Abstract
Debio 025 is a potent inhibitor of hepatitis C virus (HCV) replication (J. Paeshuyse et al., Hepatology 43:761-770, 2006). In phase I clinical studies, monotherapy (a Debio 025 dose of 1,200 mg twice a day) resulted in a mean maximal decrease in the viral load of 3.6 log10 units (R. Flisiak et al., Hepatology 47:817-826, 2008), whereas a reduction of 4.6 log10 units was obtained in phase II studies when Debio 025 was combined with interferon (R. Flisiak et al., J. Hepatol., 48:S62, 2008). We here report on the particular characteristics of the in vitro anti-HCV activities of Debio 025. The combination of Debio 025 with either ribavirin or specifically targeted antiviral therapy for HCV (STAT-C) inhibitors (NS3 protease or NS5B [nucleoside and nonnucleoside] polymerase inhibitors) resulted in additive antiviral activity in short-term antiviral assays. Debio 025 has the unique ability to clear hepatoma cells from their HCV replicon when it is used alone or in combination with interferon and STAT-C inhibitors. Debio 025, when it was used at concentrations that have been observed in human plasma (0.1 or 0.5 μM), was able to delay or prevent the development of resistance to HCV protease inhibitors as well as to nucleoside and nonnucleoside polymerase inhibitors. Debio 025 forms an attractive drug candidate for the treatment of HCV infections in combination with standard interferon-based treatment and treatments that directly target the HCV polymerase and/or protease.
Hepatitis C virus (HCV) represents a major health burden. An estimated 170 million to 180 million people worldwide are chronically infected with this virus and are at increased risk of developing liver cirrhosis and/or hepatocellular carcinoma (64). The current standard of care for chronic hepatitis C consists of pegylated alpha interferon (IFN) in combination with ribavirin (RBV) (12). This therapy is, however, associated with serious side effects and results in a sustained virological response in only 50 to 60% of patients (depending on the genotype). There is thus an urgent need for more effective and better-tolerated drugs.
Selective inhibitors of HCV replication that target the NS3 protease and the NS5B RNA-dependent RNA polymerase (RdRp) in particular have been pursued as potential new therapies (38). BILN 2061 (culprivir), a peptidomimetic inhibitor of the HCV NS3 protease (the first selective inhibitor of HCV to be administered to patients), resulted in a rapid and pronounced decline in the level of viral replication in patients chronically infected with HCV genotype 1. Its clinical development was, however, halted because of cardiotoxicity (22, 31). Currently, four NS3 protease inhibitors are in clinical development, i.e., VX-950 (telaprevir), SCH-503034 (boceprevir), ITMN-191, and TMC435350 (2, 51, 56). Telaprevir and boceprevir are at the most advanced stages of development and are being evaluated in combination with the standard therapy in phase III and phase II clinical trials, respectively (3, 39, 45, 50). Several nucleoside polymerase inhibitors (NIs) and nonnucleoside polymerase inhibitors (NNIs) are or have been in development. Nucleoside analogue inhibitors of HCV replication basically act as chain terminators of the polymerization process once they have been phosphorylated to their 5′-triphosphate metabolite (13). 2′-C-Methylcytidine (2′-C-MeCyt), the active component of valopicitabine, was the first polymerase inhibitor to enter clinical trials, but this compound is not being further pursued because of limited efficacy and side effects (6, 46). R1626, the prodrug of 4′-azidocytidine (R1479), and R7128, the prodrug of 2′-C-methyl-2′-fluoro-cytidine, have been shown to have efficacy against HCV in clinical studies (1, 28, 29, 52, 53). However, the development of R1626 has been put on hold because of hematological side effects in patients. Also, treatment with the NNI HCV 796, a benzofuran, resulted in a marked reduction in the viral load. However, the clinical development of this compound has been stopped because of hepatotoxicity (42). Several other NNIs that target different allosteric sites of the enzyme have been discovered, such as the Japan Tobacco (JT) benzimidazole derivatives, which act as allosteric inhibitors that block the polymerase before elongation. However, limited data on these drug candidates have been released (14).
Due to the low fidelity of RdRp and the high rate of turnover of the viral RNA, drug resistance is and will continue to be a major problem in the development of specific HCV inhibitors (55). Indeed, drug-resistant variants have readily been selected in vitro and in clinical studies with several protease inhibitors as well as with NIs and NNIs (32-34). Alternatively, host cell factors essential for viral replication can be envisaged as antiviral agents. It may be anticipated that drugs that prevent the interaction of host cell factors with the viral replication machinery may possibly have a higher genetic barrier to the development of resistance. Cyclosporine (Cs), a widely used immunosuppressive drug, was shown to exert anti-HCV activity in vitro (40, 59). In one controlled clinical trial, the use of Cs was reported to result in a higher sustained virological response rate when it was combined with IFN in comparison with the response rate of IFN monotherapy (25). Cs binds to cyclophilins and inhibits their cis-trans peptidyl-prolyl isomerase activity (18). The compound forms a complex with cyclophilin A to inhibit calcineurin and, thus, the activation of T cells. Tacrolimus (FK506), an immunosuppressive drug that interacts with calcineurin but not with cyclophilins, exerts no anti-HCV activity, which indicates that immunosuppressive activity is not a prerequisite for potency against HCV (41).
We recently reported on the potent anti-HCV activity of the cyclophilin inhibitor Debio 025 (previously named UNIL025), a nonimmunosuppressive Cs analogue (43). The lack of immunosuppressive activity is explained by the inability of the compound to bind to calcineurin. Debio 025 is at least 10-fold more potent as an anti-HCV agent than Cs (43). In virus-infected chimeric mice, Debio 025 was better tolerated than Cs, and the anti-HCV effect of Debio 025 appeared to be synergistic when it was used in combination with pegylated IFN (26). During a 15-day phase 1b study in which patients coinfected with human immunodeficiency virus (HIV) and HCV received 1,200 mg of Debio 025 or placebo twice daily, Debio 025 resulted in a mean maximal decrease in the viral load of 3.6 log10 units (19). When Debio 025 was combined with pegylated IFN alpha 2a during phase IIa studies, a reduction in the viral load of 4.6 log units was obtained (17). NIM811 (37) and SCY-635 (23), two other nonimmunosuppressive Cs analogues, have also been shown to specifically inhibit HCV replication. Akin to Cs and NIM811, Debio 025 is also endowed with anti-HIV activity (8, 48, 49). We here report on the particular characteristics of the anti-HCV activities of Debio 025 in vitro.
MATERIALS AND METHODS
Compounds.
The preparation of Debio 025 was based on the strategy used for the synthesis of d-methyl-Ala3-ethyl-Xaa4-cyclosporine analogues described previously (24). Cs was purchased from Fluka Chemie GmbH (Buchs, Switzerland). The reference compounds used (VX-950 [54], BILN 2061 [16], 2′-C-MeCyt [7, 10, 15], R1479 [15, 29, 52], the benzofuran HCV 796 [20], and the benzimidazole JT-16 [21]) were synthesized as reported previously. RBV was obtained from ICN Pharmaceuticals (Costa Mesa, CA).
Replicon-containing cell lines.
Huh 7 cells containing subgenomic HCV replicons I389/luc-ubi-neo/NS3-3′/5.1 (Huh 5-2 cells), I377/NS3-3′/wild type (Huh 9-13 cells), or I389/hygro-ubi-NS3-3′/5.1 (Huh mono cells) (5, 35, 36, 58), as well as human hepatoblastoma cells (HuH6 cells) (60) containing a genotype 1b subgenomic HCV replicon derived from the Con1 isolate, were kindly provided by Ralf Bartenschlager from the University of Heidelberg. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, Merelbeke, Belgium) supplemented with 10% heat-inactivated fetal bovine serum (Integro, Zaandam, The Netherlands), 1× nonessential amino acids (Gibco), 100 IU/ml penicillin (Gibco), and 100 μg/ml streptomycin (Gibco), as well as with 250 μg/ml Geneticin (G418; Gibco) for Huh 5-2 cells, 1 mg/ml G418 for Huh 9-13 cells and HuH6 cells, or 25 μg/ml hygromycin (Gibco) for Huh mono cells.
