | Q | M | E | H | M | G | K | T | N |
| GC 2400 | TEM-43 | L | Q | M | K | H | T | G | E | T | N |
| GAR 2830 | TEM-1 | L | Q | M | E | R | M | G | E | T | N |
| GAR 3593 | TEM-1 (50),b TEM-2 (50) | L | Q/K | M | E | R | M | G | E | T | N |
| GAR 4649 | TEM-29 | L | Q | M | E | H | M | G | E | T | N |
| GAR 10941 | TEM-155 | L | K | M | E | S | M | G | K | T | N |
Amino acid numbering according to Ambler et al. (1). Boldface and underlining indicate amino acid residue substitutions identified by pyrosequence analysis.
Values in parentheses are the percentage of blaTEM alleles determined on the basis of the percentage of each SNP at amino acid 39 codon (Fig. 2).
TABLE 5.
Pyrosequence analysis of blaSHV enzymes
| Strain | β-lactamase | Amino acid at positiona:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 35 | 43 | 119 | 156 | 179 | 205 | 238 | 240 | ||
| GAR 10711 | SHV-1 | I | L | R | V | G | D | R | G | E |
| GAR 11829 | SHV-2 | I | L | R | NDb | G | D | R | S | E |
| GC 2871 | SHV-4 | I | L | R | ND | G | D | L | S | K |
| GAR 9191 | SHV-5 | I | L | R | ND | G | D | R | S | K |
| GC 2904 | SHV-6 | I | L | R | ND | G | A | R | G | E |
| GC 2337 | SHV-7 | F | L | S | ND | G | D | R | S | K |
| GC 5901 | SHV-8 | I | L | R | ND | G | N | R | G | E |
| GAR 5951 | SHV-11 | I | Q | R | ND | G | D | R | G | E |
| GC 6326 | SHV-12 | I | Q | R | ND | G | D | R | S | K |
| GC 1709 | SHV-11 | I | Q | R | ND | G | D | R | G | E |
| GAR 12767 | SHV-27 | I | L | R | ND | D | D | R | G | E |
| GC 7668 | SHV-48 | I | L | R | I | G | D | R | G | E |
| GC 7644 | SHV-1 (50),c SHV-5 (50) | I | L | R | ND | G | D | R | G/S | E/K |
| GAR 2332 | SHV-11 (60), SHV-12 (40)d | I | Q | R | ND | G | D | R | G/S | E/K |
| GC 7991 | SHV-1, SHV-5, SHV-105d | I/F (30/70)e | L | R/S (30/70) | ND | G/D (40/60) | D | R | G/S (45/55) | E/Kf |
| GAR 8110g | SHV-1, SHV-5, SHV-27, SHV-45 | I | L | R | ND | G/D (60/40) | D | R | G/S (40/60) | E/K |
| GAR 11218h | SHV-1, SHV-5, SHV-11, SHV-12 | I | L/Q (70/30) | R | ND | G | D | R | G/S (50/50) | E/K |
Amino acid numbering according to Ambler et al. (1). Boldface and underlining indicate amino acid residue substitutions identified by pyrosequence analysis.
ND, not determined.
Values in parentheses are the percentage of blaSHV alleles determined on the basis of the percentage of each SNP at amino acid 238 codon.
The presence of the blaSHV alleles listed was confirmed by cloning and standard sequence analysis.
The percentage of each SNP is indicated in parentheses.
Due to the presence of two SNPs in the sequence encoding position 240, the percentage of SNPs at codon 240 cannot be calculated.
The presence of all alleles listed was not confirmed. The alleles listed represent the possible spectrum of alleles on the basis of the pyrosequencing result. The presence of SHV-1 and SHV-45 was confirmed by cloning and standard sequence analysis.
The presence of all alleles listed was not confirmed. The alleles listed represent the possible spectrum of alleles on the basis of the pyrosequencing result.
SNP analysis of clinical isolates.
