Abstract
Mechanisms of antibiotic resistance were examined in nalidixic acid-resistant Salmonella enterica serovar Enteritidis field isolates displaying decreased susceptibility to ciprofloxacin and in in vitro-derived ciprofloxacin-resistant mutants (104-cip and 5408-cip). All field isolates harbored a single gyrA mutation (D87Y). Deletion of acrB and complementation with wild-type gyrA increased quinolone susceptibility. Selection for ciprofloxacin resistance was associated with the development of an additional gyrA (S83F) mutation in 104-cip, novel gyrB (E466D) and parE (V461G) mutations in 5408-cip, overexpression of acrB and decreased susceptibility to nonquinolone antibiotics in both mutants, and decreased OmpF production and altered lipopolysaccharide in 104-cip. Complementation of mutated gyrA and gyrB with wild-type alleles restored susceptibility to quinolones in 104-cip and significantly decreased the ciprofloxacin MIC in 5408-cip. Complementation of parE had no effect on quinolone MICs. Deletion of acrB restored susceptibility to ciprofloxacin and other antibiotics tested. Both soxS and marA were overexpressed in 104-cip, and ramA was overexpressed in 5408-cip. Inactivation of each of these global regulators lowered ciprofloxacin MICs, decreased expression of acrB, and restored susceptibility to other antibiotics. Mutations were found in soxR (R20H) and in soxS (E52K) in 104-cip and in ramR (G25A) in 5408-cip. In conclusion, both efflux activity and a single gyrA mutation contribute to nalidixic acid resistance and reduced ciprofloxacin sensitivity. Ciprofloxacin resistance and decreased susceptibility to multiple antibiotics can result from different genetic events leading to development of target gene mutations, increased efflux activity resulting from differential expression of global regulators associated with mutations in their regulatory genes, and possible altered membrane permeability.
Salmonella enterica serovar Enteritidis is the most common etiological agent of food-borne salmonellosis worldwide. Ciprofloxacin is the antibiotic of choice for the treatment of severe Salmonella infections when therapeutic intervention is warranted. To date, fluoroquinolone resistance (MIC of ciprofloxacin, ≥4 μg/ml) remains relatively uncommon in Salmonella. However, the incidence of nalidixic acid resistance in Salmonella isolates from humans and food animals has increased, with these isolates showing decreased susceptibility to fluoroquinolones (6, 27). Of paramount concern to public health are reports of therapeutic failure of ciprofloxacin in cases of invasive salmonellosis associated with isolates displaying reduced fluoroquinolone susceptibility (26, 30, 48).
Currently well-recognized mechanisms of quinolone resistance in Salmonella include target gene mutations, increased efflux pump activity, and plasmid-mediated protection of target topoisomerases (13, 18). The contribution of changes in the cell envelope, including porin loss or alterations of the lipopolysaccharide (LPS), to quinolone resistance is currently unclear (14, 29, 36). Nalidixic acid resistance and decreased susceptibility to ciprofloxacin have been largely associated with single gyrA mutations at codon S83 or D87 (7, 18, 36). Double mutations at both residues 83 and 87 have been found in fluoroquinolone-resistant strains, often in association with mutations in other topoisomerase genes (3, 4, 7). Overexpression of the multidrug efflux pump AcrAB-TolC has been shown to directly contribute to fluoroquinolone and multidrug resistance (MDR) in Salmonella (3-5).
Much of our knowledge on the regulation of expression of AcrAB comes from work carried out in Escherichia coli (34). At a local level acrAB expression is modulated by the local repressor AcrR, and at a global level it is modulated by MarA, SoxS, and Rob, which belong to the AraC/XylS family of transcriptional regulators (2). In addition to activating tolC and acrAB genes, these transcriptional activators activate transcription of micF, an antisense RNA that inhibits synthesis of the OmpF outer membrane porin (34). Mutations within the local repressor AcrR have also been shown to contribute to acrB overexpression (33). The mar locus consists of two transcription units, marC and marRAB, which are divergently transcribed from a central putative operator-promoter region, marO (10, 45). MarA regulates its own transcription as well as regulating the expression of the mar regulon, whereas MarR acts by repressing marRAB transcription. The functions of MarB and MarC are unknown. marRAB transcription can also be activated by the MarA homologs SoxS and Rob (2, 10, 45). SoxS is the effector of the soxRS global superoxide response regulon. SoxR is a constitutively expressed homodimeric transcriptional regulator that contains redox-active iron-sulfur clusters [2Fe-2S]. Oxidation of these clusters activates SoxR to trigger transcription of the soxS gene (22, 50). Increased expression of these global regulators may be associated with mutations in the regulatory genes of the operons (20, 21, 32) or the selective binding of inducers (38, 39). As in E. coli, increased expression of marA and soxS has been associated with fluoroquinolone resistance and MDR in Salmonella (7, 12). However, the contribution of these global regulators to overexpression of acrAB in fluoroquinolone resistance and MDR Salmonella phenotypes is still currently unclear. Furthermore, little is known about rob and its contribution to antibiotic resistance in Salmonella. More recently, RamA, which displays close homology to MarA and is absent from E. coli, has been implicated in MDR in Salmonella and other bacteria (1, 16, 19, 42). Overexpression of ramA has been associated with increased expression of acrB in Salmonella and other Enterobacteriaceae (1, 19, 42).
In this study we examined the mechanisms of quinolone resistance in nalidixic acid-resistant serovar Enteritidis field isolates showing decreased susceptibility to ciprofloxacin. We generated in vitro ciprofloxacin-resistant mutants from two of these isolates in order to assess the contribution of target gene mutations, altered membrane permeability, and active efflux to the development of fluoroquinolone resistance and MDR. The role of the global regulators marA, soxS, rob, and ramA in upregulation of acrB and consequently in efflux-mediated MDR was investigated. Local and global regulatory genes of AcrAB-TolC were examined for the presence of mutations.
MATERIALS AND METHODS
Bacterial strains.
