Abstract
The CCR100136 (EPIC) study evaluated the antiviral activity of the novel CCR5 entry inhibitor aplaviroc in combination with lopinavir-ritonavir in drug-naïve human immunodeficiency virus type 1-infected subjects. Although the trial was stopped prematurely due to idiosyncratic hepatotoxicity, 11 subjects met the protocol-defined virologic failure criteria. Clonal analyses of the viral envelope tropism, aplaviroc susceptibility, and env sequencing were performed on plasma at day 1 and at the time of virologic failure. Molecular evolutionary analyses were also performed. Treatment-emergent resistance to aplaviroc or lopinavir-ritonavir was not observed at the population level. However, aplaviroc resistance was detected prior to therapy at both the clonal and population levels in one subject with virologic failure and in six subjects in a minority (<50%) of clones at day 1 or at the time of virologic failure. Reduced aplaviroc susceptibility manifested as a 50% inhibitory concentration curve shift and/or a plateau. Sequence changes in the clones with aplaviroc resistance were unique to each subject and scattered across the envelope coding region. Clones at day 1 and at the time of virologic failure were not phylogenetically distinct. Two subjects with virologic failure had a population tropism change from CCR5- to dual/mixed-tropic during treatment. Virologic failure during a regimen of aplaviroc and lopinavir-ritonavir may be associated with aplaviroc resistance, only at the clonal level, and/or, infrequently, tropism changes.
Although highly active antiretroviral therapy results in a profound and sustained reduction in plasma human immunodeficiency virus type 1 (HIV-1) RNA in many individuals, the side effects of currently available antiretroviral therapy (ART) and the emergence of multidrug-resistant viral strains continue to represent major challenges for the management of HIV infection (7, 29). The discovery that the chemokine receptors CCR5 and CXCR4 function as coreceptors that mediate HIV-1 entry into CD4+ host cells (2) has led to the development of coreceptor inhibitors, which are currently being tested for use in ART.
Unlike other drug classes, resistance to CCR5 entry inhibitors (CCR5 EIs) could involve the use of drug-bound CCR5 or a change in coreceptor usage. Much of the data on CCR5 EI resistance come from in vitro passage and short-term monotherapy studies. An analysis of virus envelopes (Env) during passage studies with CCR5 EIs demonstrated the maintenance of R5 tropism in a setting where the cells utilized expressed both CCR5 and CXCR4 (17, 34, 38). Resistance or reduced susceptibility to aplaviroc (APL) was slow to emerge in passage studies; the characterization of CCR5-tropic (R5-tropic) HIV-1 isolates from long-term passage (>48 weeks) showed marginal increases in the change (n-fold) in the 50% inhibitory concentration (FCIC50) values, the standard measure by which resistance to other antiretroviral classes is detected (16, 17).
Another measure of drug susceptibility that has been suggested for CCR5 EIs is a decreased maximum calculated percent inhibition (MPI) value (25). An MPI value of <100% indicates that the virus is unable to be fully suppressed by high concentrations of drug, implying the ability to use a drug-bound receptor. In vitro studies with other CCR5 EIs have found a plateau in the MPI value in some instances (15, 31, 38). Short-term monotherapy studies with CCR5 EIs demonstrated that while the majority of individuals maintained R5-tropic virus, tropism readout changes from R5 to dual/mixed (DM)-tropic at the population level were observed in a few subjects in the absence of a change in FCIC50 or MPI values (15, 37). Phylogenetic studies on clones from these subjects found evidence in favor of the emergence of a preexisting CXCR4-using env population rather than a switch from a CCR5- to a CXCR4-using phenotype during the course of therapy.
Aplaviroc was developed for the treatment of HIV-1 infection in combination with other ARTs. In vitro data demonstrated that APL is a specific CCR5 antagonist that inhibits R5-tropic HIV-1 replication at subnanomolar concentrations (4, 21). A 10-day study of APL monotherapy in HIV-1-positive subjects demonstrated acceptable short-term safety and potency, with a mean 1.66 log10 copies/milliliter (c/ml) decrease at the nadir in the viral load from the baseline in the highest dosage arm (18).