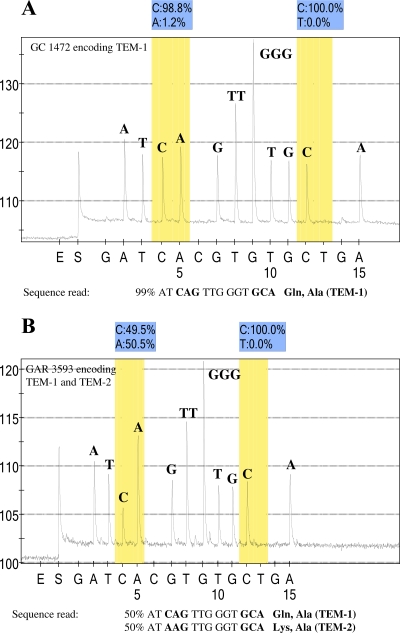
Pyrosequencing was employed to determine the sequence at relevant positions for four blaTEM genes and eight blaSHV genes from clinical isolates determined to encode an ESBL through phenotypic and preliminary PCR testing (Tables 4 and 5). The value of the SNP protocol for pyrosequencing can be seen with clinical isolate GAR 3593, which encodes both TEM-1 and TEM-2. By standard sequence analysis, analysis of this isolate would result in a mixed base; however, by the SNP protocol, the polymorphism is resolved and the mixed base (50:50) is noted on the pyrogram (Fig. 2). The interpretation of the pyrogram is performed automatically by the pyrosequencing software and is based on the dispensation order (shown as the x axis in Fig. 2 and 3). For example, as shown in Fig. 2A (TEM-1), 99% of the time the incorporated base is C at the fourth cycle of synthesis. By contrast, as shown in Fig. 2B (a mixture of TEM-1 and TEM-2), the fourth cycle of synthesis incorporates a C residue 50% of the time and an A residue 50% of the time. In TEM-1 (Fig. 2A), the peaks at the fourth and fifth cycles of synthesis are similar in size because the fifth cycle of synthesis incorporates an A residue (100%), whereas in Fig. 2B (TEM-1 and TEM-2 mixture), the fifth cycle of synthesis produces a larger peak representative of the incorporation of two A residues 50% of the time for the TEM-2 allele and a single A residue for the TEM-1 allele. The presence of both TEM-1 and TEM-2 in this isolate was confirmed by cloning and standard sequence analysis (data not shown).
FIG. 2.
Pyrosequence analysis of blaTEM derivatives. (A) Pyrogram of sequence analysis of PCR products with primer TEM_up_39 and the template from isolate GC 1472 (TEM-1). This primer was designed to sequence through codons 39 and 42, which encode glutamine and alanine, respectively. The pyrogram indicates that the codons at positions 39 and 42 conform to the TEM-1 sequence. (B) Pyrogram of sequence analysis of PCR products with primer TEM_up_39 and the template from isolate GAR 3593. The pyrogram indicates an SNP in the codon at position 39, such that 50% of the sequences read CAG, which encodes glutamine, and 50% of the sequences read AAG, which encodes lysine. In contrast, the codon encoding position 42 reads GCA with 100% C residues at the polymorphic position of interest. The y axis represents the relative light units as a measure of nucleotide incorporation, and the x axis is the nucleotide dispensation order that is programmed into the instrument. Peaks indicate the level of the respective nucleotide incorporated. E, enzyme-only control; S, substrate-only control. The initial nucleotide injected (in this case, G) is irrelevant to the sequence in order to provide a baseline. The shaded bars indicate the potentially polymorphic position in the sequence, and the relative percentage of each nucleotide at that position is indicated directly above the pyrogram. An irrelevant nucleotide is programmed into the dispensation order following each polymorphic position of interest. In panel A, 1.2% of the A residues at the polymorphic position are below the level of background acceptable for SNP sequence analysis, according to the manufacturer (Biotage AB).
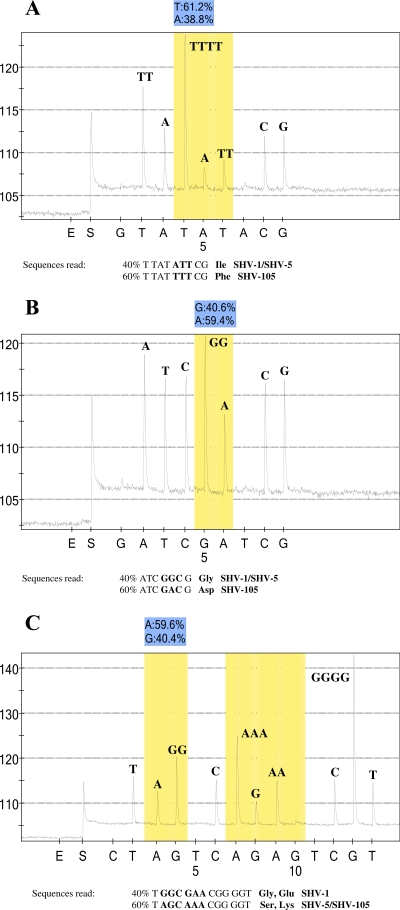
FIG. 3.