Antiviral assays.
HuH6 cells were seeded at a density of 1.5 × 104 cells and Huh 9-13 and Huh mono cells were seeded at a density of 5 × 103 cells per well in 96-well cell culture plates in complete culture medium supplemented with G418 or hygromycin. Following incubation for 24 h at 37°C (5% CO2), the cell culture medium was removed and serial dilutions of the test compounds were added in culture medium without G418 or hygromycin. After a 3-day incubation period at 37°C, the culture supernatant was removed, the monolayers were washed once with phosphate-buffered saline, and the cells were lysed in 100 μl Cells-to-cDNA lysis buffer (Ambion, Cambridgeshire, United Kingdom), according to the manufacturer's instruction. The lysates were transferred to 48-well PCR plates, and the plates were incubated for 15 min at 75°C, after which the plates were placed on ice and samples were diluted with 100 μl water. These lysates were used to determine the amount of HCV replicon RNA by means of quantitative real-time PCR (RT-qPCR). The 50% effective concentration (EC50) was calculated as the concentration of compound that caused a 50% reduction in HCV RNA levels compared to that of the untreated control. Serial dilutions of known quantities of a plasmid containing the neomycin gene were used to generate the standard curve. The amount of viral RNA produced in treated cultures was expressed as the percentage of that in the untreated control culture.
Anti-HCV assay with Huh 5-2 cells.
Huh 5-2 cells were seeded at a density of 5 × 103 per well in a tissue culture-treated white 96-well view plate (Canberra, Zellik, Belgium) in complete DMEM supplemented with 250 μg/ml G418. Following incubation for 24 h at 37°C (5% CO2), the medium was removed and threefold serial dilutions of the test compounds in complete DMEM (without G418) were added in a total volume of 100 μl. After 4 days of incubation at 37°C, the cell culture medium was removed and the luciferase activity was determined with a Steady-Glo luciferase assay system (Promega, Leiden, The Netherlands); the luciferase signal was measured with a Luminoskan Ascent system (Thermo, Vantaa, Finland). The EC50 was defined as the concentration of compound that reduced the luciferase signal by 50%.
RT-qPCR.
A 25-μl RT-qPCR mixture contained 12.5 μl 2× reaction buffer (Eurogentec, Seraing, Belgium), 6.3 μl H2O, and 5 μl total cellular RNA extract. In addition, for the Huh 9-13 and HuH6 cell samples, the reaction mixture contained 300 nmol/liter neo-forward primer (5′-CCG GCT ACC TGC CCA TTC-3′), 300 nmol/liter neo-reverse primer (5′-CCA GAT CAT CCT GAT CGA CAA G-3′), and 300 nmol/liter neo-probe (5′-6-carboxyfluorescein-ACA TCG CAT CGA GCG AGC ACG TAC-6-carboxytetramethylrhodamine-3′). For the Huh mono samples, the reaction mixture contained 300 nmol/liter UTR-forward primer (5′-ACG CAG AAA GCG TCT AGC CAT GGC GTT AGT-3′), 300 nmol/liter UTR-reverse primer (5′-TCC CGG GGC ACT CGC AAG CAC CCT ATC AGG-3′), and 300 nmol/liter UTR-probe (5′-6-carboxyfluorescein-TGG TCT GCG GAA CCG GTG AGT ACA CC-6-carboxytetramethylrhodamine-3′). The reverse transcription step was performed at 48°C for 30 min and 15 min at 95°C, and the subsequent PCR amplification consisted of 40 cycles of denaturation at 94°C for 20 s and annealing and extension at 60°C for 1 min in an ABI 7000 sequence detector.
Cytotoxicity assay.
To determine the cytotoxic activity of the compounds alone or in combination, the cells were handled in the same way as described for the antiviral assays. The cells were allowed to proliferate for 3 days at 37°C, after which the cell number was determined by means of the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium)/phenazinemethosulfate method (Promega). The 50% cytotoxic concentration was defined as the concentration that inhibited the proliferation of exponentially growing cells by 50%.
Analysis of combined drug efficacy.
The effects of drug-drug combinations were evaluated by the method of Prichard and Shipman (47). In brief, the theoretical additive effect was calculated from the dose-response curves for the individual compounds by the equation Z = X + Y(1 - X), where X represents the inhibition produced by either RBV, 2′-C-MeCyt, VX-950, or JT-16 alone and Y represents the inhibition by Debio 025 alone. Z represents the effect of either combination of compounds. The theoretical additive surface is subtracted from the actual experimental surface, resulting in a horizontal surface that equals the zero plane when the combination has an additive effect, a surface that lies above the zero plane when the combination has a synergistic effect, and a surface that lies below the zero plane when the combination has an antagonistic effect. The antiviral assay was carried out essentially as described above for HuH6 cells, except that the compounds were added in a checkerboard format.
HCV replicon clearance and rebound.
Huh 9-13 cells were seeded in a 25-cm2 T-flask at a density of 3 × 105 cells per culture flask in complete DMEM (without G418) that contained either no antiviral compounds or the compounds alone or in combination. The cells were grown until they reached 90% confluence (on average, about 4 days later). Subsequently, the cells were trypsinized and counted with a Coulter counter, after which 3 × 105 cells were seeded in a new 25 cm2 T-flask containing the same concentration of compound; 1.5 × 105 cells from each flask were lysed in RLT buffer (RNeasy Minikit; Qiagen, Venlo, The Netherlands) and were stored at −80°C until further use. In total, the cells were passaged seven consecutive times in the presence or the absence of compound. After all samples had been collected, the RNA was extracted according to the manufacturer's instructions and the samples were analyzed by RT-qPCR for their replicon contents.
Delayed resistance selection.
A total of 7.1 × 104 Huh 9-13 cells were plated in each well of a 12-well plate under G418 selection (1 mg/ml) in the presence of Debio 025 (0.5 μM or 0.1 μM) alone or in combination with BILN 2061, VX-950, JT-16, or R1479 at various concentrations or no antiviral compound. The culture medium, including the antiviral drug, was changed at least once a week. At the time that the cultures became confluent or a sufficiently large number of colonies had developed, the cells were further passaged under the same experimental conditions. At the end of the experiment (on average, after 4 weeks of culture) the cells were fixed and stained with Giemsa.
Selection of resistant replicon cell lines.
Drug-resistant replicons were generated by passaging HCV subgenomic replicon cells (Huh 9-13 cells) under G418 selection (1 mg/ml G418) in the presence of gradually increasing concentrations of the protease inhibitors BILN 2061 and VX-950, the NIs 4′-azidocytidine and 2′-C-MeCyt, Debio 025, or Cs.
RESULTS
Comparative in vitro activities of Debio 025 and other selective anti-HCV inhibitors.