CCR100136, a larger phase IIb study, was designed to evaluate a novel two-drug regimen comprising several doses of APL administered in combination with a boosted protease inhibitor (PI) (lopinavir-ritonavir [LPV-r]; Kaletra) in treatment-naïve subjects with either R5- or DM-tropic virus (40). CCR100136 was prematurely terminated due to treatment-emergent, idiosyncratic hepatotoxicity that occurred among some subjects receiving APL (24). The reason(s) for the observed hepatotoxicity remains to be determined; hepatotoxicity did not appear to be associated with the APL dose or mechanism of action. Despite the early termination of the study, 11 of the 191 subjects across all dosing groups met the criteria for protocol-defined virologic failure. Phenotypic and genotypic analyses of the HIV-1 reverse transcriptase (RT), protease (PRO), and envelope revealed that while no subjects with virologic failure developed resistance to LPV-r, the majority of the subjects with failure on APL-containing regimens had resistance to APL detected at the clonal but not the population level. Furthermore, the majority of the subjects had stable tropism readouts, suggesting that changes in coreceptor tropism were not a primary determinant of virologic failure in this study. Despite the premature termination of the APL clinical development program, the virologic analyses described here inform the use of CCR5 EIs with a ritonavir-boosted PI (LPV-r) in the ART-naïve population.
(This work was presented in part at the XV International Drug Resistance Workshop, Sitges, Spain, 13 to 17 June 2006, and the 46th ICAAC, San Francisco, CA, 27 to 30 September 2006.)
MATERIALS AND METHODS
Study population.
ART-naïve subjects (n = 191) were enrolled in CCR100136; the full clinical data set is described elsewhere (40). The presence of only R5 tropism was not a requirement for enrollment in CCR100136; plasma was taken from 173 subjects that tested as R5-tropic and 18 as DM-tropic at enrollment. The subjects received 400/100 mg LPV-r twice daily (BID) in combination with either 200 mg APL BID, 400 mg APL BID, 800 mg APL once daily (QD), or 150 mg/300 mg lamivudine-zidovudine (Combivir) BID. Informed consent was obtained from all patients or their parent/guardian, and human experimentation guidelines in accordance with GlaxoSmithKline policies and standard operating procedures were followed. HIV-1 RNA levels in plasma were assessed by the Roche COBAS monitor Amplicor standard PCR assay (lower limit of detection, 400 c/ml) and the Roche PCR UltraSensitive assay (lower limit of detection, 50 c/ml).
Definition of virologic failure.
Confirmed virologic failure was defined as one of the following: (i) incomplete virological response (the subject did not achieve a 1 log10 c/ml decrease in plasma HIV-1 RNA by week 4, relative to the baseline value) or (ii) virological rebound (the subject's plasma HIV-1 RNA rebounded to ≥400 c/ml on two consecutive measurements at least 2 to 4 weeks apart after being previously <400 c/ml on or after week 4, or the subject had two consecutive viral load determinations at least 2 to 4 weeks apart that were >0.5 log10 c/ml plasma HIV-1 RNA from the nadir value of the study, where the nadir was the lowest HIV-1 value of ≥400 c/ml on or after week 4).
Study samples.
All subjects had plasma samples collected for analysis at screening, day 1, weeks 2 and 4, and every 4 weeks thereafter. Tropism testing was performed on all samples for which the viral load exceeded the validated cutoff of the assay (≥1,000 c/ml). All other testing (RT/PRO genotype and phenotype, APL susceptibility, and HIV-1 env clonal analysis) was performed on plasma samples at the day 1 and confirmed virologic failure time points. Additional time points were tested if an interesting phenotype was observed.
RT/PRO genotype and phenotype.
HIV RT/PRO genotype and phenotype data were generated using the Monogram Biosciences PhenoSense GT assay (San Francisco, CA) (26). Briefly, a pseudovirus is created which contains the relevant viral sequence from the sample of interest (pol) in the context of a backbone of the remaining sequence of HIV. The pseudovirus is then used to evaluate the efficacy of the antiviral compound(s) in a single cycle assay as defined by the FCIC50 value, comparing the susceptibility of the patient RT/PRO to that of the assay control virus.
HIV envelope tropism and APL susceptibility.
HIV envelope tropism and APL susceptibility data were generated by the Monogram Biosciences PhenoSense HIV entry assay (12, 39) by the creation of a pseudovirus containing the env sequence from the sample of interest in the context of a backbone of the remaining sequence of HIV. A single-cycle infection of U87 cells expressing CD4 in combination with either CCR5 or CXCR4 was assessed by luciferase activity. Drug susceptibility was assessed using serial dilutions of APL.
Two measures are currently utilized for the characterization of phenotypic resistance to an HIV EI, FCIC50, and MPI. As IC50s can fluctuate from assay to assay, the FCIC50 value compares the IC50 of the patient sample to that of a control virus included in each assay. Thus, the FCIC50 normalizes data across assays run at different points in time and is a more reliable analysis of changes in drug susceptibility over time. As the clinically relevant FCIC50 cut point for APL has not been determined, a cut point of threefold, based on the intrinsic assay variability, was used. MPI is a measure of virus suppression, indicating complete (100%) or no (0%) suppression in this assay system. Values of <100% suggest incomplete virus suppression and the ability of the viral envelope to utilize a drug-bound receptor. The clinically relevant MPI value for APL has not been determined.