Pyrosequence analysis of a novel blaSHV derivative, SHV-105, encoded by isolate GC 7991. (A) Pyrogram of SNP analysis of PCR products with primer SHV_up_8 and the template from isolate GC 7991. This primer was designed to sequence through codons 7 and 8, which encode tyrosine and isoleucine, respectively. The pyrogram indicates an SNP at codon 8, with approximately 40% of the sequence reading ATT, which encodes isoleucine, and 60% of the sequence reading TTT, which encodes phenylalanine. The sequence at codon 7, TAT, which encodes tyrosine, is as expected. (B) Pyrogram of SNP analysis of PCR products with sequencing primer SHV_up_156n and the template from isolate GC 7991. The pyrogram indicates an SNP in the codon at position 156, such that 40% of the sequences read GGC, which encodes glycine, and 60% of the sequences read GAC, which encodes aspartic acid. (C) Pyrogram of SNP analysis of PCR products with sequencing primer SHV_up_238n and the template from isolate GC 7991. The pyrogram indicates an SNP at both positions 238 and 240. At position 238, the sequence is 60% AGC, which encodes serine, and 40% GGC, which encodes glycine. Similarly, the codon at position 240 shows a mixture of sequences with GAA, which encodes glutamic acid, and AAA, which encodes lysine. Interestingly, there is a second naturally occurring polymorphism at position 240 with an alternate codon, GAG, which encodes glutamic acid. Due to the proximity of the two SNPs, the pyrosequencing software is unable to quantitate the percentage of each of the SNPs present at position 240. The y axis represents relative light units as a measure of nucleotide incorporation, and the x axis is the nucleotide dispensation order that is programmed into the instrument. Peaks indicate the level of the respective nucleotide incorporated. E, enzyme-only control; S, substrate-only control. The initial nucleotide injected (in this case, G) is irrelevant to the sequence in order to provide a baseline. The shaded bars indicate the potentially polymorphic position in the sequence, and the relative percentage of each nucleotide at that position is indicated directly above the pyrogram. An irrelevant nucleotide is programmed into the dispensation order following each polymorphic position of interest.
While screening for potential ESBL-encoding pathogens from the tigecycline phase III clinical trials (22), a novel blaTEM ESBL was identified in a P. mirabilis isolate and has been given the designation TEM-155 (23). This novel blaTEM, as determined by pyrosequencing, encodes three amino acid substitutions (Q39K, R164S, and E240K), a combination that had not been described before (Table 4). Sequence changes were confirmed by standard cloning and sequence analysis (data not shown).
Two novel blaSHV variants were examined in the present study. The first isolate, GC 7668, collected during clinical trials with piperacillin-tazobactam, encodes SHV-48 (www.lahey.org/studies) and contains a substitution of isoleucine for valine at position 119. This novel blaSHV was originally identified due to the novel susceptibility pattern of GC 7668, which is suggestive of an inhibitor-resistant enzyme (Table 6). Standard cloning and sequence analysis was utilized to define the nucleotide change in the gene (data not shown). In the present study, primers were designed to determine if this novel substitution could be detected by the SNP protocol. As shown in Table 5, the SNP protocol was able to identify this novel substitution. The second isolate, GC 7991 (a gift of L. Saiman, New York Presbyterian Hospital), was associated with an outbreak of ESBL-producing K. pneumoniae isolates in a neonatal intensive care unit (17). Upon pyrosequencing, a mixture of blaSHV sequences could clearly be detected by SNP analysis (Fig. 3). This isolate encodes a novel enzyme that has been designated SHV-105 (www.lahey.org/studies). The substitutions at position 8 (phenylalanine for isoleucine; Fig. 3A), position 43 (serine for arginine), position 156 (aspartic acid for glycine; Fig. 3B), position 238 (serine for glycine), and position 240 (lysine for glutamic acid; Fig. 3C) were all confirmed by cloning and standard sequence analysis (data not shown). The presence of both SHV-1 and SHV-5 in this isolate was also confirmed by cloning and standard sequence analysis (data not shown).
TABLE 6.
Susceptibility data determined by microdilution methods
| Isolate | bla gene | MIC (μg/ml)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIP | TZP | AMP | AMC | FOX | CTX | CAZ | CRO | FEP | IPM | ATM | ||
| GC 8130 | SHV-1 | 64 | 1 | >128 | 16 | 16 | ≤0.125 | 0.5 | 0.25 | ≤0.125 | 0.25 | 0.25 |
| GC 7668 | SHV-48 | >128 | >128 | >128 | 32 | 4 | ≤0.125 | 1 | ≤0.125 | 0.5 | 0.25 | 0.5 |
| GC 8128 | SHV-105 | >128 | 1 | >128 | 16 | 8 | 128 | >128 | 128 | 16 | 0.25 | >128 |
| GC 8133 | TEM-155 | >128 | 2 | >128 | 32 | 8 | 32 | >128 | 64 | 128 | 0.25 | >128 |
| ATCC 25922 | None | 2 | 2 | 4 | 8 | 4 | ≤0.125 | 0.25 | ≤0.125 | ≤0.125 | ≤0.125 | 0.25 |
| DH5α | None | 0.5 | 0.5 | 8 | 8 | 8 | ≤0.125 | 0.25 | ≤0.125 | ≤0.125 | 0.25 | ≤0.125 |
Antibiotic abbreviations: PIP, piperacillin; TZP, piperacillin-tazobactam; AMP, ampicillin; AMC, amoxicillin-clavulanic acid; FOX, cefoxitin; CTX, cefotaxime; CAZ, ceftazidime; CRO, cefuroxime; FEP, cefepime; IPM, imipenem; ATM, aztreonam.