Nine serovar Enteritidis strains of animal (54, 58, 51, and 104) and human (CUH48, CUH52, CUH60, 4931, and 5408) origin were used in this study (Table 1). The serovar Enteritidis reference strain PT4 NCTC 13349 was also included. Ciprofloxacin-resistant mutants (104-cip and 5408-cip) were selected from serovar Enteritidis strains 104 and 5408. Salmonella enterica serovar Typhimurium knockout strains L110 acrB::aph, L130 marA::aph, L133 ramA::aph, and L135 soxS::aph derived from SL1344 (12, 41) and donor E. coli strains carrying the plasmids pBP513 gyrA+, pBP548 gyrB+, and pBP568 parE+ were also used and kindly provided to L. Piddock by P. Heisig (15).
TABLE 1.
Phenotypic and genotypic characteristics of the serovar Enteritidis strains used in this study
| Strain | MIC (μg/ml) of druga,b:
|
Amino acid substitution(s)c,d
|
Fold change in expression of genee:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NAL | CIP | gyrA | gyrB | parE | acrB | soxS | marA | ramA | rob | |
| CUH48 | 6 (0.5) | 0.016 (0.012) | * | * | * | |||||
| CUH52 | 3 (0.75) | 0.016 (0.008) | * | * | * | |||||
| NCTC 13349 | 8 (0.38) | 0.023 (0.008) | * | * | * | |||||
| 54 | >256 (16) | 0.19 (0.064) | D87Y | * | * | |||||
| 58 | >256 (24) | 0.125 (0.064) | D87Y | * | * | |||||
| 51 | >256 (24) | 0.19 (0.094) | D87Y | * | * | |||||
| 51 acrB::aph | 32 | 0.032 | D87Y | * | * | |||||
| CUH60 | >256 (32) | 0.19 (0.094) | D87Y | * | * | |||||
| CUH60 acrB::aph | 24 | 0.032 | D87Y | * | * | |||||
| 4931 | >256 (16) | 0.19 (0.064) | D87Y | * | * | |||||
| 4931 acrB::aph | 32 | 0.047 | D87Y | * | * | |||||
| 104 | >256 (24) | 0.19 (0.094) | D87Y | * | * | |||||
| 104 gyrA+ | 4 | 0.047 | * | * | * | |||||
| 104 acrB::aph | 32 | 0.032 | D87Y | * | * | |||||
| 104-cip | >256 (>256) | >32 (2) | D87Y, S83F | * | * | 6.1 ± 1.5 | 26.1 ± 4.0 | 8.9 ± 0.6 | 1.2 ± 0.2 | −4.6 ± 0.2 |
| 104-cip gyrA+ | 8 | 0.094 | * | * | * | |||||
| 104-cip acrB::aph | >256 | 0.38 | D87Y, S83F | * | * | |||||
| 104-cip soxS::aph | >256 | 1.5 | D87Y, S83F | * | * | −4.2 ± 1.0 | −0.6 ± 1.6 | |||
| 104-cip marA::aph | >256 | 4 | D87Y, S83F | * | * | 1.4 ± 0.3 | 1.4 ± 0.2 | |||
| 5408 | >256 (24) | 0.19 (0.094) | D87Y | * | * | |||||
| 5408 gyrA+ | 4 | 0.032 | * | * | * | |||||
| 5408 acrB::aph | 32 | 0.047 | D87Y | * | * | |||||
| 5408-cip | >256 (16) | >32 (2) | D87Y | E466D | V461G | 5.4 ± 1.6 | −3.4 ± 0.5 | 1.3 ± 0.2 | 33.7 ± 4.0 | −2.3 ± 1.0 |
| 5408-cip gyrA+ | >256 | 1 | * | E466D | V461G | |||||
| 5408-cip gyrB+ | >256 | 3 | D87Y | * | V461G | |||||
| 5408-cip parE+ | >256 | >32 | D87Y | E466D | * | |||||
| 5408-cip acrB::aph | >256 | 0.5 | D87Y | E466D | V461G | |||||
| 5408-cip ramA::aph | >256 | 4 | D87Y | E466D | V461G | 1.6 ± 0.1 | ||||
NAL, nalidixic acid; CIP, ciprofloxacin.
Values in parentheses are the MICs determined in the presence of PAβN at 80 μg/ml.
*, wild-type allele (no mutation).
D, aspartic acid; Y, tyrosine; S, serine; F, phenylalanine; E, glutamic acid; V, valine; G, glycine.
Gene expression data represent the means ± standard deviations of three independent total RNA extractions. Changes in gene expression are relative to the parental strain.
Selection of ciprofloxacin-resistant Salmonella mutants in vitro.
Ciprofloxacin-resistant mutants (104-cip and 5408-cip) were obtained by seven serial passages on tryptone soy agar (Oxoid, Hampshire, United Kingdom) containing doubling concentrations of ciprofloxacin (0.25 to 16 μg/ml; Sigma-Aldrich, Ireland). Colonies from the highest-concentration selecting plates were subcultured five times on antibiotic-free medium before antibiotic sensitivities were determined. Mutants were stored on beads in cryopreservation fluid at −80°C (Technical Service Consultants Ltd., Lancashire, England).
Antimicrobial susceptibility testing.
MICs of nalidixic acid, ciprofloxacin, ampicillin, chloramphenicol, tetracycline, and sulfamethoxazole-trimethoprim were determined by Etest on Mueller-Hinton agar following the manufacturer's instructions (AB-Biodisk, Solna, Sweden) and according to the Clinical and Laboratory Standards Institute (CLSI) (9). E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control organisms. To assess the contribution of efflux pump activity to intrinsic and acquired antibiotic resistance, Etests were performed in the presence and absence of the efflux pump inhibitor Phe-Arg-β-naphthylamide (PAβN [80 μg/ml]; Sigma-Aldrich). MICs of PAβN were >640 μg/ml in all isolates.
PCR amplification and sequencing of QRDRs of quinolone target genes and local and global regulators of AcrAB-TolC.
Sequences of the primers used in the PCR amplifications are given in Table 2. Genomic DNA was extracted from overnight cultures in tryptone soy broth (Oxoid) at 37°C using a Wizard genomic DNA purification kit (Promega, Madison, WI). PCR mixtures contained 100 ng of template DNA, 100 pmol of each quinolone resistance-determining region (QRDR) primer or 10 pmol of each regulator primer (MWG-Biotech AG, Ebersberg, Germany), 200 μM deoxynucleoside triphosphates (Promega), 1 U Taq DNA polymerase (New England Biolabs, Ipswich, MA), and 1× PCR buffer containing 2.5 mM MgCl2. After an initial denaturation step of 3 min at 94°C, amplification was performed over 30 cycles, with each cycle consisting of 1 min at 94°C, 1 min at appropriate annealing temperature (Table 2), and 1 min at 72°C, with a final extension step of 10 min at 72°C. PCR products were purified with the Qiaquick spin PCR purification kit (Qiagen, West Sussex, United Kingdom) and sequenced commercially (Qiagen, Hilden, Germany).