HIV-1 env clonal analysis.
A clonal analysis of HIV-1 env was performed at Monogram Biosciences as previously described (12). Forty-eight clones were screened to determine the relative frequency of HIV envelope tropism. Twelve representative clones were chosen from the 48 screened clones for an analysis of the full-length HIV env genotype and APL sensitivity; tropism was also confirmed. The number of clones with particular tropism readouts in the 12-clone analysis may not accurately reflect the relative percent of these clones in the viral quasispecies as the clones were not necessarily chosen in proportion to the larger 48-clone tropism screening, although selection did represent all of the minority tropic species.
Molecular evolutionary analyses.
Homologous nucleotide sequence alignments of full-length envelope clones were generated for each patient using ClustalX version 1.83 (33) with manual refinement in GeneDoc version 2.6.0.2 (23). Distance- and maximum likelihood (ML)-based phylogenetic reconstructions were performed using PAUP* 4.0b (32), incorporating the best-fitting model of sequence evolution and the corresponding values for the rate matrix, shape of the gamma distribution, and proportion of invariant sites as estimated by Modeltest (27). Bootstrap support values were obtained with 100 replicates of ML bootstrapping. A total of 1,000 replicates of neighbor-joining bootstrapping were also performed to corroborate the ML findings. The neighbor-joining and ML values were highly concordant, and thus, only ML trees and bootstrap values are reported.
In order to evaluate the nucleotide diversity across time points in the envelope clones from each subject, nucleotide diversity calculations were performed using DnaSP4.10 (30). Nucleotide diversity is defined as the average number of nucleotide differences per site between two sequences drawn at random from the population of sequences (22). If there was a change in env nucleotide diversity over time or a separation of time points on the phylogenetic tree, then statistical tests for genetic differentiation (1, 10, 11) were performed using DnaSP4.10. Due to small sample sizes and in order to be conservative in the determination of temporal structure, the test statistics were considered significant at P < 0.001. A rejection of the null hypothesis of no genetic differentiation between subpopulations indicates a partial or complete turnover in the env population between the time points sampled.
Amino acid sequences for the envelope proteins were translated from nucleotide sequence alignments for each subject using GeneDoc version 2.6.0.2 (23). Differences in amino acid composition unique to certain clones, compared to those of the nearest-neighbor APL-sensitive clone, were identified by manual inspection of the conservation at each site as displayed by GeneDoc.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the sequences reported here are FJ653314 to FJ653619.
RESULTS
Characteristics of virologic failures.
Eleven of the 191 subjects (6%) met the protocol definition of virologic failure, with two (4%) subjects in the group receiving 200 mg APL BID, two (4%) in the group receiving 400 mg APL BID, six (12%) in the group receiving 800 mg APL QD, and one (4%) in the lamivudine-zidovudine group (Table 1). Ten of the subjects with virologic failure tested as having R5-tropic virus at screening; one subject (P10) tested as having DM-tropic virus at the screening. Most of the subjects met the protocol definition of virologic failure at or after week 12 (range, 4 to 20 weeks). Three subjects exhibited incomplete virologic responses, while eight exhibited virologic rebounds. There was no consistent pattern in the HIV-1 RNA levels or CD4+ counts between day 1 and the time of virologic failure. In addition, the duration of exposure for those subjects exhibiting virologic failure was similar to that for all subjects across the treatment arms (∼14 weeks mean duration; Table 1 and reference 40).
TABLE 1.