Finally, an interesting observation regarding plasmid R1010, which has been the quintessential control plasmid for SHV-1 (50), was made during the course of this study. In attempts to validate the pyrosequencing protocol for the identification of SHV-type ESBLs, plasmid R1010 was examined with the expectation of verifying the sequence of blaSHV-1. However, pyrosequencing, followed by complete nucleotide sequencing, revealed that R1010 did not express SHV-1 but, rather, expressed SHV-11 (a leucine substitution for glutamine at position 35). Because of the narrow spectra of activity and the identical pIs of both SHV-1 and SHV-11, it is likely that these were assumed to be the same enzyme in the years before nucleotide sequencing became commonplace and the β-lactamase on R1010 was misidentified.
Susceptibility studies.
The novel TEM-type (TEM-155) and SHV-type (SHV-48, SHV-105) enzyme variants identified by the pyrosequencing effort were shotgun cloned into pCLL2300 (5) and sequenced by the standard methodology to confirm the SNP results (data not shown). The in vitro susceptibilities of recombinant strains encoding each of the novel β-lactamases to a panel of β-lactams were determined by the microdilution format (Table 6) as well as the Etest format (Table 7). TEM-155 and SHV-105 had the expected extended spectrum of activity, whereas the non-ESBL SHV-48 showed a broad spectrum of activity and may be an inhibitor-resistant SHV; additional work will be required to further explore the spectrum of activity of this novel enzyme.
TABLE 7.
Susceptibility data determined by Etest
| Isolate | bla gene | MIC (μg/ml)a
|
Interpretationb | |||
|---|---|---|---|---|---|---|
| CTX | CTX + CLA | CAZ | CAZ + CLA | |||
| GC 8130 | SHV-1 | <0.25 | 0.023 | <0.5 | 0.125 | No |
| GC 7668 | SHV-48 | <0.25 | 0.023 | <0.5 | 0.125 | No |
| GC 8128 | SHV-105 | >16 | 0.032 | >32 | 0.25 | Yes |
| GC 8133 | TEM-155 | 2 | 0.023 | >32 | 0.5 | Yes |
| ATCC 25922 | None | <0.25 | 0.047 | <0.5 | 0.125 | No |
| DH5α | None | <0.25 | 0.023 | <0.5 | 0.094 | No |
Antibiotic abbreviations: CTX, cefotaxime; CLA, clavulanic acid; CAZ, ceftazidime.
Determination of the presence of an ESBL by Etest, according to the instructions of the manufacturer.
DISCUSSION
Although the TEM- and SHV-type ESBLs have fallen behind the CTX-M family enzymes in terms of their global prevalence, these enzymes are widespread globally and remain the most prevalent ESBLs in the United States (22). In addition, worldwide reports of novel TEM-type and SHV-type enzymes are commonplace (29, 35, 44, 49, 58). The recent reports of novel blaTEM enzymes that have both an extended spectrum of activity and resistance to β-lactamase inhibitors suggest that the TEM-type enzymes are continuing to evolve in the face of current therapies (11, 49). Moreover, in the study described in this report, three novel β-lactamases were identified, two of which have extended spectra of activity.
ESBLs are an increasing problem in the clinic, as strains expressing multiple ESBLs can show clinical resistance to cephamycins, thereby reducing the therapeutic options to a single broad-spectrum agent, such as imipenem, and in some cases resulting in resistance to all β-lactams, leaving the physician with few treatment options (48, 56). Moreover, a number of studies have demonstrated a poorer clinical outcome as well as increased mortality in patients infected with ESBL-encoding pathogens (42, 59). Two recent reports show that the utility of cefepime for the treatment of E. coli, K. pneumoniae, and Enterobacter cloacae infections was compromised by the presence of SHV-type ESBLs (26, 54). In addition, an ESBL combined with the reduction in the level of expression of an outer membrane porin can result in a broadening of the resistant phenotype to include the cephamycins (33).
The identification of ESBLs and epidemiological studies performed to track ESBLs within the hospital setting as well as countrywide or worldwide are dependent on innovative methodologies for determination of the identity of a given ESBL in a timely manner. The rapid identification of an ESBL would result in the initiation of the appropriate therapeutic intervention sooner and may result in fewer clinical failures. Current methodologies based on susceptibility tests suffer, as resistance to expanded-spectrum cephalosporins does not always present in vitro (31, 40). A number of molecular-based detection approaches have been presented (2, 32, 46, 55); however, the large number of mutations described in TEM- and SHV-type ESBLs requires that sequence analysis be employed in order to provide a definitive result. Real-time PCR and pyrosequencing have recently been utilized to define the five subtypes of CTX-M-type β-lactamases by reading through a small region of sequence divergence among the five subtypes (37).