TABLE 2.
Primers used in this study
| Primer purpose and gene | Primer sequence (5′-3′) | Annealing temp (°C) | Amplicon size (bp) | Reference or accession no. |
|---|---|---|---|---|
| QRDR | ||||
| gyrA | TGTCCGAGATGGCCTGAAGC | 55 | 470 | Modified from reference 6 |
| CGTTAATCACTTCCGTCAG | ||||
| gyrB | GAAATGACCCGTCGTAAAGG | 54 | 710 | AE008878 |
| TACAGTCTGCTCATCAGAAAG | ||||
| parC | ATGAGCGATATGGCAGAGCG | 52 | 413 | 6 |
| TGACCGAGTTCGCTTAACAG | ||||
| parE | GACCGAGCTGTTCCTTGTGG | 52 | 493 | 6 |
| GCGTAACTGCATCGGGTTCA | ||||
| RT-PCR | ||||
| 16S rRNA | GCGGCAGGCCTAACACAT | 59/60 | 182 | X80681 |
| GCAAGAGGCCCGAACGTC | ||||
| acrB | TTTTGCAGGGCGCGGTCAGAATAC | 59 | 184 | This study |
| TGCGGTGCCCAGCTCAACGAT | ||||
| soxS | AAATCGGGCTACTCCAAGTG | 59 | 217 | 9 |
| CTACAGGCGGTGACGGTAAT | ||||
| marA | ATCCGCAGCCGTAAAATGAC | 59 | 180 | 9 |
| TGGTTCAGCGGCAGCATATA | ||||
| rob | CATTACGGCTGGGCGAGTTTACC | 60 | 180 | This study |
| CTGGCGGAATAGTTGGCGAATGAC | ||||
| ramA | CGTCATGCGGGGTATTCCAAGTG | 60 | 107 | This study |
| CGCGCCGCCAGTTTTAGC | ||||
| Regulation | ||||
| acrR | CAGTGGTTCCGTTTTTAGTG | 58 | 992 | 40 |
| ACAGAATAGCGACACAGAAA | ||||
| soxRS | CGAACAGGGCGTCGTCGCTT | 60 | 1,199 | 40 |
| CTGGTTGCTAAAACGCGGCG | ||||
| marORAB | ACGGTGGTTAGCGGATTGGC | 58 | 1,329 | 40 |
| AGCGGCGGACTTGTCATAGC | ||||
| ramA | TTGCCGCTTCCAGTAATGCTTGTT | 60 | 724 | NC_003197 |
| CTTTATCTGGCGGCGCTGGTTTTC | ||||
| ramR | CGTGTCGATAACCTGAGCGG | 60 | 934 | 1 |
| AAGGCAGTTCCAGCGCAAAG | ||||
| Gene knockout verification | ||||
| acrB | GGATCACACCTTATTGCCAG | 52 | 3,541a/2,107b | 15 |
| CGGCCTTATCAACAGTGAGC | ||||
| marA | GCGGACTTGTCATAGCCAGA | 52 | 1,037a/1,818b | 49 |
| GCTGGATATCACCGCAACAC | ||||
| soxS | TACCGGCTATTCGAACTTGC | 50 | 989a/1,912b | 49 |
| CTCGCTTAACGTATGTCCTT | ||||
| ramA | CCGCTTCCAGTAATGCTTGT | 50 | 918a/1,755b | 49 |
| GAATCATTGATGACCGCTGC |
Without cassette.
With cassette.
Sequence analysis was carried out online using the programs BLAST (http://www.ncbi.nlm.nih.gov/BLAST/), CLUSTALW (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and Transeq (http://www.ebi.ac.uk/emboss/transeq/).
Accumulation of ciprofloxacin.
Ciprofloxacin accumulation was measured by a fluorometric method described by Mortimer and Piddock (31). Accumulation experiments were performed with and without the addition of efflux pump inhibitor PAβN (final concentration, 80 μg/ml) 6 min after the addition of ciprofloxacin (final concentration, 10 μg/ml). Fluorescence was measured with a spectrofluorimeter (spectraMax Gemini; Molecular Devices, Sunnyvale, CA) at excitation and emission wavelengths of 279 and 447 nm, respectively. The amount of ciprofloxacin accumulated was calculated by comparison with a standard curve for ciprofloxacin (0.02 to 2.5 μg/ml) in 0.1 M glycine hydrochloride (pH 3.0). Results are expressed as nanograms of ciprofloxacin incorporated per milligram (dry weight) of bacteria. All experiments were performed at least three times to ensure reproducibility.
Expression analyses of efflux transporter gene acrB and global regulators.
Reverse transcription-PCR (RT-PCR) was used to assess gene expression of acrB and global regulators soxS, marA, ramA, and rob. Overnight cultures were diluted 1 in 100 in prewarmed LB broth and grown to mid-logarithmic phase (optical density at 600 nm [OD600], 0.6) with shaking at 37°C. A 1-ml aliquot of each culture was pelleted by centrifugation at 15,339 × g for 10 min, and RNA was extracted immediately using a RiboPure-Yeast (Ambion, Texas) kit. Contaminating genomic DNA was eliminated by two DNase I treatments according to the manufacturer's instructions (Ambion), and its absence was confirmed by including a reverse transcriptase-minus control on each RNA sample. Total RNA concentration was estimated by OD260 using a Nanodrop ND-1000 spectrophotometer (Thermoscientific, Delaware). Real-time quantification of RNA templates by real-time One-Step RT-PCR was performed in a Rotor Gene 3000 thermocycler (Corbett Research, Sydney, Australia) using a QuantiTect SYBR Green RT-PCR kit (Qiagen, Hilden, Germany). The RT-PCR was carried out in a 25-μl reaction mixture containing 12.5 μl 2× QuantiTect SYBR Green RT-PCR Master Mix, 10 pmol of each primer (Table 2), 5 ng of purified RNA template, and 0.25 μl of QuantiTect RT mix. Initial reverse transcription at 50°C for 30 min was followed by a denaturation step of 15 min at 95°C. Amplification was then performed over 35 cycles, with each cycle consisting of 15 s at 94°C, 30 s at appropriate annealing temperature (Table 2), and 30 s at 72°C, with a final melting step. Within bacterial cells, the level of 16S rRNA was assumed to be transcribed at a constant rate throughout the growth conditions in this study. Relative gene expression was calculated using the threshold cycle method (24).
