Abstract
Chromosomally encoded ß-lactamases from the Burkholderia cepacia complex species (formerly Pseudomonas cepacia) were characterized. Cloning and sequencing identified an Ambler class A ß-lactamase (PenB) from B. cenocepacia. It shares 82% amino acid identity with the PenA ß-lactamases previously identified from B. multivorans 249. Its expression was dependent upon a LysR-type regulatory protein. Its narrow-spectrum hydrolysis activity mostly included penicillins but also included expanded-spectrum cephalosporins and aztreonam at lower levels. In that study, Pen-like ß-lactamases (PenC, PenD, PenE, PenF) that shared 63 to 92% identity with PenB from B. cenocepacia were identified from other Burkholderia species. The corresponding ß-lactamase genes might be used as genetic tools for accurate Burkholderia species identification.
Over the last two decades, Burkholderia cepacia (previously Pseudomonas cepacia) has been recognized as an ubiquitous and opportunistic pathogen of increasing importance, particularly in nosocomial infections in immunocompromised hosts and cystic fibrosis (CF) patients (20, 30, 32, 35). The Burkholderia cepacia complex is divided into at least 10 different closely related species: B. cepacia, B. multivorans, B. cenocepacia, B. stabilis, B. vietnamiensis, B. dolosa, B. ambifaria, B. anthina, B. pyrrocinia, and B. ubonensis (genomovars I, II, III, IV, V, VI, VII, VIII, IX, and X, respectively) (21, 31, 40). Some epidemic clones have been described as sources of human infections in Canada, the United Kingdom, and France (12, 14, 15, 16, 33); and B. cepacia complex strains are also commonly found in the environment in soil (10, 18), water (39), and onions (6). The prevalence of isolation of Bulkholderia sp. strains in CF patients in France is about 3.1%, with B. cenocepacia (genomovar III) and B. multivorans (genomovar II) being the most frequently isolated bacterial species (3, 21).
The treatment of B. cepacia infections is difficult, since B. cepacia species often have high-level intrinsic resistance to many antibiotics, including ticarcillin, most cephalosporins, aminoglycosides, fosfomycin, and the polymyxins (5, 41). However, Nzula et al. have noticed a heterogeneity of intrinsic antibiotic resistance patterns among the members of the B. cepacia complex that is likely related to the genomovar type (25).
Resistance to β-lactam antibiotics in isolates of the B. cepacia complex has been related to a chromosomal and inducible β-lactamase which has been falsely identified as an AmpC enzyme (28). Then, Trépanier et al. (36) described a chromosomal Ambler class A β-lactamase (PenA) from B. cepacia 249. The PenA ß-lactamase possesses a narrow-spectrum profile, and it is regulated by a LysR-type transcriptional regulator, PenR. This negative regulator is responsible for the inducibility of PenA expression (36).
The aim of the study described here was to identify the putative ß-lactamases produced by strains belonging to the B. cepacia complex and, in particular, to characterize the ß-lactamase determinants from B. cenocepacia, which is the main Burkholderia species identified from CF patients in France. The newly identified ß-lactamase, PenB, was studied for (i) its hydrolysis activity toward ß-lactams, (ii) the inducibility of its expression, and (iii) the distribution of its gene among B. cepacia complex isolates.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Eight B. cenocepacia clinical isolates obtained from CF patients and belonging to genomovar III were obtained from C. Segonds, Toulouse, France. They had been recovered from distinct hospitals in distinct cities in France. Two Burkholderia multivorans (genomovar II), one Burkholderia stabilis (genomovar IV), one Burkholderia pyrrocinia (genomovar IX), and two Burkholderia vietnamiensis (genomovar V) isolates identified by the use of molecular tools (38), recovered from French CF patients, and provided by the collection Observatoire Cepacia (Laboratoire de Bacteriologie-Virologie-Hygiene, Hôpital Rangueil, Toulouse, France) were included in the study. Escherichia coli TOP10 (Invitrogen, Life Technologies, Cergy-Pontoise, France) was used as the host for the cloning and expression experiments. Kanamycin-resistant plasmid pBK-CMV was used as the cloning vector. Bacterial cultures were grown in Trypticase soy (TS) broth at 30°C and 37°C for 18 h for the Burkholderia spp. and E. coli, respectively.
Antimicrobial agents and MIC determinations.
The antimicrobial agents used in this study were obtained from pure laboratory powders and were used immediately after their solubilization. The agents and their sources were as follows: amoxicillin (amoxicilline), clavulanic acid, ceftazidime, and ticarcillin, GSK (Nanterre, France); aztreonam and cefepime, Bristol-Myers Squibb (Paris-La-Défense, France); cephalothin (cefalotin) and moxalactam (latamoxef), Eli Lilly (Saint-Cloud, France); piperacillin and tazobactam, Lederle (Oullins, France); sulbactam, Pfizer (Orsay, France); and cefotaxime and cefpirome, Hoechst-Roussel (Paris, France). MICs were determined by the microdilution and Etest techniques, as described previously (26). The results of susceptibility testing were interpreted according to the guidelines of the CLSI (8).