Pyrosequence analysis by use of the SNP protocol is a tool well suited to the characterization of multiple alleles of genes, such as ESBLs and non-ESBLs, in clinical isolates. This is of particular value when isolates are being investigated for blaTEM as well as blaSHV genes: multiple alleles of these important resistance genes in pathogens are commonplace (5, 28, 41). For instance, in a recent survey of the ESBL genes found in clinical isolates from the worldwide tigecycline clinical trials, 52% of the isolates encoded multiple SHV-type enzymes, including both ESBLs and non-ESBLs (22). In the present study, isolates with three and, potentially, four alleles of blaSHV were resolved by use of the SNP protocol (Table 5). Furthermore, pyrosequencing enabled the correction of the identity of the β-lactamase on resistance plasmid R1010, which for many years has mistakenly been reported to express SHV-1.
By way of comparison, Haanpera et al. used pyrosequencing to type a collection of blaSHV genes; however, those investigators used the SQA protocol for standard sequence analysis (18). Although the chemistries used for both pyrosequencing protocols (SNP and SQA) are identical, the SNP methodology is ideal for the sequencing of mixed templates and determination of SNPs at the positions of interest. The SQA protocol, which provides a sequence output as a pyrogram from which the relative peak size at each dispensation is used to determine the sequence at each nucleotide position, is less well suited for the analysis of samples with mixtures of enzymes. This methodology, as employed by the authors of the study, requires interpretation of the peak size in order to confirm the presence of multiple blaSHV genes in the sample. Due to the fact that during the programming step for the SNP protocol the operator provides the software with the expected output sequence and indicates the positions of polymorphism, the interpretation of the sequence data by the software is more robust. In our hands, the SNP protocol had greater reproducibility than the SQA protocol and provided a superior ability to discriminate the sequence; it had the added benefit of calculating the percentage of the allele at each position of interest. Our work is also differentiated from that of Haanpera et al. due to the fact that we investigated SNPs at nine positions in blaSHV, whereas they investigated SNPs at three positions in their study (18), which allowed us to capture many more SHV-type derivatives. The methodology as designed will identify 86 (54%) of the 160 TEM variants and 49 (47%) of the 115 SHV variants currently listed at the Lahey site (www.lahey.org/studies). It should be noted that the method was designed to identify ESBL variants and not all variants of these important resistance determinants; therefore, pyrosequencing was focused on residues critical to the ESBL phenotype (e.g., substitutions at positions 238 and 240).
In an environment in which ESBLs are becoming an increasing threat in the clinic, tools for the rapid identification of these rapidly evolving β-lactamases are needed both to aid in clinical practice and for epidemiological studies. Pyrosequencing is a real-time sequencing-by-synthesis approach that has clear value for the detection of SNPs for TEM- and SHV-type ESBL identification. From a practical perspective, this technology is very similar to PCR and is instrument driven. While certain expertise is required at the front end and some troubleshooting is necessary to define the appropriate reagents (primers) and conditions needed to achieve optimal outcomes, pyrosequence analysis represents a robust tool for rapid sequence determination that may have a place in the clinical setting.
Footnotes
Published ahead of print on 15 December 2008.
REFERENCES
- 1.Ambler, R. P., A. F. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276(Pt 1):269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arlet, G., G. Brami, D. Decre, A. Flippo, O. Gaillot, P. H. Lagrange, and A. Philippon. 1995. Molecular characterisation by PCR-restriction fragment length polymorphism of TEM β-lactamases. FEMS Microbiol. Lett. 134:203-208. [DOI] [PubMed] [Google Scholar]