Phenotype microarray.
Ciprofloxacin-sensitive parents and ciprofloxacin-resistant mutants were examined for cellular phenotypes using Omnilog phenotype microarrays (PM11-20) (Biolog, Inc., Hayward, CA). Briefly, bacteria were grown on blood agar overnight at 37°C. Colonies were picked with a sterile cotton swab and suspended in 10 ml IF-0a (Biolog), and the cell density was adjusted to an OD600 of 0.035 on a spectrophotometer (Biomate 5; Thermospectronic, Cambridge, United Kingdom). A 750-μl aliquot of this cell suspension was added to 150 ml IF-10 (Biolog). Microtiter plates were inoculated with 100 μl of cell suspension per well, incubated at 37°C for 48 h in the Omnilog, and monitored continuously for color changes in the wells. Kinetic data were analyzed with Omnilog PM software.
Complementation.
The relevance of target gene mutations in 104-cip and 5408-cip was evaluated by complementation assays. Plasmids containing the wild-type alleles were introduced into the mutant cells by electrotransformation, and transformants were selected on LB agar supplemented with 50 μg/ml kanamycin and incubated at 37°C overnight. Quinolone MICs of transformants were compared to those of parent and mutant strains using Etest strips.
P22 transduction.
Gene deletions (acrB, marA, soxS, and ramA) from SL1344 mutants were transduced into bacterial isolates by using P22 phage according to standard procedures. The resulting deletional mutants were selected on LB agar containing 50 μg/ml kanamycin, and insertion of the kanamycin resistance gene into mutants was confirmed by PCR using primers listed in Table 2.
Analysis of LPS: cell lysis and proteinase K digestion.
LPS was isolated by proteinase K treatment of bacterial cells as described by Hitchcock and Brown (17). LPS preparations were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis with a 4% (wt/vol) stacking gel and a 15% (wt/vol) separating gel. LPS from Salmonella enterica serotype Minnesota (Sigma-Aldrich) was used as a smooth LPS control. A full-range rainbow molecular weight marker was used as the size standard (Amersham, England). Following electrophoresis, the LPS was visualized by silver staining as previously described by Tsai and Frasch (47).
SDS-polyacrylamide gel electrophoresis analyses and immunocharacterization.
Exponential-phase bacteria in LB broth were pelleted and solubilized in boiling buffer at 96°C as previously described (25). Equal amounts of total cell protein (OD600 = 0.01) were loaded onto an SDS-polyacrylamide gel (10% polyacrylamide, 0.1% SDS). Gels were stained with Coomassie brilliant blue R-250 (0.25% [wt/vol]). For Western blots, proteins were electrotransferred onto nitrocellulose membranes in transfer buffer (25). An initial saturating step was performed overnight at 4°C with Tris-buffered sodium (TBS; 50 mM Tris-HCl [pH 8.0], 150 mM NaCl) containing skim milk powder (10%). The nitrocellulose sheets were then incubated in TBS containing skim milk powder (10%) and Triton X-100 (0.2%) for 2 h at room temperature in the presence of polyclonal antibodies (1:2,000 dilution) directed against denatured OmpF porin or with F4 polyclonal antibody directed against the L3 internal loop of E. coli porins (11). These antibodies directed against denatured OmpF and the L3 internal porin loop recognized the denatured enterobacterial porins including Salmonella F and D porins (44). The detection of antigen-antibody complexes was performed with alkaline phosphatase-conjugated AffinitiPure goat anti-rabbit immunoglobulin G antibodies as previously reported (25).
RESULTS
Antimicrobial susceptibility.
Two field isolates (CUH48 and CUH52) and the reference strain (NCTC 13349) were fully susceptible to nalidixic acid (MICs, ≤8 μg/ml) and ciprofloxacin (MICs, ≤0.023 μg/ml). Seven strains showing high-level nalidixic acid resistance (MIC, ≥256 μg/ml) displayed decreased susceptibility to ciprofloxacin (MICs between 0.125 and 0.19 μg/ml). The ciprofloxacin-selected mutants, 104-cip and 5408-cip, displayed high-level quinolone resistance (nalidixic acid MICs, ≥256 μg/ml; ciprofloxacin MICs, ≥32 μg/ml). Additionally, 104-cip showed decreased susceptibility to tetracycline, ampicillin, and chloramphenicol and 5408-cip showed decreased susceptibility to sulfamethoxazole-trimethoprim, ampicillin, and chloramphenicol (Table 3). Phenotype microarray analysis showed that 104-cip tested resistant to 36 antimicrobials including beta-lactams, narrow- and expanded-spectrum cephalosporins, chloramphenicol, tetracycline, fungicides, and biocides. Similarly, 5408-cip tested resistant to 23 antimicrobials, including beta-lactams, narrow- and expanded-spectrum cephalosporins, chloramphenicol, folate synthesis inhibitors, macrolides, aminoglycosides, and chelators (data not shown).
TABLE 3.
Antimicrobial susceptibilities of wild-type serovar Enteritidis strains, their isogenic ciprofloxacin-resistant mutants, and deletional mutants
| Strain | MIC (μg/ml) of druga:
|
|||
|---|---|---|---|---|
| TC | TS | AM | CL | |
| 104 | 1 (0.19) | 0.094 (0.023) | 1 (0.75) | 2 (0.75) |
| 104-cip | 12 (3) | 0.19 (0.023) | 16 (1) | 96 (1) |
| 104-cip acrB::aph | 0.25 | 0.008 | 0.125 | 0.75 |
| 104-cip marA::aph | 1.5 | 0.064 | 1.5 | 3 |
| 104-cip soxS::aph | 0.75 | 0.016 | 0.5 | 1.5 |
| 5408 | 1 (0.19) | 0.064 (0.023) | 1 (1) | 3 (0.5) |
| 5408-cip | 1.5 (0.094) | 0.25 (0.032) | 4 (2) | 16 (0.38) |
| 5408-cip acrB::aph | 0.094 | 0.023 | 0.094 | 0.5 |
| 5408-cip ramA::aph | 1 | 0.064 | 1 | 3 |
AM, ampicillin; CL, chloramphenicol; TC, tetracycline; TS, trimethoprim-sulfamethoxazole. Values in parentheses are disk diffusion results in the presence of efflux pump inhibitor PAβN at 80 μg/ml. Values represent means of three separate determinations.