Cloning experiments and PCR experiments.
For each PCR experiment, 500 ng of total DNA was used in a standard PCR mixture. By using the total DNA of the different Burkholderia sp. isolates, PCR amplifications of the blaPen-like genes were performed with either external primers Pre-PenA-1 and Pre-PenA-2 or internal primers PenA-1 and PenA-2 (Table 1), designed from the published penA sequence. In a second step, other PCR amplifications were performed with either external primers Pre-PenB-1 and Pre-PenB-2 or internal primers PenB-1 and PenB-2 (Table 1), which were newly designed from the identified penB sequence.
TABLE 1.
Primers used in this study
| Primer | Gene, primer type | Sequence (5′ to 3′) |
|---|---|---|
| Pre-PenA-1 | blaPEN-A, external | GCGTTCCGCATTCGTTTCGC |
| Pre-PenA-2 | blaPEN-A, external | TGGCCGCTCACGCGAGCGTC |
| PenA-1 | blaPEN-A, internal | TTACACCATTCTCGGCACGG |
| PenA-2 | blaPEN-A, internal | AGAAACAAGGAACTGTTGGC |
| Pre-PenB-1 | blaPEN-B, external | CTCATCGAAACGTCGAACCC |
| Pre-PenB-2 | blaPEN-B, external | CGTCTGCGTGTAGTACACGG |
| PenB-1 | blaPEN-B, internal | GTCTCGATAGTGCTGTCTCG |
| PenB-2 | blaPEN-B, internal | TTACGTGCCAGTTGCTGACG |
Total DNAs of the B. cepacia strains were partially digested with the Sau3AI restriction enzyme, ligated into the BamHI site of linearized plasmid pBK-CMV, and transformed into reference strain E. coli TOP10, as described previously (13). Recombinant plasmids were selected on TS agar plates containing amoxicillin (50 μg/ml) and kanamycin (30 μg/ml).
DNA sequencing and protein analysis.
The PCR-generated fragments, purified by using QIAquick PCR purification spin columns (Qiagen, Courtaboeuf, France), and both strands of the inserts from the recombinant plasmids were sequenced on an ABI 3100 automated sequencer (Applied Biosystems, Les Ulis, France). The nucleotide and deduced protein sequences were analyzed with software available over the Internet at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov). Dendrograms were derived from a multiple-sequence alignment by a parsimony method with the phylogeny package PAUP (Phylogenetic Analysis Using Parsimony), version 3.0 (34).
IEF analysis and induction studies.
Isoelectric focusing (IEF) analysis was performed with a pH 3.5 to 9.5 Ampholine polyacrylamide gel (GE Healthcare, Orsay, France) with culture extracts of the different B. cepacia complex isolates and of recombinant E. coli TOP10 harboring plasmid pBcSau13. The inducibility of the ß-lactamase was tested in TS broth at 37°C with imipenem (0.6 μg/ml) as the ß-lactam inducer, and hydrolysis was measured with 100 μM benzylpenicillin as the substrate. The ß-lactamase activity was defined as the hydrolysis of 1 μmol of benzylpenicillin per min by 1 U of enzyme. The total protein content was measured with bovine serum albumin as the standard (DC protein assay kit; Bio-Rad).
ß-Lactamase purification and IEF analysis.
Cultures of recombinant E. coli TOP10(pBcSau13) were grown overnight at 37°C in 4 liters of Trypticase soy broth containing amoxicillin (100 μg/ml) and kanamycin (30 μg/ml). ß-Lactamase was purified by ion-exchange chromatography, as described previously (26). Briefly, the ß-lactamase extract was sonicated, cleared by ultracentrifugation, treated with DNase, and dialyzed against 20 mM bis-Tris buffer (pH 8). This extract was loaded onto a Q-Sepharose column, and the ß-lactamase-containing fractions were eluted with a linear NaCl gradient from 0 to 0.5 mM. The fractions containing the highest ß-lactamase activity were dialyzed against 20 mM bis-Tris buffer (pH 5.5) and were subsequently reloaded onto the preequilibrated Q-Sepharose column. The ß-lactamase activity was recovered in the flowthrough, and then the extract was concentrated with an ultrafiltration filter tip (Sartorius, Göttingen, Germany). The purity of the enzyme was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (11).
Kinetic studies.