- 3.Arlet, G., M. Rouveau, and A. Philippon. 1997. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum β-lactamase. FEMS Microbiol. Lett. 152:163-167. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, P. A., C. E. Cherubin, V. Idemyor, B. A. Rasmussen, and K. Bush. 1994. Multiply resistant Klebsiella pneumoniae strains from two Chicago hospitals: identification of the extended-spectrum TEM-12 and TEM-10 ceftazidime-hydrolyzing β-lactamases in a single isolate. Antimicrob. Agents Chemother. 38:761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, P. A., N. V. Jacobus, N. S. Bhachech, and K. Bush. 1996. TEM-28 from an Escherichia coli clinical isolate is a member of the His-164 family of TEM-1 extended-spectrum ß-lactamases. Antimicrob. Agents Chemother. 40:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, P. A., and C. C. Sanders. 1995. Development of test panel of β-lactamases expressed in a common Escherichia coli host background for evaluation of new β-lactam antibiotics. Antimicrob. Agents Chemother. 39:308-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, P. A., C. Urban, A. Jaiswal, N. Mariano, B. A. Rasmussen, S. J. Projan, J. J. Rahal, and K. Bush. 1995. SHV-7, a novel cefotaxime-hydrolyzing β-lactamase, identified in Escherichia coli isolates from hospitalized nursing home patients. Antimicrob. Agents Chemother. 39:899-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canton, R., and T. M. Coque. 2006. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9:466-475. [DOI] [PubMed] [Google Scholar]
- 11.Canton, R., M. I. Morosini, O. M. de la Maza, and E. G. de la Pedrosa. 2008. IRT and CMT β-lactamases and inhibitor resistance. Clin. Microbiol. Infect. 14(Suppl. 1):53-62. [DOI] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing: M100-S17, 17th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 13.Corkill, J. E., L. E. Cuevas, R. Q. Gurgel, J. Greensill, and C. A. Hart. 2001. SHV-27, a novel cefotaxime-hydrolysing β-lactamase, identified in Klebsiella pneumoniae isolates from a Brazilian hospital. J. Antimicrob. Chemother. 47:463-465. [DOI] [PubMed] [Google Scholar]
- 14.Datta, N., and P. Kontomichalou. 1965. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208:239-241. [DOI] [PubMed] [Google Scholar]
- 15.Diggle, M. A., and S. C. Clarke. 2004. Pyrosequencing: sequence typing at the speed of light. Mol. Biotechnol. 28:129-137. [DOI] [PubMed] [Google Scholar]
- 16.Gniadkowski, M. 2008. Evolution of extended-spectrum β-lactamases by mutation. Clin. Microbiol. Infect. 14(Suppl. 1):11-32. [DOI] [PubMed] [Google Scholar]
- 17.Gupta, A., P. Della-Latta, B. Todd, P. San Gabriel, J. Haas, F. Wu, D. Rubenstein, and L. Saiman. 2004. Outbreak of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a neonatal intensive care unit linked to artificial nails. Infect. Control Hosp. Epidemiol. 25:210-215. [DOI] [PubMed] [Google Scholar]
- 18.Haanpera, M., S. D. Forssten, P. Huovinen, and J. Jalava. 2008. Typing of SHV extended-spectrum β-lactamases by pyrosequencing in Klebsiella pneumoniae strains with chromosomal SHV β-lactamase. Antimicrob. Agents Chemother. 52:2632-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haanpera, M., P. Huovinen, and J. Jalava. 2005. Detection and quantification of macrolide resistance mutations at positions 2058 and 2059 of the 23S rRNA gene by pyrosequencing. Antimicrob. Agents Chemother. 49:457-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helfand, M. S., and R. A. Bonomo. 2003. β-Lactamases: a survey of protein diversity. Curr. Drug Targets Infect. Disord. 3:9-23. [DOI] [PubMed] [Google Scholar]
- 21.Huletsky, A., J. R. Knox, and R. C. Levesque. 1993. Role of Ser-238 and Lys-240 in the hydrolysis of third-generation cephalosporins by SHV-type β-lactamases probed by site-directed mutagenesis and three-dimensional modeling. J. Biol. Chem. 268:3690-3697. [PubMed] [Google Scholar]
- 22.Jones, C. H., M. Tuckman, D. Keeney, A. Ruzin, and P. A. Bradford. Characterization and sequence analysis of extended-spectrum β-lactamase-encoding genes from Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates collected during tigecycline phase 3 clinical trials. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 23.Keeney, D., M. Tuckman, S. Mandiyan, P. Petersen, C. H. Jones, and P. A. Bradford. 2006. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-51.