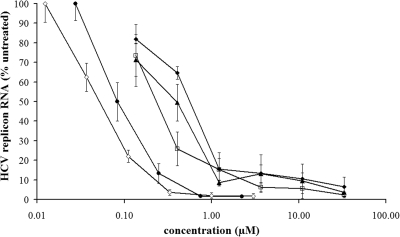
We previously reported that Debio 025 is a potent inhibitor of in vitro HCV replication and has antiviral activity that is on average 10-fold more pronounced than that of Cs (43). The in vitro anti-HCV activity of Debio 025 was compared to that of a number of reference anti-HCV molecules (in an HCV genotype 1b subgenomic replicon-containing cell line [Huh 9-13 cells]). Replicon-containing cells were treated with various concentrations of either Debio 025, Cs, the protease inhibitor VX-950, or the polymerase inhibitors 2′-C-MeCyt (an NI) or HCV 796 (an NNI). All compounds inhibited HCV replicon replication in a dose-dependent manner (Fig. 1). The mean EC50 of Debio 025 was 0.04 ± 0.03 μM, which is comparable to the antiviral activity of the benzofuran NNI HCV 796 (EC50, 0.09 ± 0.06 μM). Debio 025 proved markedly more potent than the protease inhibitor telaprevir (VX-950; EC50, 0.58 ± 0.10 μM) and the NI 2′-C-MeCyt, which exhibits antiviral activity (EC50) of 0.41 ± 0.16 μM. These data were confirmed with three other HCV subgenomic replicon systems (Huh 5-2, HuH6, and Huh mono cells); Debio 025 proved equipotent as HCV 796 and 10-fold more potent than telaprevir and 2′-C-MeCyt (data not shown).
FIG. 1.
Dose-dependent inhibition of HCV subgenomic replicon replication in Huh 9-13 cells by Debio 025 (open diamonds), Cs (open squares), VX-950 (closed diamonds), 2′-C-MeCyt (closed triangles), and HCV 796 (closed circles). Huh 9-13 cells were treated for 72 h with various concentrations of the compounds. The levels of HCV RNA were quantified by RT-qPCR and were expressed as the percentage of the level of HCV RNA for the untreated control cells. Data are the mean values of at least three independent experiments.
Drug combinations.
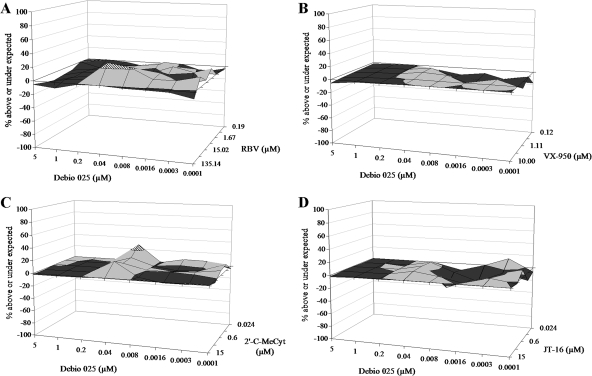
We previously reported an additive to slightly synergistic effect when Debio 025 was combined with IFN (43). In the present study, we also evaluated the effect of Debio 025 in combination with RBV in a 3-day antiviral assay. Because (i) RBV is a cytostatic agent and (ii) replicon replication is dependent on the proliferation of the host cells, in this analysis we employed HuH6 HCV replicon-containing cells. In this cell line, replicon replication is independent of ongoing cell proliferation (60). Thus, (semi)confluent HuH6 cells that are not (very) sensitive to the cytostatic activity of RBV (unlike exponentially proliferating Huh 5-2 or Huh 9-13 cells) were used for these experiments with Debio 025 in combination with RBV. Compounds were added in a checkerboard format, the RNA content was measured after 3 days by means of RT-qPCR, and the data were analyzed by the method of Prichard and Shipman (47). A slightly synergistic effect was noted at the highest concentrations of RBV, but overall, the antiviral activity of this combination appeared to be additive (Fig. 2A). The combination of Debio 025 with VX-950 (protease inhibitor), 2′-C-MeCyt (an NI), or JT-16 (an NNI) was studied next. Each combination resulted in an additive antiviral effect (with a slight tendency toward synergistic activity found at some concentrations) (Fig. 2B to D). No cytotoxicity was observed at the concentrations of the different compounds used, either alone or in combination (data not shown).
FIG. 2.
Antiviral effect of the combination of Debio 025 with RBV (A), the protease inhibitor VX-950 (B), the NI 2′-C-MeCyt (C), or the NNI JT-16 (D) in HuH6 cells. The zero plane on the z axis represents an additive effect, the volume above the zero plane indicates synergistic activity, and the volume below the zero plane indicates an antagonistic effect. The different shades represent different ranges of values: black, −20% to 0%; gray, 0% to 20%; dashed line, 20% to 40%. Data for the combinations Debio 025-RBV and Debio 025-2′-C-MeCyt are mean values for three independent experiments; data for the combinations of Debio 025-VX-950 and Debio 025-JT-16 are mean values for two independent experiments.
HCV replicon clearance and rebound.
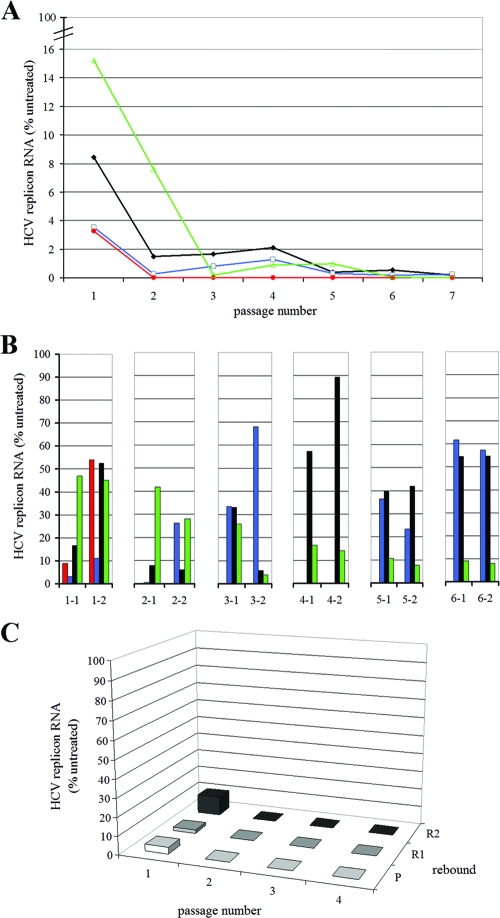
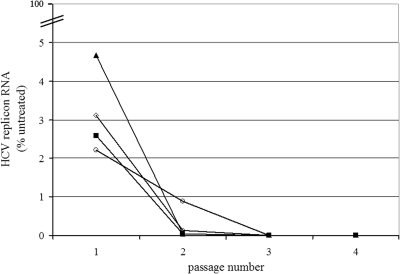
Since anti-HCV therapy, even with combinations of potent HCV inhibitors, will require or does require weeks or months of treatment, we studied the impact of prolonged Debio 025 treatment, either alone or in combination, on the replicon content of replicon-containing cells. For this purpose, Huh 9-13 subgenomic replicon cells were cultured (without G418 pressure) in the presence of the indicated concentrations of Debio 025, IFN, RBV, or VX-950 or were left untreated for seven consecutive passages (Fig. 3A). One parallel culture was grown in the absence of antiviral drug but under the selective pressure of G418. After seven passages, no significant difference in the HCV replicon content was noted between Huh 9-13 cells grown with G418 selective pressure and those grown without G418 selective pressure. After G418 selection was reinitiated, both cultures continued to grow normally, which indicates that the culturing of Huh 9-13 for seven passages without G418 pressure does not result in a drop in the replicon content. At every passage, a sample of the cells was taken to extract and quantify the total HCV RNA by means of RT-qPCR. The amount of RNA is expressed as a percentage of the amount of RNA from the control cells (the same passage of the untreated cells without G418 selection).
FIG. 3.