Purified ß-lactamase was used for determination of kinetic parameters (kcat, Km), which was performed at 30°C in a reaction buffer made of 100 mM phosphate (pH 7.0) (18, 24). The initial rates of hydrolysis of the ß-lactams were determined with a UV spectrophotometer, as described previously (13). The 50% inhibitory concentration (IC50) was determined as the clavulanate or tazobactam concentration that reduced the rate of hydrolysis of 100 μM piperacillin by 50% under conditions in which the enzyme was preincubated with various concentrations of inhibitor for 3 min at 30°C before addition of the substrate (13).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to the EMBL/GenBank nucleotide sequence databases under the following accession numbers: EU872211 for PenB1, FJ386399 for PenB2, FJ386401 for PenB3, FJ386402 for PenB4, FJ457906 for PenC, FJ457907 for PenD, and FJ386400 for PenE.
RESULTS AND DISCUSSION
Susceptibility testing.
The eight B. cenocepacia isolates showed various profiles of resistance to ß-lactams. However, they were all highly resistant to aminoglycosides, tetracycline, fosfomycin, and trimethoprim. B. cenocepacia strains 07-34, 09-54, 212, and 5007902 showed resistance to amoxicillin and ticarcillin but various levels of susceptibility to piperacillin, expanded-spectrum cephalosporins, and imipenem (Table 2). Addition of clavulanic acid and tazobactam did not restore the ß-lactam susceptibilities for amoxicillin and piperacillin, respectively (Table 2). Induction experiments with two B. cenocepacia strains (strains 212 and 5007902) showed a ca. 40-fold increase in the ß-lactamase activity by using imipenem as the inducer and cephalothin as the substrate. IEF analysis of cultures of all B. cenocepacia strains revealed a single ß-lactamase with activity and pI values that ranged from 7.5 to 8.5; the exception was strain 09-54, which coexpressed an additional ß-lactamase with a pI value of 6.0. This suggested the probable production of a similar ß-lactamase among those isolates.
TABLE 2.
MICs of β-lactams for B. cepacia complex isolates 07-34, 09-54, 212, and 5007902; E. coli TOP10 harboring recombinant plasmid pBcSau13 expressing the PenB1 β-lactamase; and the E. coli TOP10 reference strain
| β-Lactam(s)a | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| B. cenocepacia 07-34 (PenB1) | B. cenocepacia 09-54 (PenB2) | B. cenocepacia 212 (PenB3) | B. cenocepacia 5007902 (PenB4) | E. coli TOP10(pBcSau13) (PenB1) | E. coli TOP10 | |
| Amoxicillin | 512 | 512 | 512 | 512 | 512 | 4 |
| Amoxicillin+ CLA | 512 | 512 | 512 | 512 | 16 | 4 |
| Ticarcillin | 512 | 512 | 512 | 512 | 512 | 4 |
| Piperacillin | 4 | 512 | 128 | 4 | 8 | 2 |
| Piperacillin + TZB | 4 | 256 | 128 | 4 | 4 | 2 |
| Cefuroxime | 16 | 512 | 512 | 32 | 16 | 8 |
| Ceftazidime | 4 | 32 | 16 | 0.25 | 1 | 0.5 |
| Cefotaxime | 1 | 512 | 512 | 4 | 0.25 | 0.06 |
| Cefepime | 32 | 64 | 64 | 1 | 0.125 | 0.06 |
| Cefoxitin | 512 | 512 | 512 | 512 | 8 | 8 |
| Aztreonam | 4 | 512 | 512 | 16 | 0.5 | 0.06 |
| Imipenem | 8 | 16 | 16 | 8 | 0.5 | 0.25 |
CLA, clavulanic acid at a fixed concentration of 4 mg/ml; TZB, tazobactam at a fixed concentration of 4 mg/ml.
Screening of PenA-like producers.
Surprisingly, PCR assays with internal and external primers failed to identify a penA-like gene among the B. cenocepacia (belonging to genomovar groups I and III, together with B. cepacia), B. vietnamiensis, B. pyrrocinia, and B. stabilis isolates. However, positive results were obtained for the two B. multivorans strains. B. multivorans isolates 232 and 281 expressed ß-lactamases PenA2 and PenA3, which shared 96% and 99% amino acid sequence homologies with PenA (renamed PenA1), respectively (Table 3). These results agree with the reclassification of B. cepacia 249 (in which the penA gene had primarily been identified) as B. multivorans ATCC 17616 (36, 37).
TABLE 3.
Amino acid substitutions in PenB-like proteins
| Protein | Substitution at the following allele/positiona:
|
||||||
|---|---|---|---|---|---|---|---|
| 80 | 119 | 138 | 149 | 173 | 182 | 214 | |
| PenB1 | V | K | A | T | A | A | R |
| PenB2 | I | T | S | A | V | T | L |
| PenB3 | T | V | |||||
| PenB4 | T | ||||||
Note that the position does not refer to the Ambler nomenclature but refers to the positions in the PenB-like sequences. GenBank accession numbers are as follow; EU872211 for PenB1, FJ386399 for PenB2, FJ386401 for PenB3, and FJ386402 for PenB4.
Cloning of the B. cenocepacia 212 penicillinase-encoding gene.