Virologic characteristics of the 11 subjects with virologic failurea
| Subject | APL dose (mg) | Visitb | Population tropism | RT/PRO mutations | CD4 count (cells/mm3)c | CD4 count change (cells/mm3) | Log10 HIV-1 RNA (c/ml)c | HIV-1 RNA change (c/ml) | APL FCIC50 (fold) | APL MPI (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 200 BID | Day 1 | R5 | 77 | 6.256 | 0.90 | 98.8 | |||
| Week 20 | R5 | WT | NT | NT | 4.360 | −1.896 | 1.37 | 97.5 | ||
| P2 | 400 BID | Day 1 | R5 | K103N | 365 | 4.652 | 0.53 | 95.0 | ||
| Week 8 | R5 | K103N | 606 | 241 | 4.987 | 0.335 | 0.64 | 91.6 | ||
| P3 | 800 QD | Day 1 | R5 | 124 | 5.121 | 0.76 | 97.3 | |||
| Week 16 | DM | WT | 162 | 38 | 4.260 | −0.861 | 0.92 | 93.3 | ||
| P4 | 800 QD | Day 1 | R5 | 175 | 5.885 | 1.09 | 99.5 | |||
| Week 4 | R5 | WT | 218 | 43 | 5.860 | −0.025 | 1.11 | 99.8 | ||
| P5 | 800 QD | Day 1 | R5 | 125 | 4.654 | 0.66 | 72.5 | |||
| Week 12 | DM | WT | 122 | −3 | 4.583 | −0.071 | 1.36 | 70.7 | ||
| P6 | 800 QD | Day 1 | R5 | 172 | 4.619 | 0.77 | 98.4 | |||
| Week 4 | R5 | WT | 284 | 112 | 4.356 | −0.263 | 0.67 | 99.0 | ||
| P7 | 800 QD | Day 1 | R5 | M41L | 334 | 5.481 | 0.80 | 99.6 | ||
| Week 20 | R5 | M41L | 363 | 29 | 5.405 | −0.076 | 0.77 | 99.2 | ||
| P8 | 400 BID | Day 1 | DM | NA | 187 | 4.889 | 0.30 | 99.5 | ||
| Week 4 | DM | NA | 213 | 26 | 4.918 | 0.029 | 0.18 | 99.7 | ||
| P9 | 800 QD | Day 1 | R5 | NA | 617 | 4.107 | 1.23 | 96.9 | ||
| Week 12 | R5 | NA | 1182 | 565 | 3.528 | −0.579 | 0.62 | 93.9 | ||
| P10d | 200 BID | Day 1 | DM | 109 | 5.624 | 0.75 | 96.9 | |||
| Week 12 | DM | WT | 178 | 69 | 5.667 | 0.043 | 0.73 | 98.6 | ||
| P11 | —e | Day 1 | R5 | K103N | 342 | 5.243 | 0.57 | 99.0 | ||
| Week 20 | R5 | K103N | 564 | 222 | 3.511 | −1.732 | 0.95 | 98.2 |
NA, value could not be determined; NT, not tested; WT, wild-type sequence detected.
Visits other than on day 1 indicate the virologic failure time point.
Single value per time point, determined as described in Materials and Methods.
Subject was DM-tropic at enrollment.
Subject received lamivudine-zidovudine/LPV-r without APL.
Lack of emergent RT/PRO mutations in subjects with virologic failure.
All subjects had RT/PRO phenotype and genotype testing performed at day 1 and at the time of virologic failure. Primary resistance mutations of note were classified based on the International AIDS Society treatment guidelines (13). For the nine subjects with available genotypes, no significant treatment-emergent PI, nucleoside reverse transcriptase inhibitor (NRTI), or nonnucleoside reverse transcriptase inhibitor (NNRTI) mutations were detected (Table 1). Preexisting NRTI or NNRTI mutations were detected in three subjects. Phenotypic testing confirmed the genotypic results for all subjects (data not shown).
The viral tropism remains stable in the majority of the subjects with virologic failure.
Population tropism readouts remained stable for eight virologic-failure subjects throughout the study, while three subjects exhibited one or more changes in tropism readout (Tables 1 and 2). Subject P8 had a tropism readout change from R5-tropic to DM-tropic at day 1, prior to the initiation of therapy, and remained DM-tropic throughout the study. Subject P3 had tropism readout changes from R5 tropic to nonphenotypable at week 4 to DM-tropic at week 12 to R5-tropic at week 20, after virologic failure (week 16). Subject P5 had tropism readout changes from R5-tropic to DM-tropic at week 2 to R5-tropic at week 4 to DM-tropic at week 8 and remained DM-tropic through week 12 (last visit).
TABLE 2.
Clonal analysis results for the three virologic failure subjects with tropism readout changesa
| Subject | Visit | Population tropism | No. of clones confirmed with indicated tropism
|
||
|---|---|---|---|---|---|
| R5 | R5X4 | X4 | |||
| P3 | Screening | R5 | NT | NT | NT |
| Day 1 | R5 | 12 | 0 | 0 | |
| Week 4 | NP | NT | NT | NT | |
| Week 8 | NT (HIV RNA < 1,000 c/ml) | NT | NT | NT | |
| Week 12 | DM | NT | NT | NT | |
| Week 16 (VF) | DM | 8 | 4 | 0 | |
| P5 | Screening | R5 | 11 | 1 | 0 |
| Day 1 | R5 | 12 | 0 | 0 | |
| Week 2 | DM | NT | NT | NT | |
| Week 4 | R5 | 12 | 0 | 0 | |
| Week 8 | DM | NT | NT | NT | |
| Week 12 (VF) | DM | 11 | 1 | 0 | |
| P8 | Screening | R5 | NT | NT | NT |
| Day 1 | DM | 7 | 5 | 0 | |
| Day 21 | DM | NT | NT | NT | |
| Week 4 (VF) | DM | 7 | 5 | 0 | |
NP, nonphenotypeable; NT, not tested; VF, virologic failure.