Contribution of target gene mutations to quinolone resistance.
Fully quinolone-susceptible strains had no mutations in the target genes (Table 1). A single gyrA mutation (D87Y) was present in all strains showing high-level nalidixic acid resistance. Complementation of gyrA in both 104 and 5408 reduced nalidixic acid MICs to 4 μg/ml in both isolates and decreased ciprofloxacin MICs to 0.047 μg/ml and 0.032 μg/ml, respectively. In 104-cip an additional mutation in gyrA (S83F) was associated with the development of high-level ciprofloxacin resistance. In 5408-cip novel mutations were detected in gyrB (E466D) and parE (V461G) (Table 1). Complementation of gyrA in 104-cip (S83F, D87Y) restored susceptibility to nalidixic acid (MIC, 8 μg/ml) and ciprofloxacin (MIC, 0.094 μg/ml). Complementation of gyrA and gyrB in 5408-cip reduced ciprofloxacin MICs to 1 and 3 μg/ml, respectively. Complementation of parE in 5408-cip had no effect on quinolone MICs (Table 1).
Contribution of efflux pump activity to antibiotic resistance.
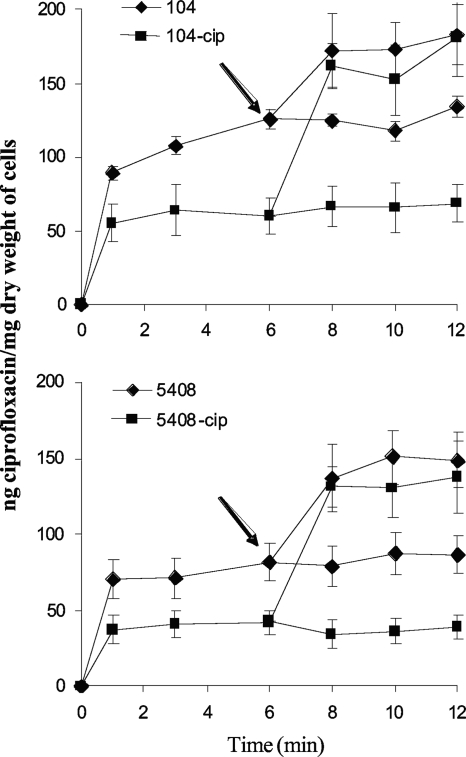
The contribution of efflux pump activity to antibiotic resistance was assessed by use of the efflux pump inhibitor PAβN. PAβN decreased the MIC of nalidixic acid and ciprofloxacin in all wild-type strains (Table 1). It decreased the MIC of ciprofloxacin in 104-cip to 2 μg/ml but had no effect on the MIC of nalidixic acid. The MICs of both nalidixic acid and ciprofloxacin in 5408-cip were reduced to 16 μg/ml and 2 μg/ml, respectively, in the presence of PAβN (Table 1). The susceptibility of ciprofloxacin-resistant mutants to other antibiotics was also restored in the presence of PAβN (Table 3). Both mutants accumulated less ciprofloxacin than did their isogenic parents (Fig. 1). Addition of PAβN increased ciprofloxacin accumulation in both mutants and parents, the two eventually reaching identical steady-state levels of cell-associated ciprofloxacin.
FIG. 1.
Accumulation of ciprofloxacin in in vitro-selected ciprofloxacin-resistant serovar Enteritidis isolates (104-cip and 5408-cip) and their isogenic parent strains (104 and 5408) in the presence and absence of PAβN (80 μg/ml). Ciprofloxacin (10 μg/ml) was added to each bacterial suspension at time zero. PAβN was added at 6 min as indicated by the arrow. Each value represents the mean ± standard error of the mean of three or four separate experiments.
Contribution of AcrAB-TolC to antibiotic resistance.
104-cip and 5408-cip showed increased expression of acrB (6.1- ± 1.5- and 5.4- ± 1.6-fold increases, respectively). Deletion of acrB decreased the MIC of ciprofloxacin from ≥32 to 0.38 μg/ml in 104-cip and from ≥32 to 0.5 μg/ml in 5408-cip and had no effect on the MICs of nalidixic acid (Table 1). Susceptibility to other classes of antibiotics was also restored in both deletional mutants (Table 3). Deletion of acrB also decreased the MICs of ciprofloxacin and nalidixic acid in isolates (51, CUH60, 4931, 104, and 5408) showing nalidixic acid resistance and decreased susceptibility to ciprofloxacin (Table 1).
Contribution of global regulators to antibiotic resistance.
In 104-cip there was a 26.1- ± 4.0-fold increase in soxS expression and an 8.9- ± 0.6-fold increase in marA expression. Deletion of soxS in 104-cip decreased the expression of acrB by −4.2- ± 1.0-fold and the expression of marA by −0.6- ± 1.6-fold and decreased the ciprofloxacin MIC to 1.5 μg/ml (Table 1). Deletion of marA in 104-cip decreased the expression of acrB by 1.4- ± 0.3-fold and the expression of soxS by 1.4- ± 0.2-fold and decreased the ciprofloxacin MIC to 4 μg/ml. Both marA and soxS deletional mutants remained nalidixic acid resistant (≥256 μg/ml) but lost their MDR phenotype (Table 3). In 5408-cip there was significantly increased expression of ramA (33.7- ± 4.0-fold). Deletion of ramA in 5408-cip reduced acrB expression by 1.6- ± 0.1-fold and decreased the MIC of ciprofloxacin to 4 μg/ml. It had no effect on the MIC of nalidixic acid (≥256 μg/ml) (Table 1). The ramA deletional mutant became susceptible to other classes of antibiotics (Table 3). Expression of rob was found to be decreased in both mutants (Table 1).
Genetic analysis of local and global regulators.