Cloning experiments were therefore performed to identify the naturally occurring ß-lactamase gene(s) of B. cenocepacia. Whole-cell DNA from B. cenocepacia isolate 212 was digested with Sau3AI and inserted in BK-CMV to give recombinant strain E. coli TOP10(pBcSau13). It expressed a penicillinase phenotype with resistance to amoxicillin and ticarcillin and reduced susceptibility to piperacillin and aztreonam. Addition of clavulanic acid and tazobactam only partially restored the ß-lactam activities of amoxicillin and piperacillin, respectively (Table 2). This phenotype agrees with the expression of a class A ß-lactamase with narrow-spectrum penicillinase activity. The recombinant strain remained susceptible to cephalosporins and imipenem, despite slight increases in their MICs (Table 2).
Identification of ß-lactamase PenB1.
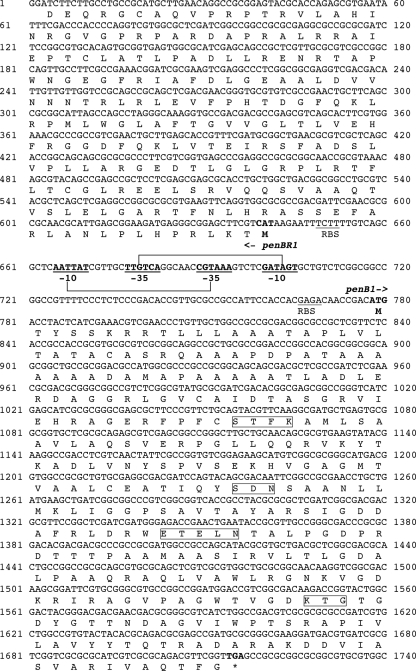
DNA sequence analysis of the 2,471-bp insert of pBcSau13 revealed two open reading frames that corresponded to β-lactamase PenB1 and a LysR-type regulator, PenR-B, respectively (Fig. 1). Two putative −10 and −35 promoter boxes were identified in the 140-bp intercistronic region extending between the two corresponding genes. Two potential ribosome-binding sites (RBSs) were also identified in that intergenic region; these were located 9 bp upstream of the penB gene and 8 bp upstream of the penR-B gene. Both RBS sequences were identical to those found upstream of the penA and penR genes in B. multivorans 249 (Fig. 1).
FIG. 1.
Nucleotide sequence of the 1,740-bp fragment of pBcSau13 containing the penB1- and penRB-coding regions. The deduced amino acid sequences are designated in single-letter code. The putative and overlapped promoter sequences are represented by −35 and −10 regions (in boldface and underlined). The start and stop codons of these genes are also in boldface, and the putative RBS sequence is underlined. The critical motifs for class A ß-lactamases are bracketed.
The penB1 ß-lactamase gene identified was 936 bp and coded for a 312-amino-acid preprotein named PenB1 with a relative molecular mass of 32 kDa. The G+C content of this penB1 gene was 69%, which is within the G+C content of the B. cepacia genome (66%). A search with the Signalp program (24) showed the presence of a signal peptide with a putative cleavage site between positions 33 and 34 of the N-terminal region. The resulting 279-amino-acid protein had a calculated molecular size of 29 kDa. PenB1 contains the four conserved motifs 70S*XXK73 (where 70S* is the active-site serine), 130SDN133, 166EXXXN170, and 234KTG236 of class A ß-lactamases (by use of the numbering scheme of Ambler et al. [1]) (Fig. 1). Analysis of the insert sequence of recombinant plasmid pBsSau13 evidenced a gene encoding a putative LysR regulator that we named PenR-B, that is located upstream of penB1, and that is transcribed in an orientation opposite that of penB1 (Fig. 1).
ß-Lactamase PenB1 shared 82% amino acid identity with PenA from B. multivorans 249 (formerly B. cepacia) (Table 4). In addition, ß-lactamase PenB1 shared 85, 84, 71, and 69% amino acid identities with putative class A ß-lactamases identified from the genomes of Burkholderia vietnamiensis strain 383, Burkholderia dolosa AUO158, Burkholderia thailandensis E264, and Burkholderia pseudomallei 668, respectively (in silico analysis). The closest amino acid identities with plasmid-mediated class A ß-lactamases were 59% and 51% with CTX-M-14 and CTX-M-2, respectively.
TABLE 4.