Clearance and rebound experiment with Huh 9-13 cells. (A) Cells were treated for seven consecutive passages with either 1 μg/ml (0.82 μM) Debio 025 (red), 5 μg/ml (7.06 μM) VX-950 (blue), 30 μg/ml (122.85 μM) RBV (green), or 100 IU/ml IFN (black) in the absence of selective G418 pressure. The HCV RNA content was quantified at every passage by means of RT-qPCR and is expressed as the percentage of the HCV RNA content of the untreated (control) sample at the same passage number. (B) During rebound, the compounds were omitted from the culture medium but the cells were again cultured under the selective pressure of G418 (1,000 μg/ml). Following every passage, two rebounds were inserted; on the x axis, the first number refers to the passage number, while the second number stands for the rebound number. Again, the HCV RNA content was quantified and expressed as a percentage of the HCV RNA content of the untreated (control) sample at the same passage and rebound number. (C) Cells were treated for four consecutive passages with 1 μg/ml (0.82 μM) Debio 025, 30 μg/ml (122.85 μM) RBV, and 100 IU/ml IFN in the absence of selective G418 pressure. During rebound, the compounds were omitted from the culture medium but the cells were again cultured under the selective pressure of G418 (1,000 μg/ml). Following every passage, two rebounds were inserted. P, passage; R1, rebound 1; R2, rebound 2.
Following one passage in the presence of either antiviral compound, a marked reduction of the replicon content of the cells was noted. The most pronounced reduction of the HCV replicon content was obtained with Debio 025 and VX-950 (on average, 96.50% inhibition). After the second passage and, for RBV, after the third passage, there was no marked further reduction in the replicon contents of the cells. To verify whether Huh 9-13 cells that had been treated with these compounds were indeed cleared of their replicons (and, if so, at which passage number), G418 selection (rebound) was initiated for every condition at every passage (Fig. 3B). Only cells that still carried the HCV replicon would be able to proliferate under G418 pressure. Only the Debio 025-treated Huh 9-13 cells died in the presence of G418 after three passages, indicating that complete clearance was obtained after three passages of treatment with Debio 025. All other cultures were still able to proliferate in the presence of G418 at every passage number, indicating that the cells were not cleared of their replicons. No significant cytotoxicity was noted during these experiments. When we combined Debio 025 with IFN and RBV at the same concentrations used in the previous experiment, complete clearance was already obtained after the second passage (Fig. 3C).
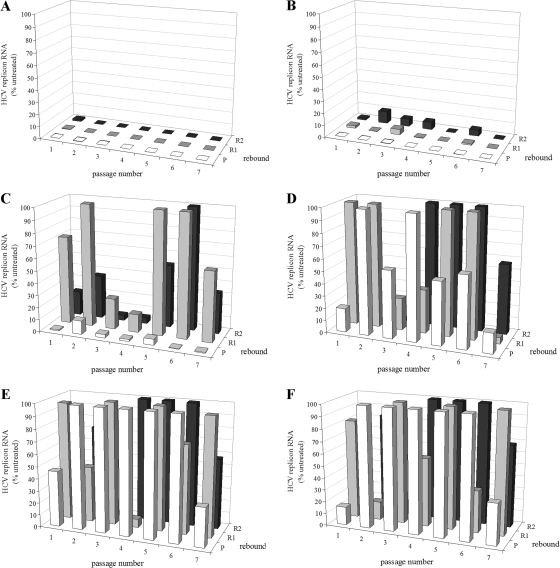
To determine the lowest possible concentration at which Debio 025 is still able to completely clear the cells of their replicons, clearance-rebound experiments were carried out with Debio 025 at concentrations ranging from 0.50 to 0.02 μg/ml (0.41 to 0.01 μM). Replicon-containing cells were cultured for seven consecutive passages in the presence of Debio 025. A concentration of 0.25 μg/ml (0.21 μM) was identified as that which was still able to result in complete clearance after six passages (Fig. 4). At passage 8, the viral RNA levels were also below the detection limit and no rebound was possible (data not shown).
FIG. 4.
Clearance experiment with Huh 9-13 cells and various concentrations of Debio 025. Cells were treated for seven consecutive passages, in the absence of G418 selective pressure, with the following twofold dilutions of Debio 025: 0.50 μg/ml (0.41 μM) (A), 0.25 μg/ml (0.21 μM) (B), 0.125 μg/ml (0.10 μM) (C), 0.063 μg/ml (0.05 μM) (D), 0.031 μg/ml (0.03 μM) (E), or 0.016 μg/ml (0.01 μM) (F). During rebound, the compounds were omitted from the culture medium but the cells were again cultured under the selective pressure of G418 (1,000 μg/ml). Following every passage, two rebounds were inserted. P, passage; R1, rebound 1; R2, rebound 2. The levels of the remaining HCV RNA were determined by means of RT-qPCR at every passage (and the accompanying rebound passages) and are expressed as a percentage of the amount of total HCV RNA in the untreated (control) cells of the same passage number.
The effect of the combination of Debio 025 with telaprevir on clearance was studied next. The cells were treated with various concentrations of Debio 025 (1 to 0.125 μg/ml [0.82 to 0.10 μM]) and a fixed concentration of VX-950 (5 μg/ml [7.06 μM]). The use of the lowest concentration of Debio 025 tested (i.e., 0.125 μg/ml [0.10 μM]) together with 5 μg/ml (7.06 μM) VX-950 still resulted in complete clearance of the replicon from the host cells after only two passages (Fig. 5) (the rebound of replicon replication was not possible from this culture [data not shown]), whereas either compound alone was not able to completely clear the replicon from the host cells at the indicated concentrations, not even following seven passages under antiviral pressure.
FIG. 5.
Clearance experiment with Huh 9-13 cells and various concentrations of Debio 025 in combination with VX-950. Cells were treated for four consecutive passages with 5 μg/ml (7.06 μM) VX-950 in combination with Debio 025 at either 1 μg/ml (0.82 μM) (open diamonds), 0.5 μg/ml (0.41 μM) (closed squares), 0.25 μg/ml (0.21 μM) (open circles), or 0.125 μg/ml (0.10 μM) (closed triangles) in the absence of G418 selective pressure. The quantity of HCV RNA was measured at every passage by means of RT-qPCR and was normalized against the amount of total RNA extracted from the untreated (control) sample at the same passage number.
Delayed resistance selection.
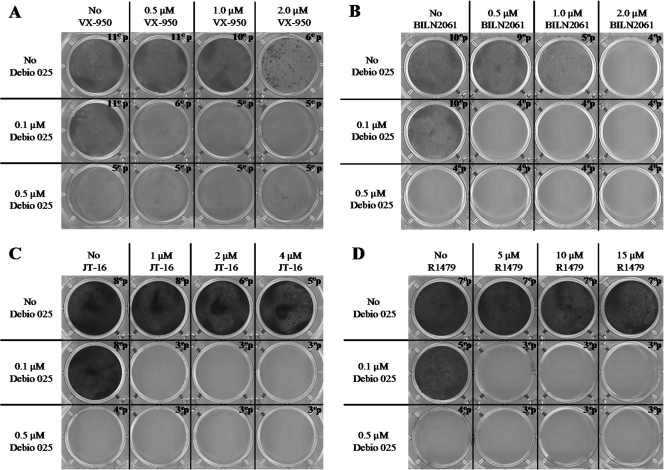
To study whether Debio 025 is able to delay or prevent the emergence of development of HCV replicon resistance to HCV protease inhibitors (BILN 2061 and VX-950), an HCV NI (R1479), or an NNI (JT-16), Huh 9-13 cells were cultured in the presence of BILN 2061, VX-950, R1479, or JT-16 at the indicated concentrations alone or in combination with either 0.1 μM or 0.5 μM Debio 025 (Fig. 6). After several passages, Huh 9-13 cells that replicated in the presence of VX-950 at a concentration of 2 μM (∼4-fold the EC50) or BILN 2061 at a concentration of 1 μM (∼50-fold the EC50) were selected (six and five passages, respectively). Following seven and five passages, Huh 9-13 cells that replicated in the presence of 15 μM R1479 (∼8-fold the EC50) or 4 μM JT-16 (∼3-fold the EC50) were selected, respectively. No colonies of cells were able to grow under the selective pressure of 0.5 μM Debio 025. Replicon-containing cells continued to replicate in the presence of 0.1 μM Debio 025. However, under none of the conditions in which 0.1 μM Debio 025 was combined with BILN 2061, VX-950, R1479, or JT-16 were drug-resistant replicons selected.