In an attempt to better understand the population tropism readout changes, clonal analyses of full-length env were performed at the day 1 and virologic-failure time points. The clonal analysis revealed that subjects P5 and P8 had preexisting dual (R5X4)-tropic clones present at day 1 or at screening (Table 2). Phylogenetic analyses of P8 sequences also suggested that the R5X4-tropic clones observed at the time of virologic failure were preexisting, because they clustered with the day 1 R5X4-tropic clones (Fig. 1). Interestingly, the ratio of R5- and R5X4-tropic clones at the time of virologic failure was not altered compared to that at the pretreatment time points for either subject. All the R5X4-tropic clones isolated from subjects P5 and P8 had high X4 relative light units (RLUs), suggesting the efficient utilization of CXCR4. These clones appear to be comparable to the dual-X clones described by Huang et al. (9).
FIG. 1.
Phylogenetic analysis of full-length env clones from subject P8. Subject P8 exhibited a change in tropism readout from R5-tropic to DM-tropic prior to the initiation of therapy (screening to day 1). Clonal sequences are designated by time point and clone number. R5X4-tropic clones are indicated by a §, and all other clones are R5-tropic. ML phylogeny was reconstructed with support values estimated via 100 replicates of ML bootstrapping. Only bootstrap values of ≥70% are reported on the tree.
The clonal analysis for subject P3 revealed no preexisting R5X4-tropic clones at day 1. A phylogenetic analysis demonstrated that R5X4-tropic clones at the time of virologic failure were intermingled with the R5-tropic clones and did not form a distinct lineage. An analysis of the env sequences revealed that the R5X4-tropic clones did not have any unique amino acid changes compared to those of the intrapatient R5-tropic clones in the V3 region, the primary determinant of coreceptor usage (3, 5, 6, 8), or in the rest of the envelope coding region. Furthermore, the X4 RLUs for the R5X4-tropic clones were close to the assay threshold (<900 RLUs). In total, these data suggest that the R5X4-tropic clones derived from the subject P3 plasma virus may be inefficiently utilizing CXCR4. These clones appear to be comparable to the dual-R clones described by Huang et al. (9).
APL susceptibility is detected at the clonal but not the population level for most virologic-failure subjects.
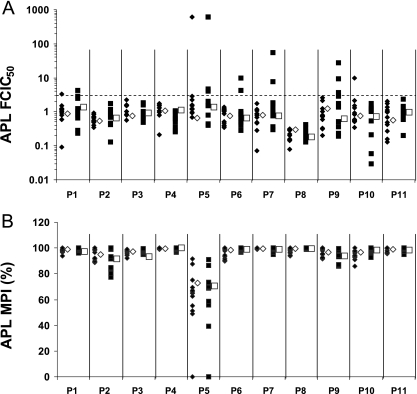
APL susceptibility was assessed for all subjects at day 1 and at the time of virologic failure. Two measures were utilized to characterize APL-reduced susceptibility, FCIC50 and MPI. The majority of the subjects with virologic failure had virus populations sensitive to APL, with 11 subjects having FCIC50 values of <3-fold (Fig. 2A) and 10/11 subjects having MPI values of >90% at all time points tested (Fig. 2B). Subject P5 had decreased MPI values (∼70%) at both day 1 and the time of virologic failure.
FIG. 2.
APL FCIC50 (n-fold) (A) and MPI (B) values at the population and clonal levels for the 11 subjects with virologic failure. Population values are represented by open symbols: diamond, day 1; square, time of virologic failure. Clonal values are represented by shaded symbols: diamond, day 1; square, time of virologic failure.
Reduced susceptibility to APL was detected at the clonal level for seven subjects (Fig. 2A and B). Clones with reduced susceptibility to APL were detected at day 1 for subject P10; at the time of virologic failure for subjects P6, P7, and P9; and at both day 1 and the time of virologic failure for subjects P1, P2, and P5. Interestingly, the three subjects (P6, P7, and P9) with clones exhibiting reduced susceptibility to APL only at the time of virologic failure were all from the group receiving 800 mg QD. For six of the seven subjects with clones exhibiting reduced susceptibility to APL (all but subject P5), the clones made up less than 50% of the population.