Amino acid identities between chromosome-encoded Pen-like proteins
| Bacterial species | Pen homologue | % Amino acid sequence identitya
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PenB1 | PenA | PenC | PenD | PenE | PenF | PenG | PenH | PenI | PenJ | PenK | PenL | ||
| B. cenocepacia | PenB1 | 82 | 92 | 91 | 85 | 83 | 84 | 73 | 69 | 64 | 63 | 71 | |
| B. multivorans | PenA | 82 | 84 | 85 | 85 | 82 | 84 | 78 | 63 | 64 | 64 | 66 | |
| B. stabilis | PenC | 92 | 84 | 86 | 85 | 82 | 78 | 76 | 62 | 62 | 61 | 66 | |
| B. pyrrocina | PenD | 91 | 85 | 86 | 85 | 81 | 78 | 75 | 63 | 61 | 60 | 65 | |
| B. vietnamiensis | PenE | 85 | 85 | 85 | 85 | 85 | 80 | 74 | 62 | 63 | 62 | 64 | |
| B. ambifaria | PenF | 83 | 82 | 82 | 81 | 85 | 83 | 75 | 59 | 59 | 60 | 62 | |
| B. dolosa | PenG | 84 | 84 | 78 | 78 | 80 | 83 | 78 | 67 | 66 | 67 | 69 | |
| B. ubonensis | PenH | 73 | 78 | 76 | 75 | 74 | 75 | 78 | 64 | 61 | 63 | 64 | |
| B. pseudomallei | PenI | 69 | 63 | 62 | 63 | 62 | 59 | 67 | 64 | 83 | 98 | 89 | |
| B. oklahomensis | PenJ | 64 | 64 | 62 | 61 | 63 | 59 | 66 | 61 | 83 | 82 | 87 | |
| B. mallei | PenK | 63 | 64 | 61 | 60 | 62 | 60 | 67 | 63 | 98 | 82 | 88 | |
| B. thailandensis | PenL | 71 | 66 | 66 | 65 | 64 | 62 | 69 | 64 | 89 | 87 | 88 | |
The amino acid identities of the Pen-like proteins obtained in this study are indicated in boldface. The PenB1 (EU872211), PenC (FJ457906), PenD (FJ457907), and PenE (FJ386400) sequences were obtained from the strains used in this study (GenBank accession numbers are indicated in parentheses). PenA, PenF (YP_776246), PenG (EAY70884), PenH (ZP_02376965), PenI (YP_001062378), PenJ (ZP_02365625), PenK (YP_105905), and PenL (ZP_02384846) were obtained from the sequences deposited in the NCBI database (GenBank accession numbers are indicated in parentheses).
The PenR-B regulatory protein shared 97% amino acid identity with the LysR transcriptional regulator PenR from Burkholderia sp. strain 383 and 95% and 92% amino acid identities with the PenR proteins from B. dolosa AUO158 and B. multivorans 249, respectively. The PenR-B sequence also showed some identity with ß-lactamase transcriptional regulators of the LysR family, such as the AmpR proteins regulating the AmpC expression of Pseudomonas aeruginosa PAO1 (59%) and Proteus vulgaris B317 CumR (58.7%) (9, 19). A search for a peptide motif, which was performed with the Genetics Computer Group program Motifs and the database Prosite, identified a helix-turn-helix motif in the N-terminal part of the PenR peptide sequence (25FTRAGLELSVTQAAVSQQVRS45), as is usually found for LysR-type transcriptional regulators.
Biochemical properties of PenB1.
IEF analysis of cultures of E. coli TOP10(pBcSau13) revealed a pI value of 8.5. After purification, the specific activity of the PenB1 ß-lactamase against 100 μM benzylpenicillin was 520 U · mg of protein−1, its purity was estimated to be >95% by SDS-PAGE analysis, and its purification coefficient was calculated to be 200. The kinetic parameters for the purified PenB1 ß-lactamase showed an hydrolysis profile that included penicillins, expanded-spectrum cephalosporins, and aztreonam. Imipenem and meropenem were hydrolyzed at low levels, whereas the hydrolysis of moxalactam and cefoxitin was not detected (Table 5).
TABLE 5.
Kinetic parameters of purified PenB1 β-lactamasea
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (mM−1 · s−1) |
|---|---|---|---|
| Benzylpenicillin | 130 | 25 | 5,200 |
| Ampicillin | 70 | 100 | 700 |
| Ticarcillin | 15 | 60 | 250 |
| Piperacillin | 5 | 10 | 500 |
| Cephalothin | 300 | 100 | 3,000 |
| Cefuroxime | 170 | 80 | 2,100 |
| Cefoxitin | NDb | ND | ND |
| Cefotaxime | 20 | 150 | 130 |
| Ceftazidime | 1.5 | 3,000 | 0.5 |
| Cefepime | 10 | 1,000 | 10 |
| Aztreonam | 30 | 2,500 | 10 |
| Imipenem | 0.1 | 60 | 2 |
| Meropenem | 1 | 2,500 | 0.5 |
The data are the means of three independent experiments. Standard deviations were within 10% of the means.
ND, no detectable hydrolysis (<0.01 s−1) with a maximum amount of 5 μg of purified enzyme and up to 200 nmol of substrate.
Inhibition studies, as measured by determination of the IC50s, showed that the activity of PenB1 was inhibited by tazobactam (0.5 μM) but that its activity was inhibited very poorly by clavulanic acid (10 μM), in accordance with what has been found for PenA, which was previously considered clavulanic acid resistant (27, 28).