FIG. 6.
Colony formation assay. Huh 9-13 cells were treated with Debio 025 alone or in combination with VX-950 (A), BILN 2061 (B), JT-16 (C), or R1479 (D) at the indicated concentrations in the presence of 1,000 μg/ml G418. At the time that the cultures became confluent or a sufficiently large number of colonies had developed, the cells were further passaged under the same experimental conditions. Following 4 weeks of culture, on average, the cells were fixed and stained with Giemsa.
Debio 025 remains active against replicons resistant to various STAT-C inhibitors.
The antiviral activity of Debio 025 was determined with a panel of Huh 9-13 replicons resistant to either Debio 025, Cs, 2′-C-MeCyt, R1479, BILN 2061, or VX-950 (Table 1). Debio 025 and Cs retained wild-type activity against the various polymerase- and protease-resistant replicons. Likewise, 2′-C-MeCyt, R1479, VX-950, and BILN 2061 proved equipotent against both wild-type as well as Debio 025-resistant (Debio 025r) and Cs-resistant (Csr) replicon cell lines. No cross-resistance between Debio 025 and the selection of specifically targeted antiviral therapy for HCV (STAT-C) inhibitors tested was thus observed.
TABLE 1.
Antiviral activities of Debio 025, Cs, and STAT-C inhibitors against a set of resistant replicons
| Agent | EC50 (μM)a
|
||||||
|---|---|---|---|---|---|---|---|
| WT | Debio 025r | CsAr | 2′-C-MeCytr | R1479r | BILN 2061r | VX-950r | |
| Debio 025 | 0.04 ± 0.03 | 2.72 ± 1.22 | 0.35 ± 0.27 | 0.11 ± 0.05 | 0.12 ± 0.01 | 0.1 ± 0.003 | 0.09 ± 0.01 |
| Cs | 0.29 ± 0.05 | 3.01 ± 1.88 | 3.85 ± 0.71 | 0.21 ± 0.04 | 0.26 ± 0.05 | 0.17 ± 0.06 | ND |
| 2′-C-MeCyt | 0.41 ± 0.16 | 0.41 ± 0.53 | 0.40 ± 0.40 | 30.15 | 3.38 ± 1.07 | 0.29 ± 0.18 | 1.02 ± 0.70 |
| R1479 | 2.95 ± 0.88 | 2.76 ± 0.89 | 1.98 ± 0.50 | 1.16 ± 0.43 | >15.65 | 2.16 ± 0.27 | ND |
| BILN 2061 | 0.02 ± 0.01 | <0.004 | 0.004 | 0.04 ± 0.025 | 0.02 ± 0.01 | 1.25 ± 0.47 | 0.79 ± 0.08 |
| VX-950 | 0.58 ± 0.10 | 0.36 ± 0.04 | 0.49 ± 0.32 | 1.01 ± 0.32 | 0.69 ± 0.15 | 0.31 ± 0.05 | 13.97 ± 1.43 |
Data are the mean values of at least three independent experiments. WT, wild type; ND, not determined.
DISCUSSION
We previously reported that the nonimmunosuppressive Cs analogue Debio 025 is a potent inhibitor of HCV replication (43). In the present study, we elaborate on the specific characteristics of the anti-HCV effect of this molecule.
The development of STAT-C inhibitors has so far mainly focused on the NS3 protease and the NS5B polymerase. As shown here, Debio 025 proved to be an exquisitely potent inhibitor of HCV replication in vitro; the drug was roughly 10-fold more potent than telaprevir and 2′-C-MeCyt and proved to be equipotent as the NNI HCV 796.
It has now become generally accepted that STAT-C inhibitors, in the first years after they become available, will likely have to be combined with the current standard therapy (30). At a later stage, two or more potent STAT-C inhibitors with different resistance profiles may, it is hoped, be combined without further need for IFN-RBV combination therapy. It is important to obtain information about the antiviral efficacies of particular combinations. For this reason, we studied the antiviral activity of Debio 025 when it was combined with several other HCV inhibitors. To this end, two different assay systems were employed. First, the antiviral activities of the combinations were evaluated in regular antiviral assays, i.e., with a readout obtained after 3 days of incubation of the replicon-containing cells with the antiviral drug(s). We reported earlier on the combined activity of Debio 025 and IFN (43). In the present study, we combined Debio 025 with RBV, the protease inhibitor VX-950, the NI 2′-C-MeCyt, and the NNI JT-16. All the combinations resulted in additive antiviral activities, and this result was in line with our expectation that the use of compounds that (most likely) do not interfere with each others’ metabolism or mechanism of action should result in an additive effect. The use of combinations of compounds that interfere with each others’ biological activity may result in either synergistic or antagonistic antiviral activity. For example, RBV is able to potentiate the antiherpesvirus and anti-HBV activities of purine-based deoxynucleoside analogues, but the combination of RBV and pyrimidine ribonucleoside analogues results in an antagonistic effect against HIV and HCV replication (4, 11, 44, 57, 63). The specific interference of RBV with purine and pyrimidine metabolism could explain these observations. The mechanism by which RBV exerts its activity against HCV remains elusive. Several hypotheses have been proposed, such as (i) the direct inhibition of the HCV RdRp by the 5′-triphosphate metabolite of RBV, (ii) depletion of intracellular GTP pools through the inhibition of IMP dehydrogenase by the 5′-monophosphate metabolite of RBV, (iii) mutagenic properties that lead to “error catastrophe,” or (iv) immunomodulatory properties which alter the T-helper cytokine balance from a Th2 profile to a more antiviral Th1 profile (9). The previously described antagonistic or synergistic activities were not noted when Debio 025 was combined with either RBV or other selective inhibitors of HCV replication. However, a short antiviral assay may not necessarily predict the in vivo antiviral effect of the compounds used either alone or in combination. We therefore conducted clearance-rebound assays. We already reported that Debio 025 alone is able to clear the replicon from replicon-containing cells (43). We now mapped in more detail the potential of Debio 025 to clear cells of their replicons. Even though IFN, RBV, and VX-950 all produced a drop in the HCV replicon content after the first passages, only Debio 025 succeeded in completely curing the replicon-containing cells following only three passages of antiviral pressure. The lowest concentration of Debio 025 at which clearance (at passage 6) was obtained was 0.25 μg/ml (0.21 μM). Debio 025 in combination with the other anti-HCV drugs studied was also particularly efficient in curing replicon-containing cells of their replicons. Debio 025 at a concentration as low as 0.125 μg/ml (0.10 μM) was able to clear the cells when it was combined with VX-950 at a fixed concentration of 5 μg/ml (7.06 μM), whereas these concentrations of each compound used alone were not able to result in a complete clearance. Surprisingly, and despite the fact that no synergistic activity was noted in a regular 3-day antiviral assay, this combination efficiently cleared the cells of their replicons. Clearance-rebound experiments may possibly have predictive value for estimating the potential of drugs (or combinations thereof) to clear liver cells of replicating virus. The use of combinations of Debio 025 with the standard of care (IFN and/or RBV) or with the specifically targeting anti-HCV inhibitor VX-950 may have the potential to achieve rapid clearance. Our data also indicate that regular short-term (in this study, 3 days) antiviral combination assays (in replicon-based systems) may not necessarily predict synergistic antiviral effects following long-term culture.