- 24.Kliebe, C., B. A. Nies, J. F. Meyer, R. M. Tolxdorff-Neutzling, and B. Wiedemann. 1985. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob. Agents Chemother. 28:302-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knothe, H., P. Shah, V. Krcmery, M. Antal, and S. Mitsuhashi. 1983. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11:315-317. [DOI] [PubMed] [Google Scholar]
- 26.Kotapati, S., J. L. Kuti, C. H. Nightingale, and D. P. Nicolau. 2005. Clinical implications of extended spectrum β-lactamase (ESBL) producing Klebsiella species and Escherichia coli on cefepime effectiveness. J. Infect. 51:211-217. [DOI] [PubMed] [Google Scholar]
- 27.Kurokawa, H., N. Shibata, Y. Doi, K. Shibayama, K. Kamachi, T. Yagi, and Y. Arakawa. 2003. A new TEM-derived extended-spectrum β-lactamase (TEM-91) with an R164C substitution at the omega-loop confers ceftazidime resistance. Antimicrob. Agents Chemother. 47:2981-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, Y. H., B. Cho, I. K. Bae, C. L. Chang, and S. H. Jeong. 2006. Klebsiella pneumoniae strains carrying the chromosomal SHV-11 β-lactamase gene produce the plasmid-mediated SHV-12 extended-spectrum β-lactamase more frequently than those carrying the chromosomal SHV-1 β-lactamase gene. J. Antimicrob. Chemother. 57:1259-1261. [DOI] [PubMed] [Google Scholar]
- 29.Ling, B. D., G. Liu, Y. E. Xie, Q. X. Zhou, T. K. Zhao, C. Q. Li, X. Yu, and J. Lei. 2006. Characterisation of a novel extended-spectrum β-lactamase, SHV-70, from a clinical isolate of Enterobacter cloacae in China. Int. J. Antimicrob. Agents 27:355-356. [DOI] [PubMed] [Google Scholar]
- 30.Livermore, D. M., R. Canton, M. Gniadkowski, P. Nordmann, G. M. Rossolini, G. Arlet, J. Ayala, T. M. Coque, I. Kern-Zdanowicz, F. Luzzaro, L. Poirel, and N. Woodford. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165-174. [DOI] [PubMed] [Google Scholar]
- 31.Livermore, D. M., and M. Yuan. 1996. Antibiotic resistance and production of extended-spectrum β-lactamases amongst Klebsiella spp. from intensive care units in Europe. J. Antimicrob. Chemother. 38:409-424. [DOI] [PubMed] [Google Scholar]
- 32.Mabilat, C., and P. Courvalin. 1990. Development of “oligotyping” for characterization and molecular epidemiology of TEM β-lactamases in members of the family Enterobacteriaceae. Antimicrob. Agents Chemother. 34:2210-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Martinez, L., S. Hernandez-Alles, S. Alberti, J. M. Tomas, V. J. Benedi, and G. A. Jacoby. 1996. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum-cephalosporins. Antimicrob. Agents Chemother. 40:342-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthew, M., R. W. Hedges, and J. T. Smith. 1979. Types of β-lactamase determined by plasmids in gram-negative bacteria. J. Bacteriol. 138:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minarini, L. A., A. C. Gales, I. C. Palazzo, and A. L. Darini. 2007. Prevalence of community-occurring extended spectrum β-lactamase-producing Enterobacteriaceae in Brazil. Curr. Microbiol. 54:335-341. [DOI] [PubMed] [Google Scholar]
- 36.Monstein, H., S. Nikpour-Badr, and J. Jonasson. 2001. Rapid molecular identification and subtyping of Helicobacter pylori by pyrosequencing of the 16S rDNA variable V1 and V3 regions. FEMS Microbiol. Lett. 199:103-107. [DOI] [PubMed] [Google Scholar]
- 37.Naas, T., C. Oxacelay, and P. Nordmann. 2007. Identification of CTX-M-type extended-spectrum-β-lactamase genes using real-time PCR and pyrosequencing. Antimicrob. Agents Chemother. 51:223-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nyberg, S. D., M. Osterblad, A. J. Hakanen, P. Huovinen, J. Jalava, and T. F. Resistance. 2007. Detection and molecular genetics of extended-spectrum beta-lactamases among cefuroxime-resistant Escherichia coli and Klebsiella spp. isolates from Finland, 2002-2004. Scand. J. Infect. Dis. 39:417-424. [DOI] [PubMed] [Google Scholar]
- 39.Page, M. G. 2008. Extended-spectrum β-lactamases: structure and kinetic mechanism. Clin. Microbiol. Infect. 14(Suppl. 1):63-74. [DOI] [PubMed] [Google Scholar]
- 40.Paterson, D. L., and R. A. Bonomo. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paterson, D. L., K. M. Hujer, A. M. Hujer, B. Yeiser, M. D. Bonomo, L. B. Rice, and R. A. Bonomo. 2003. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type β-lactamases. Antimicrob. Agents Chemother. 47:3554-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paterson, D. L., W. C. Ko, A. Von Gottberg, S. Mohapatra, J. M. Casellas, H. Goossens, L. Mulazimoglu, G. Trenholme, K. P. Klugman, R. A. Bonomo, L. B. Rice, M. M. Wagener, J. G. McCormack, and V. L. Yu. 2004. Antibiotic therapy for Klebsiella pneumoniae bacteremia: implications of production of extended-spectrum β-lactamases. Clin. Infect. Dis. 39:31-37. [DOI] [PubMed] [Google Scholar]