Mutations were found in soxR (R20H) and in soxS (E52K) in 104-cip. The soxR mutation mapped to the helix-turn-helix region of the SoxR protein. A mutation was also found in ramR (G25A) in 5408-cip, which is found upstream of ramA. No mutations were found in the local regulator acrR or the global regulators marORAB and ramA.
Porin expression and LPS profiles.
Immunodetection of OmpF with polyclonal antibodies directed against the denatured OmpF porin and with an F4 polyclonal antibody directed against the L3 internal loop of E. coli porins revealed a decrease in the production of OmpF in 104-cip (Fig. 2). All isolates displayed a smooth LPS phenotype. The LPS profile of 104-cip showed significant loss of short and intermediate O-chain LPSs compared to its isogenic parent (data not shown). No changes were observed in the LPS or OmpF profile of 5408-cip.
FIG. 2.
The detection of porins was carried out using the polyclonal antibodies directed against denatured OmpF porin (A) or the F4 polyclonal antibody directed against the L3 internal loop of E. coli porins (B). Lanes 1, 104; lanes 2, 104-cip; lanes 3, 5408; lanes 4, 5408-cip, lanes 5, NCTC 13349. Arrows indicate the migration of F and D porins, respectively.
DISCUSSION
In this study, we confirmed that both AcrAB-TolC efflux pump activity and a D87 mutation in gyrA contribute to quinolone resistance in serovar Enteritidis field isolates displaying high-level nalidixic acid resistance and decreased susceptibility to fluoroquinolones (7). Ciprofloxacin-resistant and MDR mutants were readily selected in vitro from two of these field isolates, highlighting the ease with which resistance to fluoroquinolones and other clinically important antibiotics could potentially emerge during prolonged fluoroquinolone therapy in infected patients. Selection for high-level ciprofloxacin resistance was associated with the development of an additional gyrA mutation in 104-cip and hitherto-undocumented mutations in gyrB and parE in 5408-cip, overexpression of acrB and the development of an MDR phenotype in both isolates, and a decrease in OmpF expression and altered LPS in 104-cip.
Double mutations in gyrA have been widely reported in ciprofloxacin-resistant Salmonella isolates, whereas mutations in gyrB and parE in Salmonella are rarely detected (18). By complementation, we defined a significant role for the double gyrA mutations (D87Y and S83F) in quinolone resistance in 104-cip and for the single gyrA (D87Y) and the novel gyrB (E466D) mutation in ciprofloxacin resistance in 5408-cip. Although complementation with wild-type parE was without effect, the possibility that parE mutations may indirectly contribute to high-level fluoroquinolone resistance by increasing the level of resistance in isolates already harboring target gene mutations cannot be excluded (23).
Consistent with increased expression of an RND multidrug efflux pump, both mutants displayed an MDR phenotype. The increase in acrB expression was reflected in the decrease observed in ciprofloxacin accumulation, which increased following addition of PAβN. Inactivation of acrB restored susceptibility to ciprofloxacin and other nonquinolone antibiotics tested in accordance with previous reports that overexpression of AcrAB-TolC contributes to fluoroquinolone resistance and MDR in Salmonella (3-5). Similar effects were observed with PAβN, highlighting its utility as a pharmacological tool to screen for efflux-mediated antibiotic resistance in Salmonella. Interestingly, we observed a reproducible discrepancy between the effects of PAβN and deletion of acrB on nalidixic acid resistance in 5408-cip. The possibility of the contribution of an unidentified efflux pump, sensitive to PAβN, to nalidixic acid resistance in this isolate cannot be excluded.
The increased expression of acrB was associated with differential expression of global regulators, with 104-cip showing increased expression of both soxS and marA and 5408-cip showing increased expression of ramA. By deleting each of these regulators, we provided direct evidence for their involvement in fluoroquinolone resistance and MDR in serovar Enteritidis through acrB activation. To the best of our knowledge, this is the first report to document a direct contribution of marA and soxS to AcrAB-mediated MDR in Salmonella. Compared to the marA deletional mutant, the soxS deletional mutant showed greater downregulation of acrB and displayed lower MICs of ciprofloxacin and other antibiotics tested. These data suggest that soxS plays a greater role than marA in MDR in 104-cip. Expression of marA decreased following deletion of soxS and vice versa, highlighting the cross-regulation that exists between these transcriptional factors (12, 43). Recently, ramA has been reported to contribute to fluoroquinolone resistance and MDR in serovar Typhimurium through activation of acrB (1). Similarly our data clearly define a regulatory role for ramA in AcrAB-mediated MDR in serovar Enteritidis and furthermore show that ramA activates the MDR cascade independently of marA. As overexpression of rob has been shown to confer MDR in E. coli through activation of acrB (46), it is reasonable to assume a lack of involvement of this regulator in the development of MDR in this study. The decreased expression of rob in 104-cip is likely due to downregulation by soxS and marA (28, 43). The nature and extent of the cross talk between ramA and other global regulators are currently unknown. However, based on our data it would be interesting to speculate that it may downregulate both soxS and rob.
Increased expression of soxS and marA in E. coli has been attributed to mutations in the soxR gene that render soxR active independent of oxidative stress or mutations in marR that alleviate its repression of marA (21, 32, 49). To date, there is only one report documenting the contribution of a mutation in soxR to increased soxS expression and MDR in Salmonella (20). We identified mutations in both soxR and soxS in 104-cip. The same soxR mutation has been linked to increased soxS expression and multiple antibiotic resistance in E. coli (21). Mutations in soxS have been reported in E. coli isolates overexpressing soxS, but their significance was not determined (49). Similar to other studies of Salmonella (7, 12), we found no mutations in the marR or marO region that could explain the increased expression of marA. Therefore, it most likely resulted from feed-forward activation by SoxS. Sequence analysis also revealed a hitherto-unreported mutation within the recently defined local repressor ramR (G25A) of ramA in 5408-cip. Mutations in ramR have been reported to play a role in upregulation of ramA and AcrAB and consequently the efflux-mediated MDR phenotype in serovar Typhimurium (1). The significance of these mutations in soxR, soxS, and ramR is currently being investigated.