In the development of STAT-C inhibitors, drug resistance forms a major issue. The use of combinations of antiviral compounds that have different targets can be effective in preventing the emergence of drug-resistant viruses, as has been very well documented for HIV (62). Our data suggest that Debio 025 may have a high barrier to the development of resistance in patients. Resistance does occur in the replicon, but it is a slow process (our unpublished data). Use of the combination of Debio 025 with standard therapy or new STAT-C inhibitors should help to prevent the emergence of resistance. In vitro, Debio 025 remained active against various HCV protease- and polymerase (nucleoside and nonnucleoside)-resistant replicons. Conversely, these STAT-C inhibitors retained the activity that they had against wild-type cell lines against Debio 025-resistant replicon cell lines. No cross-resistance between Debio 025 and the selection of protease and polymerase inhibitors used in this study was observed. Remarkably, concentrations of Debio 025 as low as 0.1 μM were able to delay or prevent the development of resistance to NS3 protease inhibitors (BILN 2061 and VX-950) and polymerase (nucleoside and nonnucleoside) inhibitors. These data suggest that Debio 025 may be an excellent drug for use in combination therapy. The in vitro selection of Debio 025-resistant replicon cell lines proved to be a lengthy process (our unpublished data), which can likely be explained by the fact that host cell factors (cyclophilins) may be involved in the mechanism by which Debio 025 inhibits HCV replication. Evidence that cyclophilin A has a crucial role in HCV replication has been reported (27, 61). Since Debio 025 binds to cyclophilins, including cyclophilin A (48), it may be hypothesized that Debio 025 prevents a crucial interaction between cyclophylin A and the HCV replication complex.
Our data indicate that Debio 025 is endowed with exquisite potency against HCV, particularly in terms of its ability to clear the host cell of its HCV replicon. This curative capacity can be further exploited by the use of Debio 025 in combination with other STAT-C inhibitors. Combinations of low concentrations of Debio 025 with specific STAT-C inhibitors also prevent the development of STAT-C inhibitor-resistant variants. Furthermore, resistance to Debio 025 appeared to be a slow process and Debio 025 was not cross-resistant with NS3 protease inhibitors or various polymerase inhibitors. These data, together with the very promising clinical data, prove that Debio 025 is an attractive candidate for the treatment of HCV infection.
Acknowledgments
We thank Katrien Geerts for excellent technical assistance.
This work is part of the activities of the VIRGIL European Network of Excellence on Antiviral Drug Resistance, supported by a grant (LSHM-CT-2004-503359) from the Priority 1 Life Sciences, Genomics and Biotechnology for Health Programme in the 6th Framework Programme of the EU. Jan Paeshuyse is supported by a postdoctoral position of the Fonds voor Wetenschappelijk Onderzoek, Vlaanderen.
Footnotes
Published ahead of print on 22 December 2008.
REFERENCES
- 1.Anonymous. 2008. Pharmasset announces R7128 achieves 85% rapid virologic response in a 4-week combination study for the treatment of chronic hepatitis. Pharmasett, Princeton, NJ. http://investor.pharmasset.com/releasedetail.cfm?ReleaseID=284920.
- 2.Anonymous. 2008. InterMune announces continuing progress on ITMN-191 (R7227). InterMune, Brisbane, CA. http://phx.corporate-ir.net/phoenix.zhtml?c=100067&p=irol-newsArticle&ID=1092155&highlight=.
- 3.Anonymous. 2007. Initial results of phase II study with HCV protease inhibitor boceprevir in treatment-naive hepatitis C patients show a high rate of early virologic response. Schering-Plough, Kenilworth, NJ. http://www.schering-plough.com/schering_plough/news/release.jsp?releaseID=1064540.
- 4.Baba, M., R. Pauwels, J. Balzarini, P. Herdewijn, E. De Clercq, and J. Desmyter. 1987. Ribavirin antagonizes inhibitory effects of pyrimidine 2′,3′-dideoxynucleosides but enhances inhibitory effects of purine 2′,3′-dideoxynucleosides on replication of human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 31:1613-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, S. S., and D. B. Olsen. 2006. Nucleoside analog inhibitors of hepatitis C virus replication. Infect. Disord. Drug Targets 6:17-29. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De-Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 8.Chatterji, U., M. D. Bobardt, R. Stanfield, R. G. Ptak, L. A. Pallansch, P. A. Ward, M. J. Jones, C. A. Stoddart, P. Scalfaro, J. M. Dumont, K. Besseghir, B. Rosenwirth, and P. A. Gallay. 2005. Naturally occurring capsid substitutions render HIV-1 cyclophilin A independent in human cells and trim-cyclophilin resistant in owl monkey cells. J. Biol. Chem. 280:40293-40300. [DOI] [PubMed] [Google Scholar]
- 9.Chung, R. T., M. Gale, Jr., S. J. Polyak, S. M. Lemon, T. J. Liang, and J. H. Hoofnagle. 2008. Mechanisms of action of interferon and ribavirin in chronic hepatitis C: summary of a workshop. Hepatology 47:306-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, J. L., J. C. Mason, L. Hollecker, L. J. Stuyver, P. M. Tharnish, T. R. McBrayer, M. J. Otto, P. A. Furman, R. F. Schinazi, and K. A. Watanabe. 2006. Synthesis and antiviral activity of 2′-deoxy-2′-fluoro-2′-C-methyl purine nucleosides as inhibitors of hepatitis C virus RNA replication. Bioorg. Med. Chem. Lett. 16:1712-1715. [DOI] [PubMed] [Google Scholar]
- 11.Coelmont, L., J. Paeshuyse, M. P. Windisch, E. De Clercq, R. Bartenschlager, and J. Neyts. 2006. Ribavirin antagonizes the in vitro anti-hepatitis C virus activity of 2′-C-methylcytidine, the active component of valopicitabine. Antimicrob. Agents Chemother. 50:3444-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craxi, A., and A. Licata. 2003. Clinical trial results of peginterferons in combination with ribavirin. Semin. Liver Dis. 23(Suppl. 1):35-46. [DOI] [PubMed] [Google Scholar]
- 13.De Clercq, E., and J. Neyts. 2009. Antiviral agents acting as DNA or RNA chain terminators, p. 53-84. In H. G. Kräusslich and R. Bartenschlager (ed.), Handbook of experimental pharmacology, vol. 189. Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 14.De Francesco, R., and G. Migliaccio. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436:953-960. [DOI] [PubMed] [Google Scholar]