The phenotypic characterization of reduced susceptibility to APL varied. Subjects P1, P6, P7, P9, and P10 had clones with only elevated FCIC50 values, subjects P2 and P5 had clones with only reduced MPI values, and subject P5 had clones with both elevated FCIC50 values and reduced MPI values. Intrasubject sequence comparisons were performed between clones exhibiting reduced APL susceptibility (≥3-fold FCIC50) and sensitive clones. Subject P5 was not analyzed because 20/24 clones exhibited elevated FCIC50 values and/or reduced MPI values. These comparisons revealed that clones with reduced susceptibility to APL had relatively low numbers of amino acid changes relative to APL sensitive clones, and the mutations were scattered across the envelope (Fig. 3). No common amino acid change was observed within or across subjects. These data suggest that the genotypic changes conferring reduced susceptibility to APL are context dependent.
FIG. 3.
Intrasubject sequence comparison results. Sequence comparisons were performed between clones exhibiting reduced APL susceptibility (≥3-fold FCIC50) and nearest-neighbor APL-sensitive clones. The amino acid changes in the clones with reduced susceptibility to APL are noted above the cartoon envelope for each clone. On the left-hand side of each envelope cartoon is the clone name, as well as the FCIC50 and MPI values. The amino acid changes are noted with one-letter symbols: the amino acid for the sensitive clone is noted first, followed by the envelope position (using HXB2 numbering), followed by the amino acid for the clone with reduced APL susceptibility. The variable regions of the envelope (V1 to V5) are shaded in each cartoon, as well as the membrane-spanning domain (msd).
Env population turnover.
In order to assess the impact of therapy on the env population of virologic failures, env nucleotide diversity values and phylogenetic trees were determined with the day 1 and virologic-failure clones. These analyses revealed that nine subjects had relatively no change in env nucleotide diversity over time with no separation of time points on phylogenetic trees, indicating no env population turnover (Table 3). Two subjects did exhibit a change in env nucleotide diversity: subject P10 had a decrease, while subject P5 had an increase. However, there was no phylogenetic separation between clones from different time points in these two virologic failures. Despite the lack of distinction on the phylogenetic trees, statistical tests reject the null hypothesis of no population differentiation across time points (P < 0.001), providing evidence for some population turnover in these two subjects.
TABLE 3.
env nucleotide diversity values for the 11 virologic failure subjects
| Subject | Nucleotide diversitya at day 1 | Nucleotide diversity at the time of virologic failure |
|---|---|---|
| P1 | 0.00601 | 0.00851 |
| P2 | 0.01047 | 0.01082 |
| P3 | 0.02401 | 0.02370 |
| P4 | 0.01160 | 0.01369 |
| P5b | 0.01125 | 0.01961 |
| P6 | 0.01919 | 0.02129 |
| P7 | 0.00688 | 0.00696 |
| P8 | 0.01837 | 0.02201 |
| P9 | 0.00418 | 0.00470 |
| P10b | 0.02659 | 0.01883 |
| P11 | 0.01209 | 0.01291 |
Nucleotide diversity is defined as the average number of nucleotide differences per site between two sequences drawn at random from the population of sequences (22).
All three statistical tests for population differentiation across time points are significant at a P of <0.001.
DISCUSSION
CCR5 EIs are a new class of antiretrovirals that have recently entered clinical development. Unlike other drug classes, HIV-1 can develop resistance to CCR5 EIs by two possible mechanisms: (i) by acquiring mutations that allow the virus to utilize CCR5 in its drug-bound state or (ii) by the switching or outgrowth of viruses that can utilize CXCR4. While in vitro studies with CCR5 EIs have demonstrated that viruses will develop resistance by acquiring mutations that allow them to use drug-bound CCR5, little is known regarding the in vivo setting. Despite its early termination, an in-depth evaluation of the protocol-defined virologic failures from CCR100136 provided an opportunity to examine the development of resistance in the in vivo setting.
No treatment emergent PI, NRTI, or NNRTI mutations were observed in any subjects with virologic failure. The lack of emergent PI resistance-associated mutations has been observed in other studies of subjects with first-line virologic failures with highly active antiretroviral therapy containing ritonavir-boosted PIs (14, 20, 35). Drug exposure, genetic barriers to viral resistance, and/or other factors may have contributed to the lack of treatment-emergent PI resistance observed in this study. Low drug exposure, possibly from poor adherence, could have provided insufficient antiviral pressure and allowed for the replication of nonresistant virus. Alternatively, high drug exposure might have required the acquisition of multiple mutations, which would be consistent with a greater genetic barrier producing a delay in the emergence of resistance.