OmpF expression was decreased in 104-cip, consistent with the role of marRAB and soxRS in the control of its expression (37, 40). This mutant also displayed alterations in the LPS ladder. In contrast to the findings in Enterobacter aerogenes and E. coli (8), ramA-mediated MDR in 5408-cip was not associated with downregulation of porins. Few studies have investigated OmpF expression in fluoroquinolone-resistant Salmonella, and its contribution to resistance is unclear (14, 29, 35). Only one study to date has documented alterations in the LPS profile in fluoroquinolone-resistant Salmonella, and the authors suggested that the increase in the proportion of long O-chain LPSs observed could result in a lower level of antibiotic accessibility to the porins (14). Further studies are warranted to evaluate the exact contribution of altered porin expression and LPS to antibiotic resistance in 104-cip. Nonetheless, based on our data showing that both 104-cip and its isogenic parent accumulated the same amount of ciprofloxacin at steady state following the addition of PAβN, it would appear that the decreased accumulation of ciprofloxacin observed in 104-cip was mainly due to enhanced efflux activity rather than decreased influx resulting from altered membrane permeability. Furthermore, the hypersusceptibility to ampicillin, tetracycline, and chloramphenicol (which also enter through the porin pathway) observed following deletion of acrB also suggests that active efflux is the main mechanism associated with MDR in this isolate.
Finally, this study revealed that different mechanisms were involved in the development of MDR following ciprofloxacin exposure, as the two mutants displayed different phenotypes of resistance to nonquinolone antibiotics. It is possible that these differences may be due to pleiotropic effects associated with the different expression profiles of global regulators observed in these isolates.
In summary, this study highlights that a high-level ciprofloxacin resistance and MDR phenotype in serovar Enteritidis can result from different genetic events associated with multiple resistance mechanisms. It provides direct evidence that quinolone resistance and MDR in serovar Enteritidis result from interplay between target gene mutations and increased AcrAB-TolC efflux activity and defines a role for a novel gyrB mutation in ciprofloxacin resistance. Increased AcrAB-TolC efflux activity in fluoroquinolone-resistant and MDR isolates can be due to differential expression of the global regulators soxS, marA, and ramA associated with mutations in their regulatory genes.
Acknowledgments
We thank Jonathan Caddick and Vito Ricci (University of Birmingham) for technical training in complementation and phage transduction experiments.
This work was supported by COST Action BM0701 “ATENS”.
Footnotes
Published ahead of print on 22 December 2008.
REFERENCES
- 1.Abouzeed, Y. M., S. Baucheron, and A. Cloeckaert. 2008. ramR mutations involved in efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 52:2428-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baucheron, S., E. Chaslus-Dancla, and A. Cloeckaert. 2004. Role of TolC and parC mutation in high-level fluoroquinolone resistance in Salmonella enterica serotype Typhimurium DT204. J. Antimicrob. Chemother. 53:657-659. [DOI] [PubMed] [Google Scholar]
- 4.Baucheron, S., H. Imberechts, E. Chaslus-Dancla, and A. Cloeckaert. 2002. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type DT204. Microb. Drug Resist. 8:281-289. [DOI] [PubMed] [Google Scholar]
- 5.Baucheron, S., S. Tyler, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. AcrAB-TolC directs efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium DT104. Antimicrob. Agents Chemother. 48:3729-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrique-Mas, J. J., C. Papadopoulou, S. J. Evans, A. Wales, C. J. Teale, and R. H. Davies. 2008. Trends in phage types and antimicrobial resistance of Salmonella enterica serovar Enteritidis isolated from animals in Great Britain from 1990 to 2005. Vet. Rec. 162:541-546. [DOI] [PubMed] [Google Scholar]
- 7.Chen, S., S. Cui, P. F. McDermott, S. Zhao, D. G. White, I. Paulsen, and J. Meng. 2007. Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar Typhimurium to fluoroquinolones and other antimicrobials. Antimicrob. Agents Chemother. 51:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chollet, R., J. Chevalier, C. Bollet, J. M. Pages, and A. Davin-Regli. 2004. RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob. Agents Chemother. 48:2518-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CLSI. 2004. Performance standards for antimicrobial susceptibility testing. CLSI document M100-S14. CLSI, Wayne, PA.
- 10.Cohen, S. P., H. Hachler, and S. B. Levy. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De, E., A. Basle, M. Jaquinod, N. Saint, M. Mallea, G. Molle, and J. M. Pages. 2001. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol. Microbiol. 41:189-198. [DOI] [PubMed] [Google Scholar]
- 12.Eaves, D. J., V. Ricci, and L. J. Piddock. 2004. Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica serovar Typhimurium: role in multiple antibiotic resistance. Antimicrob. Agents Chemother. 48:1145-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giraud, E., S. Baucheron, and A. Cloeckaert. 2006. Resistance to fluoroquinolones in Salmonella: emerging mechanisms and resistance prevention strategies. Microbes Infect. 8:1937-1944. [DOI] [PubMed] [Google Scholar]
- 14.Giraud, E., A. Cloeckaert, D. Kerboeuf, and E. Chaslus-Dancla. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heisig, P. 1993. High-level fluoroquinolone resistance in a Salmonella typhimurium isolate due to alterations in both gyrA and gyrB genes. J. Antimicrob. Chemother. 32:367-377. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Urzua, E., D. S. Zamorano-Sanchez, J. Ponce-Coria, E. Morett, S. Grogan, R. K. Poole, and J. Membrillo-Hernandez. 2007. Multiple regulators of the flavohaemoglobin (hmp) gene of Salmonella enterica serovar Typhimurium include RamA, a transcriptional regulator conferring the multidrug resistance phenotype. Arch. Microbiol. 187:67-77. [DOI] [PubMed] [Google Scholar]
- 17.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopkins, K. L., R. H. Davies, and E. J. Threlfall. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25:358-373. [DOI] [PubMed] [Google Scholar]