- 15.Eldrup, A. B., C. R. Allerson, C. F. Bennett, S. Bera, B. Bhat, N. Bhat, M. R. Bosserman, J. Brooks, C. Burlein, S. S. Carroll, P. D. Cook, K. L. Getty, M. MacCoss, D. R. McMasters, D. B. Olsen, T. P. Prakash, M. Prhavc, Q. Song, J. E. Tomassini, and J. Xia. 2004. Structure-activity relationship of purine ribonucleosides for inhibition of hepatitis C virus RNA-dependent RNA polymerase. J. Med. Chem. 47:2283-2295. [DOI] [PubMed] [Google Scholar]
- 16.Faucher, A. M., M. D. Bailey, P. L. Beaulieu, C. Brochu, J. S. Duceppe, J. M. Ferland, E. Ghiro, V. Gorys, T. Halmos, S. H. Kawai, M. Poirier, B. Simoneau, Y. S. Tsantrizos, and M. Llinas-Brunet. 2004. Synthesis of BILN 2061, an HCV NS3 protease inhibitor with proven antiviral effect in humans. Org. Lett. 6:2901-2904. [DOI] [PubMed] [Google Scholar]
- 17.Flisiak, R., S. V. Feinman, M. Jablkowski, A. Horban, W. Kryczka, W. Halota, J. E. Heathcote, G. Mazzella, C. Vandelli, J. S. Liz, R. Crabbé, P. Scalfaro, and H. Porchet. 2008. Efficacy and safety of increasing doses of the cyclophilin inhibitor Debio-025 in combination with pegylated interferon alpha-2a in treatment naïve chronic HCV patients. J. Hepatol. 48(Suppl. 2):S62. [Google Scholar]
- 18.Flisiak, R., J. M. Dumont, and R. Crabbe. 2007. Cyclophilin inhibitors in hepatitis C viral infection. Expert. Opin. Investig. Drugs 16:1345-1354. [DOI] [PubMed] [Google Scholar]
- 19.Flisiak, R., A. Horban, P. Gallay, M. Bobardt, S. Selvarajah, A. Wiercinska-Drapalo, E. Siwak, I. Cielniak, J. Higersberger, J. Kierkus, C. Aeschlimann, P. Grosgurin, V. Nicolas-Metral, J. M. Dumont, H. Porchet, R. Crabbe, and P. Scalfaro. 2008. The cyclophilin inhibitor Debio-025 shows potent anti-hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology 47:817-826. [DOI] [PubMed] [Google Scholar]
- 20.Gopalsamy, A., A. Aplasca, G. Ciszewski, K. Park, J. W. Ellingboe, M. Orlowski, B. Feld, and A. Y. Howe. 2006. Design and synthesis of 3,4-dihydro-1H-[1]-benzothieno[2,3-c]pyran and 3,4-dihydro-1H-pyrano[3,4-b]benzofuran derivatives as non-nucleoside inhibitors of HCV NS5B RNA dependent RNA polymerase. Bioorg. Med. Chem. Lett. 16:457-460. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto, H., K. Mizutani, and A. Yoshida. 2005. Fused cyclic compounds and medicinal use thereof. Report WO/2003/000254. World Intellectual Property Organization, Geneva, Switzerland.
- 22.Hinrichsen, H., Y. Benhamou, H. Wedemeyer, M. Reiser, R. E. Sentjens, J. L. Calleja, X. Forns, A. Erhardt, J. Cronlein, R. L. Chaves, C. L. Yong, G. Nehmiz, and G. G. Steinmann. 2004. Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology 127:1347-1355. [DOI] [PubMed] [Google Scholar]
- 23.Houck, D. R., and S. Hopkins. 2006. Preclinical evaluation of SCY-635, a cyclophilin inhibitor with potent anti-HCV activity. Hepatology 44:534A. [Google Scholar]
- 24.Hubler, F., T. Rückle, L. Patiny, T. Muamba, J. F. Guichou, M. Mutter, and R. Wenger. 2000. Synthetic routes to NEtXaa4-cyclosporin A derivates as potential anti-HIV I drugs. Tetrahedron Lett. 41:7193-7196. [Google Scholar]
- 25.Inoue, K., K. Sekiyama, M. Yamada, T. Watanabe, H. Yasuda, and M. Yoshiba. 2003. Combined interferon alpha2b and cyclosporin A in the treatment of chronic hepatitis C: controlled trial. J. Gastroenterol. 38:567-572. [DOI] [PubMed] [Google Scholar]
- 26.Inoue, K., T. Umehara, U. T. Ruegg, F. Yasui, T. Watanabe, H. Yasuda, J. M. Dumont, P. Scalfaro, M. Yoshiba, and M. Kohara. 2007. Evaluation of a cyclophilin inhibitor in hepatitis C virus-infected chimeric mice in vivo. Hepatology 45:921-928. [DOI] [PubMed] [Google Scholar]
- 27.Kaul, A., S. Stauffer, J. Schmitt, T. Pertel, J. Luban, and R. Bartenschlager. 2008. Role of cyclophilins in hepatitis C virus replication, abstr. P-218. Abstr. 15th Int. Symp. Hepatitis C Virus Related Viruses.
- 28.Klumpp, K., G. Kalayanov, H. Ma, S. Le Pogam, V. Leveque, W. R. Jiang, N. Inocencio, A. De Witte, S. Rajyaguru, E. Tai, S. Chanda, M. R. Irwin, C. Sund, A. Winqist, T. Maltseva, S. Eriksson, E. Usova, M. Smith, A. Alker, I. Najera, N. Cammack, J. A. Martin, N. G. Johansson, and D. B. Smith. 2008. 2′-Deoxy-4′-azido nucleoside analogs are highly potent inhibitors of HCV replication despite the lack of 2′-alpha hydroxyl groups. J. Biol. Chem. 283:2167-2175. [DOI] [PubMed] [Google Scholar]
- 29.Klumpp, K., V. Leveque, S. Le Pogam, H. Ma, W. R. Jiang, H. Kang, C. Granycome, M. Singer, C. Laxton, J. Q. Hang, K. Sarma, D. B. Smith, D. Heindl, C. J. Hobbs, J. H. Merrett, J. Symons, N. Cammack, J. A. Martin, R. Devos, and I. Najera. 2006. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 281:3793-3799. [DOI] [PubMed] [Google Scholar]
- 30.Kwong, A. D., S. Cowherd, and P. Mueller. 2006. Beyond interferon and ribavirin: antiviral therapies for hepatitis C virus. Drug Discov. Today Ther. Strategies 3:211-220. [Google Scholar]
- 31.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 32.Le Pogam, S., H. Kang, S. F. Harris, V. Leveque, A. M. Giannetti, S. Ali, W. R. Jiang, S. Rajyaguru, G. Tavares, C. Oshiro, T. Hendricks, K. Klumpp, J. Symons, M. F. Browner, N. Cammack, and I. Najera. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, C., C. A. Gates, B. G. Rao, D. L. Brennan, J. R. Fulghum, Y. P. Luong, J. D. Frantz, K. Lin, S. Ma, Y. Y. Wei, R. B. Perni, and A. D. Kwong. 2005. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J. Biol. Chem. 280:36784-36791. [DOI] [PubMed] [Google Scholar]
- 34.Lin, C., K. Lin, Y. P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 35.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 37.Ma, S., J. E. Boerner, C. TiongYip, B. Weidmann, N. S. Ryder, M. P. Cooreman, and K. Lin. 2006. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob. Agents Chemother. 50:2976-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manns, M. P., G. R. Foster, J. K. Rockstroh, S. Zeuzem, F. Zoulim, and M. Houghton. 2007. The way forward in HCV treatment—finding the right path. Nat. Rev. Drug Discov. 6:991-1000. [DOI] [PubMed] [Google Scholar]
- 39.Modi, A. A., and J. H. Hoofnagle. 2007. New therapies for hepatitis C. Hepatology 46:615-617. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa, M., N. Sakamoto, N. Enomoto, Y. Tanabe, N. Kanazawa, T. Koyama, M. Kurosaki, S. Maekawa, T. Yamashiro, C. H. Chen, Y. Itsui, S. Kakinuma, and M. Watanabe. 2004. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem. Biophys. Res. Commun. 313:42-47. [DOI] [PubMed] [Google Scholar]