Distribution of penB1-like genes among the B. cepacia complex.
PCR screening was performed with internal and external primers designed from the penB1 sequence and 11 strains belonging to the B. cepacia complex. A penB1-like gene was identified in all those strains, making it a feature of the B. cepacia complex.
First, sequencing showed that the seven B. cenocepacia isolates contained PenB1-like-encoding sequences, namely, PenB2, PenB3, and PenB4, which had amino acid identities that ranged from 96 to 100% compared with the amino acid sequence of PenB1 (Table 3). Screening of the non-B. cenocepacia species identified penB-like genes encoding ß-lactamases and amino acid identities that ranged from 81 to 92% compared with the amino acid sequence of PenB1 (Table 4). Altogether, those results indicate that PenB1-like ß-lactamases are broadly distributed among isolates of the B. cepacia complex and may likely contribute to the natural ß-lactam resistance pattern observed.
Second, PCR screening of penB-like genes followed by sequencing revealed homologous sequences from the species B. stabilis, B. pyrrocinia, and B. vietnamiensis. Interestingly, B. stabilis 625 (genomovar IV) expressed ß-lactamase PenC1, which shared 92% amino acid identity with the amino acid sequence of PenB1 and 84% amino acid identity with the amino acid sequence of PenA1; B. pyrrocinia 685 (genomovar IX) expressed ß-lactamase PenD1, which shared 91% amino acid identity with the amino acid sequence of PenB1 and 85% amino acid identity with the amino acid sequence of PenA1; and B. vietnamiensis isolates 189 and 764 (genomovar V) expressed ß-lactamases PenE1 and PenE2 (three substitutions), respectively, and shared 85% amino acid identity with the amino acid sequences of both PenB1 and PenA1 (Table 4).
Conclusions.
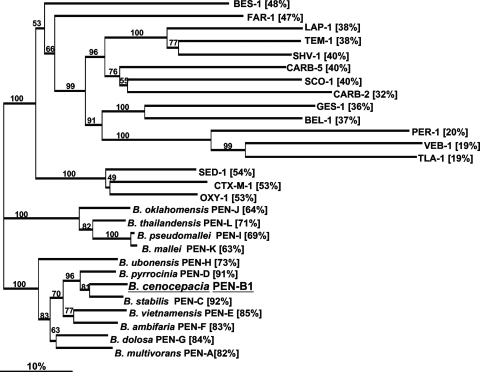
Here we showed that the Burkholderia complex possesses related but distinct, naturally occurring class A ß-lactamases very likely specific for each Burkholderia species. These findings are interesting because they may provide a useful tool for the identification of Bulkholderia isolates at the species level. We have characterized the PenB1 enzyme, which shares properties with the previously characterized PenA ß-lactamase from B. multivorans. PenB-like enzymes are naturally expressed by B. cenocepacia, which is the most prevalent Burkholderia species identified from CF patients not only in France (3) but also in Italy (4). We also identified other novel Pen-type ß-lactamases from a variety of Burkholderia sp. isolates: PenC from B. stabilis, PenD from B. pyrrocinia, and PenE from B. vietnamiensis. Finally, we took this opportunity to define a nomenclature for other Burkholderia spp. on the basis of an in silico analysis that makes PenF the class A ß-lactamase of B. ambifira, PenG that of B. dolosa, PenH that of B. ubonensis, PenI that of B. pseudomallei, PenJ that of B. oklahomensis, PenK that of B. mallei, and PenL that of B. thailandensis (Table 4; Fig. 2).
FIG. 2.
Dendrogram obtained for representative class A ß-lactamases by neighbor-joining analysis. The alignment used for tree calculation was performed with the ClustalX program. Branch lengths are drawn to scale and are proportional to the number of amino acid changes. The distance along the vertical axis has no significance. The amino acid identity of each ß-lactamase compared to the amino acid sequence of PenB1 from B. cenocepacia is indicated in parentheses. The acquired ß-lactamases of gram-negative organisms (GenBank accession numbers are indicated in parentheses) are TEM-1 (AAG47772), SHV-1 (AAD18054), CTX-M-1 (CAJ01342), LAP-1 (ABK58097), CARB-2 (Q03170), CARB-5 (AAF61417), GES-1 (AAL82589), BEL-1 (AAZ04368), BES-1 (AAF61147), SCO-1 (ABL75133), PER-1 (CAF18433), and TLA-1 (AAD37403), whereas the others are naturally occurring ß-lactamases, such as FAR-1 from Nocardia farcinica (AAB81957), SED-1 from Citrobacter sedlakii (AAK63223), and OXY-1 from Klebsiella oxytoca (P22391).