- 19.Keeney, D., A. Ruzin, and P. A. Bradford. 2007. RamA, a transcriptional regulator, and AcrAB, an RND-type efflux pump, are associated with decreased susceptibility to tigecycline in Enterobacter cloacae. Microb. Drug Resist. 13:1-6. [DOI] [PubMed] [Google Scholar]
- 20.Koutsolioutsou, A., E. A. Martins, D. G. White, S. B. Levy, and B. Demple. 2001. A soxRS-constitutive mutation contributing to antibiotic resistance in a clinical isolate of Salmonella enterica (serovar Typhimurium). Antimicrob. Agents Chemother. 45:38-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koutsolioutsou, A., S. Pena-Llopis, and B. Demple. 2005. Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob. Agents Chemother. 49:2746-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Z., and B. Demple. 1994. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J. Biol. Chem. 269:18371-18377. [PubMed] [Google Scholar]
- 23.Ling, J. M., E. W. Chan, A. W. Lam, and A. F. Cheng. 2003. Mutations in topoisomerase genes of fluoroquinolone-resistant salmonellae in Hong Kong. Antimicrob. Agents Chemother. 47:3567-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 25.Mallea, M., J. Chevalier, C. Bornet, A. Eyraud, A. Davin-Regli, C. Bollet, and J. M. Pages. 1998. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology 144:3003-3009. [DOI] [PubMed] [Google Scholar]
- 26.McCarron, B., and W. C. Love. 1997. Acalculous nontyphoidal salmonellal cholecystitis requiring surgical intervention despite ciprofloxacin therapy: report of three cases. Clin. Infect. Dis. 24:707-709. [DOI] [PubMed] [Google Scholar]
- 27.Meakins, S., I. S. Fisher, C. Berghold, P. Gerner-Smidt, H. Tschape, M. Cormican, I. Luzzi, F. Schneider, W. Wannett, J. Coia, A. Echeita, and E. J. Threlfall. 2008. Antimicrobial drug resistance in human nontyphoidal Salmonella isolates in Europe 2000-2004: a report from the Enter-net International Surveillance Network. Microb. Drug Resist. 14:31-35. [DOI] [PubMed] [Google Scholar]
- 28.Michan, C., M. Manchado, and C. Pueyo. 2002. SoxRS down-regulation of rob transcription. J. Bacteriol. 184:4733-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miro, E., C. Verges, I. Garcia, B. Mirelis, F. Navarro, P. Coll, G. Prats, and L. Martinez-Martinez. 2004. Resistance to quinolones and beta-lactams in Salmonella enterica due to mutations in topoisomerase-encoding genes, altered cell permeability and expression of an active efflux system. Enferm. Infecc. Microbiol. Clin. 22:204-211. (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 30.Molbak, K., D. L. Baggesen, F. M. Aarestrup, J. M. Ebbesen, J. Engberg, K. Frydendahl, P. Gerner-Smidt, A. M. Petersen, and H. C. Wegener. 1999. An outbreak of multidrug-resistant, quinolone-resistant Salmonella enterica serotype Typhimurium DT104. N. Engl. J. Med. 341:1420-1425. [DOI] [PubMed] [Google Scholar]
- 31.Mortimer, P. G., and L. J. Piddock. 1991. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 28:639-653. [DOI] [PubMed] [Google Scholar]
- 32.Oethinger, M., I. Podglajen, W. V. Kern, and S. B. Levy. 1998. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob. Agents Chemother. 42:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olliver, A., M. Valle, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Role of an acrR mutation in multidrug resistance of in vitro-selected fluoroquinolone-resistant mutants of Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 238:267-272. [DOI] [PubMed] [Google Scholar]
- 34.Piddock, L. J. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piddock, L. J., D. J. Griggs, M. C. Hall, and Y. F. Jin. 1993. Ciprofloxacin resistance in clinical isolates of Salmonella typhimurium obtained from two patients. Antimicrob. Agents Chemother. 37:662-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piddock, L. J., V. Ricci, I. McLaren, and D. J. Griggs. 1998. Role of mutation in the gyrA and parC genes of nalidixic-acid-resistant salmonella serotypes isolated from animals in the United Kingdom. J. Antimicrob. Chemother. 41:635-641. [DOI] [PubMed] [Google Scholar]
- 37.Pomposiello, P. J., and B. Demple. 2000. Identification of SoxS-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prouty, A. M., I. E. Brodsky, S. Falkow, and J. S. Gunn. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150:775-783. [DOI] [PubMed] [Google Scholar]
- 39.Randall, L. P., and M. J. Woodward. 2001. Multiple antibiotic resistance (mar) locus in Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 67:1190-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Randall, L. P., and M. J. Woodward. 2002. The multiple antibiotic resistance (mar) locus and its significance. Res. Vet. Sci. 72:87-93. [DOI] [PubMed] [Google Scholar]
- 41.Ricci, V., P. Tzakas, A. Buckley, and L. J. Piddock. 2006. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob. Agents Chemother. 50:38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruzin, A., M. A. Visalli, D. Keeney, and P. A. Bradford. 2005. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 49:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneiders, T., and S. B. Levy. 2006. MarA-mediated transcriptional repression of the rob promoter. J. Biol. Chem. 281:10049-10055. [DOI] [PubMed] [Google Scholar]
- 44.Simonet, V., M. Mallea, D. Fourel, J. M. Bolla, and J. M. Pages. 1996. Crucial domains are conserved in Enterobacteriaceae porins. FEMS Microbiol. Lett. 136:91-97. [DOI] [PubMed] [Google Scholar]
- 45.Sulavik, M. C., M. Dazer, and P. F. Miller. 1997. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J. Bacteriol. 179:1857-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka, T., T. Horii, K. Shibayama, K. Sato, S. Ohsuka, Y. Arakawa, K. Yamaki, K. Takagi, and M. Ohta. 1997. RobA-induced multiple antibiotic resistance largely depends on the activation of the AcrAB efflux. Microbiol. Immunol. 41:697-702. [DOI] [PubMed] [Google Scholar]
- 47.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 48.Vasallo, F. J., P. Martin-Rabadan, L. Alcala, J. M. Garcia-Lechuz, M. Rodriguez-Creixems, and E. Bouza. 1998. Failure of ciprofloxacin therapy for invasive nontyphoidal salmonellosis. Clin. Infect. Dis. 26:535-536. [DOI] [PubMed] [Google Scholar]
- 49.Webber, M. A., and L. J. Piddock. 2001. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob. Agents Chemother. 45:1550-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, J., W. R. Dunham, and B. Weiss. 1995. Overproduction and physical characterization of SoxR, a [2Fe-2S] protein that governs an oxidative response regulon in Escherichia coli. J. Biol. Chem. 270:10323-10327. [DOI] [PubMed] [Google Scholar]