- 41.Nakagawa, M., N. Sakamoto, Y. Tanabe, T. Koyama, Y. Itsui, Y. Takeda, C. H. Chen, S. Kakinuma, S. Oooka, S. Maekawa, N. Enomoto, and M. Watanabe. 2005. Suppression of hepatitis C virus replication by cyclosporin A is mediated by blockade of cyclophilins. Gastroenterology 129:1031-1041. [DOI] [PubMed] [Google Scholar]
- 42.Neyts, J. 2006. Selective inhibitors of hepatitis C virus replication. Antivir. Res. 71:363-371. [DOI] [PubMed] [Google Scholar]
- 43.Paeshuyse, J., A. Kaul, E. De Clercq, B. Rosenwirth, J. M. Dumont, P. Scalfaro, R. Bartenschlager, and J. Neyts. 2006. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology 43:761-770. [DOI] [PubMed] [Google Scholar]
- 44.Pancheva, S. N. 1991. Potentiating effect of ribavirin on the anti-herpes activity of acyclovir. Antivir. Res. 16:151-161. [DOI] [PubMed] [Google Scholar]
- 45.Perni, R. B., S. J. Almquist, R. A. Byrn, G. Chandorkar, P. R. Chaturvedi, L. F. Courtney, C. J. Decker, K. Dinehart, C. A. Gates, S. L. Harbeson, A. Heiser, G. Kalkeri, E. Kolaczkowski, K. Lin, Y. P. Luong, B. G. Rao, W. P. Taylor, J. A. Thomson, R. D. Tung, Y. Wei, A. D. Kwong, and C. Lin. 2006. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 50:899-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pierra, C., A. Amador, S. Benzaria, E. Cretton-Scott, M. D'Amours, J. Mao, S. Mathieu, A. Moussa, E. G. Bridges, D. N. Standring, J. P. Sommadossi, R. Storer, and G. Gosselin. 2006. Synthesis and pharmacokinetics of valopicitabine (NM283), an efficient prodrug of the potent anti-HCV agent 2′-C-methylcytidine. J. Med. Chem. 49:6614-6620. [DOI] [PubMed] [Google Scholar]
- 47.Prichard, M. N., and C. Shipman, Jr. 1990. A three-dimensional model to analyze drug-drug interactions. Antivir. Res. 14:181-205. [DOI] [PubMed] [Google Scholar]
- 48.Ptak, R. G., P. A. Gallay, D. Jochmans, A. P. Halestrap, U. T. Ruegg, L. A. Pallansch, M. D. Bobardt, M. P. de Bethune, J. Neyts, E. De Clercq, J. M. Dumont, P. Scalfaro, K. Besseghir, R. M. Wenger, and B. Rosenwirth. 2008. Inhibition of human immunodeficiency virus type 1 replication in human cells by Debio-025, a novel cyclophilin binding agent. Antimicrob. Agents Chemother. 52:1302-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenwirth, B., A. Billich, R. Datema, P. Donatsch, F. Hammerschmid, R. Harrison, P. Hiestand, H. Jaksche, P. Mayer, and P. Peichl. 1994. Inhibition of human immunodeficiency virus type 1 replication by SDZ NIM 811, a nonimmunosuppressive cyclosporine analog. Antimicrob. Agents Chemother. 38:1763-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarrazin, C., R. Rouzier, F. Wagner, N. Forestier, D. Larrey, S. K. Gupta, M. Hussain, A. Shah, D. Cutler, J. Zhang, and S. Zeuzem. 2007. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology 132:1270-1278. [DOI] [PubMed] [Google Scholar]
- 51.Simmen, K., O. Lenz, T. S. Lin, G. Fanning, P. Raboisson, H. de Kock, H. G. van't Klooster, A. Rosenquist, M. Edlund, M. Nilsson, L. Vrang, and B. Samuelsson. 2007. In vitro activity and preclinical pharmacokinetics of the HCV protease inhibitor, TMC435350. Hepatology 46:857A. [Google Scholar]
- 52.Smith, D. B., J. A. Martin, K. Klumpp, S. J. Baker, P. A. Blomgren, R. Devos, C. Granycome, J. Hang, C. J. Hobbs, W. R. Jiang, C. Laxton, S. Le Pogam, V. Leveque, H. Ma, G. Maile, J. H. Merrett, A. Pichota, K. Sarma, M. Smith, S. Swallow, J. Symons, D. Vesey, I. Najera, and N. Cammack. 2007. Design, synthesis, and antiviral properties of 4′-substituted ribonucleosides as inhibitors of hepatitis C virus replication: the discovery of R1479. Bioorg. Med. Chem. Lett. 17:2570-2576. [DOI] [PubMed] [Google Scholar]
- 53.Stuyver, L. J., T. R. McBrayer, P. M. Tharnish, J. Clark, L. Hollecker, S. Lostia, T. Nachman, J. Grier, M. A. Bennett, M. Y. Xie, R. F. Schinazi, J. D. Morrey, J. L. Julander, P. A. Furman, and M. J. Otto. 2006. Inhibition of hepatitis C replicon RNA synthesis by beta-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine: a specific inhibitor of hepatitis C virus replication. Antivir. Chem. Chemother. 17:79-87. [DOI] [PubMed] [Google Scholar]
- 54.Summa, V. 2005. VX-950 (Vertex/Mitsubishi). Curr. Opin. Investig. Drugs 6:831-837. [PubMed] [Google Scholar]
- 55.Timm, J., and M. Roggendorf. 2007. Sequence diversity of hepatitis C virus: implications for immune control and therapy. World J. Gastroenterol. 13:4808-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verloes, R., K. A. Farha, A. van Vliet, G. van't Klooster, F. Aharchi, K. Marien, H. de Kock, and K. Simmen. 2007. Results of a phase 1 placebo-controlled trial in healthy volunteers to examine the safety, tolerability and pharmacokinetics of the HCV protease inhibitor TMC435350 after single and repeated dosing. Hepatology 46:823A.17680645 [Google Scholar]
- 57.Vogt, M. W., K. L. Hartshorn, P. A. Furman, T. C. Chou, J. A. Fyfe, L. A. Coleman, C. Crumpacker, R. T. Schooley, and M. S. Hirsch. 1987. Ribavirin antagonizes the effect of azidothymidine on HIV replication. Science 235:1376-1379. [DOI] [PubMed] [Google Scholar]
- 58.Vrolijk, J. M., A. Kaul, B. E. Hansen, V. Lohmann, B. L. Haagmans, S. W. Schalm, and R. Bartenschlager. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201-209. [DOI] [PubMed] [Google Scholar]
- 59.Watashi, K., M. Hijikata, M. Hosaka, M. Yamaji, and K. Shimotohno. 2003. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38:1282-1288. [DOI] [PubMed] [Google Scholar]
- 60.Windisch, M. P., M. Frese, A. Kaul, M. Trippler, V. Lohmann, and R. Bartenschlager. 2005. Dissecting the interferon-induced inhibition of hepatitis C virus replication by using a novel host cell line. J. Virol. 79:13778-13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, F., J. M. Robotham, H. B. Nelson, A. Irsigler, R. Kenworthy, and H. Tang. 2008. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 82:5269-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeni, P. 2006. Update on HAART in HIV. J. Hepatol. 44(Suppl. 1):S100-S103. [DOI] [PubMed] [Google Scholar]
- 63.Ying, C., E. De Clercq, and J. Neyts. 2000. Ribavirin and mycophenolic acid potentiate the activity of guanine- and diaminopurine-based nucleoside analogues against hepatitis B virus. Antivir. Res. 48:117-124. [DOI] [PubMed] [Google Scholar]
- 64.Zoulim, F., M. Chevallier, M. Maynard, and C. Trepo. 2003. Clinical consequences of hepatitis C virus infection. Rev. Med. Virol. 13:57-68. [DOI] [PubMed] [Google Scholar]