Tropism readout changes were not observed in the majority of the subjects with virologic failure. Due to the concerns about CCR5 EIs and their impact on tropism, it is important to assess the impact of APL-containing regimens when tropism readout changes occur. The three subjects with tropism readout changes had similar phylogenetic patterns: the mixing of time points and little evidence for R5X4-tropic monophyly. Furthermore, R5X4-tropic minority populations were detected at day 1 and/or the screening in two of three subjects. Collectively, these analyses favor the hypothesis of an emergence of preexisting populations of CXCR4-using Envs. This increased proportion of CXCR4-using clones could be due to an expansion of a minority population or a sampling effect, i.e., as the R5-tropic population is diminished, there is a greater probability of sampling CXCR4-using populations. Regardless of the cause, an increased proportion of CXCR4-using virus suggests that the CCR5 EI-containing regimen was effective at reducing the R5-tropic population.
Other factors may have contributed to the tropism readout changes, including changes in the host/virus dynamic, the sampling compartment (i.e., plasma versus tissue), and/or assay limitations. The Monogram tropism assay at the time of this study had an accepted sensitivity threshold of ∼10% (39). For example, up to 10/100 clones could be R5X4-tropic and the assay would call the population R5-tropic. R5-tropic population readouts prior to therapy, when CXCR4-using viruses are present at undetectable levels, could cause misinterpretation that a subject has changed tropism in response to CCR5 EI therapy. This potential misinterpretation is of concern because clonal analysis of the envelope will likely not be a routine test. However, tropism testing has only recently been developed for clinical use and will likely improve as entry biology becomes better understood.
The identification of envs that exhibit elevated FCIC50 values to a CCR5 EI appears to be unique. In vitro (31, 38) and in vivo (19) data demonstrating decreases in MPI values have been reported for other CCR5 EIs; however, in vitro resistance passage studies with APL have only yielded populations bearing elevated FCIC50 values (16, 17). One possible explanation for this difference could be the differential binding of CCR5 EIs. Molecular modeling studies demonstrated that while all CCR5 EIs bound the second extracellular loop of CCR5, there are subtle differences in how each compound occupies the binding pocket (38). Competitive binding studies with CCR5 EIs also demonstrated differences in binding (36). Thus, a subtle difference in CCR5 binding by APL may provide env with a unique mechanism for developing resistance. As such, the appropriate phenotypic measure for CCR5 EI resistance remains an open question.
Clones exhibiting reduced susceptibility to APL made up less than 50% of the population in six of seven subjects. One possibility for why APL reduced susceptibility was not detected at the population level may involve the tissue distribution of APL. If APL concentrations were unequal, this could have caused different compartments to produce viruses with various levels of susceptibility to APL, thus resulting in the plasma virus population being comprised of viruses with a range of APL sensitivity. Another reason for why a population with reduced susceptibility to APL was not detected could relate to assay limitations. The assay used for this analysis utilizes a pseudovirus expressing Env from the sample of interest to infect cell lines engineered to express high levels of CD4 and either CCR5 or CXCR4. This artificial system may not accurately reflect the in vivo environment. Consistent with this, one study has demonstrated differences in the shape of an IC50 curve for a vicriviroc-resistant envelope when tested in a single-cycle cell assay with a cell line compared to a multicycle assay using primary cells (28). It is likely that as we come to understand more about CCR5 EI resistance, resistance assays will be modified to reflect this understanding.
In summary, protocol-defined virologic failure was relatively infrequent in CCR100136 (∼6% of subjects) and was not accompanied by a consistent viral escape mechanism; APL resistance at the clonal level, in the absence of tropism changes or population level resistance to APL and/or LPV-r, appeared to be the primary characteristic of virologic failure. The factors driving the absence of APL resistance at the population level are unknown but could include compartmentalization of APL and/or assay sensitivity. The implication of tropism readout changes in virologic failures with and without the detection of APL resistance remains to be determined.
Acknowledgments
We recognize the study subjects for their participation, the clinical investigators, our colleagues at Monogram Biosciences, and the CCR100136 Clinical Study Team. We also thank Joseph Horton for submission of sequences to GenBank.
This study was funded by GlaxoSmithKline.
All authors are current employees of GlaxoSmithKline, with the exception of K.M.K. and J.M.W.; however, it should be noted that the work described in this paper was completed before their departure. In addition, they are consenting authors. There are no other potential conflicts of interest.
Footnotes
Published ahead of print on 15 December 2008.
REFERENCES
- 1.Achaz, G., S. Palmer, M. Kearney, F. Maldarelli, J. W. Mellors, J. M. Coffin, and J. Wakeley. 2004. A robust measure of HIV-1 population turnover within chronically infected individuals. Mol. Biol. Evol. 21:1902-1912. [DOI] [PubMed] [Google Scholar]
- 2.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 3.De Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demarest, J., S. Shibayama, R. Ferris, C. Vavro, M. St. Clair, and L. Boone. 2004. A novel CCR5 antagonist, 873140, exhibits potent in vitro anti-HIV activity, abstr. WeOr1231. XV Int. AIDS Conf., Bangkok, Thailand, 16 July 2004.