- 43.Perez, F., A. Endimiani, K. M. Hujer, and R. A. Bonomo. 2007. The continuing challenge of ESBLs. Curr. Opin. Pharmacol. 7:459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perilli, M., G. Celenza, M. Fiore, B. Segatore, C. Pellegrini, F. Luzzaro, G. M. Rossolini, and G. Amicosante. 2007. Biochemical analysis of TEM-134, a new TEM-type extended-spectrum β-lactamase variant produced in a Citrobacter koseri clinical isolate from an Italian hospital. J. Antimicrob. Chemother. 60:877-880. [DOI] [PubMed] [Google Scholar]
- 45.Petit, A., L. Maveyraud, F. Lenfant, J. P. Samama, R. Labia, and J. M. Masson. 1995. Multiple substitutions at position 104 of β-lactamase TEM-1: assessing the role of this residue in substrate specificity. Biochem. J. 305(Pt 1):33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Randegger, C. C., and H. Hachler. 2001. Real-time PCR and melting curve analysis for reliable and rapid detection of SHV extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 45:1730-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rasheed, J. K., C. Jay, B. Metchock, F. Berkowitz, L. Weigel, J. Crellin, C. Steward, B. Hill, A. A. Medeiros, and F. C. Tenover. 1997. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 41:647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice, L. B. 2003. Do we really need new anti-infective drugs? Curr. Opin. Pharmacol. 3:459-463. [DOI] [PubMed] [Google Scholar]
- 49.Robin, F., J. Delmas, C. Schweitzer, O. Tournilhac, O. Lesens, C. Chanal, and R. Bonnet. 2007. Evolution of TEM-type enzymes: biochemical and genetic characterization of two new complex mutant TEM enzymes, TEM-151 and TEM-152, from a single patient. Antimicrob. Agents Chemother. 51:1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roupas, A., and J. S. Pitton. 1974. R factor-mediated and chromosomal resistance to ampicillin in Escherichia coli. Antimicrob. Agents Chemother. 5:186-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simpson, I. N., P. B. Harper, and C. H. O'Callaghan. 1980. Principal β-lactamases responsible for resistance to β-lactam antibiotics in urinary tract infections. Antimicrob. Agents Chemother. 17:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinclair, A., C. Arnold, and N. Woodford. 2003. Rapid detection and estimation by pyrosequencing of 23S rRNA genes with a single nucleotide polymorphism conferring linezolid resistance in enterococci. Antimicrob. Agents Chemother. 47:3620-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sirot, D., J. Sirot, R. Labia, A. Morand, P. Courvalin, A. Darfeuille-Michaud, R. Perroux, and R. Cluzel. 1987. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae: identification of CTX-1, a novel β-lactamase. J. Antimicrob. Chemother. 20:323-334. [DOI] [PubMed] [Google Scholar]
- 54.Szabo, D., R. A. Bonomo, F. Silveira, A. W. Pasculle, C. Baxter, P. K. Linden, A. M. Hujer, K. M. Hujer, K. Deeley, and D. L. Paterson. 2005. SHV-type extended-spectrum beta-lactamase production is associated with reduced cefepime susceptibility in Enterobacter cloacae. J. Clin. Microbiol. 43:5058-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szabo, D., M. A. Melan, A. M. Hujer, R. A. Bonomo, K. M. Hujer, C. R. Bethel, K. Kristof, and D. L. Paterson. 2005. Molecular analysis of the simultaneous production of two SHV-type extended-spectrum beta-lactamases in a clinical isolate of Enterobacter cloacae by using single-nucleotide polymorphism genotyping. Antimicrob. Agents Chemother. 49:4716-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talbot, G. H., J. Bradley, J. E. Edwards, Jr., D. Gilbert, M. Scheld, and J. G. Bartlett. 2006. Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 42:657-668. [DOI] [PubMed] [Google Scholar]
- 57.Unnerstad, H., H. Ericsson, A. Alderborn, W. Tham, M. L. Danielsson-Tham, and J. G. Mattsson. 2001. Pyrosequencing as a method for grouping of Listeria monocytogenes strains on the basis of single-nucleotide polymorphisms in the inlB gene. Appl. Environ. Microbiol. 67:5339-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vignoli, R., N. F. Cordeiro, V. Garcia, M. I. Mota, L. Betancor, P. Power, J. A. Chabalgoity, F. Schelotto, G. Gutkind, and J. A. Ayala. 2006. New TEM-derived extended-spectrum β-lactamase and its genomic context in plasmids from Salmonella enterica serovar Derby isolates from Uruguay. Antimicrob. Agents Chemother. 50:781-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong-Beringer, A., J. Hindler, M. Loeloff, A. M. Queenan, N. Lee, D. A. Pegues, J. P. Quinn, and K. Bush. 2002. Molecular correlation for the treatment outcomes in bloodstream infections caused by Escherichia coli and Klebsiella pneumoniae with reduced susceptibility to ceftazidime. Clin. Infect. Dis. 34:135-146. [DOI] [PubMed] [Google Scholar]
- 60.Yang, Y., N. Bhachech, P. A. Bradford, B. D. Jett, D. F. Sahm, and K. Bush. 1998. Ceftazidime-resistant Klebsiella pneumoniae and Escherichia coli isolates producing TEM-10 and TEM-43 β-lactamases from St. Louis, Missouri. Antimicrob. Agents Chemother. 42:1671-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]