We showed that the expression of the penB1 gene is inducible and is regulated by a LysR-type transcriptional regulator, as observed for penA from B. multivorans. This LysR-type dependence of the regulation of those class A ß-lactamases is similar to that observed with naturally occurring ß-lactamase genes from Proteus vulgaris (9), Rhodopseudomonas capsulata (7), and Citrobacter diversus (17) or with acquired blaSME-like genes from Serratia marcescens (22), blaNMC-A from Enterobacter cloacae (23),and blaIMI-like genes from E. cloacae or Enterobacter asburiae (2, 29).
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (grant UPRES-EA3539), Université Paris XI, Paris, France, and mostly by a grant from the European Community (6th PCRD, DRESP2, LSHM-CT-2005-018705). J.-M.R.-M. is funded by a postdoctoral grant from the Ministerio de Educacion y Ciencia (2007/0292).
We are grateful to C. Segonds and the French Cystic Fibrosis Association (Vaincre la Mucoviscidose) for providing genetically identified Burkholderia strains.
Footnotes
Published ahead of print on 15 December 2008.
REFERENCES
- 1.Ambler, R. P., A. F. Coulson, J.-M. Frère, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A ß-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubron, C., L. Poirel, R. J. Ash, and P. Nordmann. 2005. Carbapenemase-producing Enterobacteriaceae, U.S. rivers. Emerg. Infect. Dis. 11:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellis, G., M. De Braekeleer, and G. Chabanon. 2004. Colonisation par Burkholderia: bilan 2001 à partir des données de l'ONM complémentées et corrigées par l'observatoire B. cepacia en rapport sur la situation de la mucoviscidose en France en 2001, p. 44-45. Vaincre la Mucoviscidose-INED, Paris, France.
- 4.Bevivino, A., C. Dalmastri, and S. Tabacchioni. 2002. Burkholderia cepacia complex bacteria from clinical and environmental sources in Italy: genomovar status and distribution of traits related to virulence and transmissibility. J. Clin. Microbiol. 40:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonacorsi, S., F. Fitoussi, S. Lhopital, and E. Bingen. 1999. Comparative in vitro activities of meropenem, imipenem, temocillin, piperacillin, and ceftazidime in combination with tobramycin, rifampin, or ciprofloxacin against Burkholderia cepacia isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 43:213-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 7.Campbell, J. I., S. Scahill, T. Gibson, and R. P. Ambler. 1989. The phototrophic bacterium Rhodopseudomonas capsulata sp108 encodes an indigenous class A ß-lactamase. Biochem. J. 260:803-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2008. Performance standard for antimicrobial susceptibility testing, 18th informational supplement. Clinical Laboratory Standards Institute, Wayne, PA.
- 9.Datz, M., B. Joris, E. A. Azab, M. Galleni, J. Van Beeumen, J.-M. Frère, and H. H. Martin. 1994. A common system controls the induction of very different genes. The class-A ß-lactamase of Proteus vulgaris and the enterobacterial class-C ß-lactamase. Eur. J. Biochem. 226:149-157. [DOI] [PubMed] [Google Scholar]
- 10.Fiore, A., S. Laevens, and A. Bevivino. 2001. Burkholderia cepacia complex: distribution of genomovars among isolates from the maize rhizosphere in Italy. Environ. Microbiol. 3:137-143. [DOI] [PubMed] [Google Scholar]
- 11.Girlich, D., T. Naas, and P. Nordmann. 2004. Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:2043-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govan, J. R., P. H. Brown, and J. Maddison. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 13.Héritier, C., L. Poirel, D. Aubert, and P. Nordmann. 2003. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isles, A., T. Maclusky, and M. Corey. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 15.Jacques, I., J. Derelle, M. Weber, and M. Vidailhet. 1998. Pulmonary evolution of cystic fibrosis patients colonized by Pseudomonas aeruginosa and/or Burkholderia cepacia. Eur. J. Pediatr. 157:427-431. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, W. M., S. D. Tyler, and K. R. Rozee. 1994. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J. Clin. Microbiol. 32:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, M. E., and P. M. Bennett. 1995. Inducible expression of the chromosomal cdiA from Citrobacter diversus NF85, encoding an Ambler class A ß-lactamase, is under similar genetic control to the chromosomal ampC, encoding an Ambler class C enzyme, from Citrobacter freundii OS60. Microb. Drug Resist. 1:285-291. [DOI] [PubMed] [Google Scholar]
- 18.Lipuma, J., T. Spilker, and T. Coenye. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002-2003. [DOI] [PubMed] [Google Scholar]
- 19.Lodge, J., S. Busby, and L. Piddock. 1993. Investigation of the Pseudomonas aeruginosa ampR gene and its role at the chromosomal ampC ß-lactamase promoter. FEMS Microbiol. Lett. 111:315-320. [DOI] [PubMed] [Google Scholar]
- 20.Magalhaes, M., C. Doherty, and J. Govan. 2003. Polyclonal outbreak of Burkholderia cepacia complex bacteraemia in haemodialysis patients. J. Hosp. Infect. 54:120-123. [DOI] [PubMed] [Google Scholar]
- 21.Mahenthiralingam, E., and P. Vandamme. 2005. Taxonomy and pathogenesis of the Burkholderia cepacia complex. Chron. Respir. Dis. 2:209-217. [DOI] [PubMed] [Google Scholar]
- 22.Naas, T., D. M. Livermore, and P. Nordmann. 1995. Characterization of an LysR family protein, SmeR from Serratia marcescens S6, its effect on expression of the carbapenem-hydrolyzing ß-lactamase Sme-1, and comparison of this regulator with other ß-lactamase regulators. Antimicrob. Agents Chemother. 39:629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naas, T., and P. Nordmann. 1994. Analysis of a carbapenem-hydrolyzing class A ß-lactamase from Enterobacter cloacae and of its LysR-type regulatory protein. Proc. Natl. Acad. Sci. USA 91:7693-7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 25.Nzula, S., P. Vandamme, and J. Govan. 2002. Influence of taxonomic status on the in vitro antimicrobial susceptibility of the Burkholderia cepacia complex. J. Antimicrob. Chemother. 50:265-269. [DOI] [PubMed] [Google Scholar]
- 26.Philippon, L. N., T. Naas, A. T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum ß-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prince, A., M. S. Wood, G. S. Cacalano, and N. X. Chin. 1988. Isolation and characterization of a penicillinase from Pseudomonas cepacia 249. Antimicrob. Agents Chemother. 32:838-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proenca, R., W. W. Niu, G. Cacalano, and A. Prince. 1993. The Pseudomonas cepacia 249 chromosomal penicillinase is a member of the AmpC family of chromosomal ß-lactamases. Antimicrob. Agents Chemother. 37:667-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen, B. A., K. Bush, D. Keeney, Y. Yang, R. Hare, C. O'Gara, and A. A. Medeiros. 1996. Characterization of IMI-1 ß-lactamase, a class A carbapenem-hydrolyzing enzyme from Enterobacter cloacae. Antimicrob. Agents Chemother. 40:2080-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segonds, C., and G. Chabanon. 2001. Burkholderia cepacia: dangers of a phytopathogen organism for patients with cystic fibrosis. Ann. Biol. Clin. (Paris) 59:259-269. [PubMed] [Google Scholar]
- 31.Segonds, C., T. Heulin, N. Marty, and G. Chabanon. 1999. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J. Clin. Microbiol. 37:2201-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shehabi, A., W. Abu-Al-Soud, and A. Mahafzah. 2004. Investigation of Burkholderia cepacia nosocomial outbreak with high fatality in patients suffering from diseases other than cystic fibrosis. Scand. J. Infect. Dis. 36:174-178. [DOI] [PubMed] [Google Scholar]
- 33.Speert, D. P., D. Henry, and P. Vandamme. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swofford, D. L. 1989. PAUP (version 3.0): phylogenetic analysis using parsimony. Illinois Natural History Survey, Champaign.
- 35.Tablan, O. C., W. J. Martone, C. F. Doershuk, R. C. Stern, M. J. Thomassen, J. D. Klinger, J. W. White, L. A. Carson, and W. R. Jarvis. 1987. Colonization of the respiratory tract with Pseudomonas cepacia in cystic fibrosis. Risk factors and outcomes. Chest 91:527-532. [DOI] [PubMed] [Google Scholar]
- 36.Trépanier, S., A. Prince, and A. Huletsky. 1997. Characterization of the PenA and PenR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptionnal regulator. Antimicrob. Agents Chemother. 41:2399-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandamme, P., B. Holmes, and M. Vancanneyt. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 38.Van Pelt, C., C. M. Verduin, W. H. Goessens, M. C. Vos, B. Tümmler, C. Segonds, F. Reubsaet, H. Verbrugh, and A. Van Belkum. 1999. Identification of Burkholderia spp. in the clinical microbiology laboratory: comparison of conventional and molecular methods. J. Clin. Microbiol. 37:2158-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vermis, K., M. Brachkova, and P. Vandamme. 2003. Isolation of Burkholderia cepacia complex genomovars from waters. Syst. Appl. Microbiol. 26:595-600. [DOI] [PubMed] [Google Scholar]
- 40.Yabuuchi, E., Y. Kosako, H. Oyaizu, I. Yano, H. Hotta, Y. Hashimoto, T. Ezaki, and M. Arakawa. 1992. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia. Comb. Nov. Microbiol. Immunol. 36:1251-1275. [DOI] [PubMed] [Google Scholar]
- 41.Zhou, J., Y. Chen, S. Tabibi, L. Alba, E. Garber, and L. Saiman. 2007. Antimicrobial susceptibility and synergy studies of Burkholderia cepacia complex isolated from patients with cystic fibrosis. Antimicrob. Agents Chemother. 51:1085-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]