- 5.Fouchier, R. A., M. Brouwer, S. M. Broersen, and H. Schuitemaker. 1995. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J. Clin. Microbiol. 33:906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant, R. M., F. M. Hecht, M. Warmerdam, L. Liu, T. Liegler, C. J. Petropoulos, N. S. Hellmann, M. Chesney, M. P. Busch, and J. O. Kahn. 2002. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA 288:181-188. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman, N. G., F. Seillier-Moiseiwitsch, J. Ahn, J. M. Walker, and R. Swanstrom. 2002. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 76:3852-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, W., S. H. Eshleman, J. Toma, S. Fransen, E. Stawiski, E. E. Paxinos, J. M. Whitcomb, A. M. Young, D. Donnell, F. Mmiro, P. Musoke, L. A. Guay, J. B. Jackson, N. T. Parkin, and C. J. Petropoulos. 2007. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J. Virol. 81:7885-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudson, R. R. 2000. A new statistic for detecting genetic differentiation. Genetics 155:2011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hudson, R. R., D. D. Boos, and N. L. Kaplan. 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9:138-151. [DOI] [PubMed] [Google Scholar]
- 12.Irlbeck, D. M., H. A. Madsen, C. LaBranche, K. M. Kitrinos, and J. F. Demarest. 2008. Chemokine (C-C motif) receptor 5-using envelopes predominate in dual/mixed-tropic HIV from the plasma of drug-naive individuals. AIDS 22:1425-1431. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, V. A., F. Brun-Vézinet, B. Clotet, B. Conway, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, A. Telenti, and D. D. Richman. 2005. Update of the drug resistance mutations in HIV-1: Fall 2005. Top. HIV Med. 13:125-131. [PubMed] [Google Scholar]
- 14.Kempf, D. J., M. S. King, B. Bernstein, P. Cernohous, E. Bauer, J. Moseley, K. Gu, A. Hsu, S. Brun, and E. Sun. 2004. Incidence of resistance in a double-blind study comparing lopinavir/ritonavir plus stavudine and lamivudine to nelfinavir plus stavudine and lamivudine. J. Infect. Dis. 189:51-60. [DOI] [PubMed] [Google Scholar]
- 15.Kitrinos, K., C. LaBranche, M. Stanhope, H. Madsen, and J. Demarest. 2005. Clonal analysis detects pre-existing R5X4-tropic virus in a patient demonstrating population-level tropism shift on 873140 monotherapy. Antivir. Ther. 10:S68. [Google Scholar]
- 16.LaBranche, C., K. Kitrinos, R. Howell, C. McDanal, S. Harris, J. Jeffrey, and J. Demarest. 2005. Clonal analysis of in vitro-derived viruses with reduced susceptibility to aplaviroc (873140, APL) shows wide range in IC50 and sequence changes, abstr. 9. First Int. Workshop Target. HIV Entry, Bethesda, MD, 2 to 3 December 2005.
- 17.LaBranche, C., C. McDanal, and S. Harris. 2005. Analysis of in vitro-derived viruses exhibiting reduced susceptibility to aplaviroc, abstr. H-1098, p. 278. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., American Society for Microbiology, Washington, DC.
- 18.Lalezari, J., M. Thompson, P. Kumar, P. Piliero, R. Davey, K. Patterson, A. Shachoy-Clark, K. Adkison, J. Demarest, Y. Lou, M. Berrey, and S. Piscitelli. 2005. Antiviral activity and safety of 873140, a novel CCR5 antagonist, during short-term monotherapy in HIV-infected adults. AIDS 19:1443-1448. [DOI] [PubMed] [Google Scholar]
- 19.Landovitz, R., G. Faetkenhauer, C. Hoffman, H. Horst, J. M. Strizki, J. Whitcomb, F. Gheyas, D. Knepp, and W. Greaves. 2006. Characterization of susceptibility profiles for the CCR5 antagonist vicriviroc in treatment-naive HIV-infected subjects. Antivir. Ther. 11:S23. [Google Scholar]
- 20.MacManus, S., P. J. Yates, R. C. Elston, S. White, N. Richards, and W. Snowden. 2004. GW433908/ritonavir once daily in antiretroviral therapy-naive HIV-infected patients: absence of protease resistance at 48 weeks. AIDS 18:651-655. [DOI] [PubMed] [Google Scholar]
- 21.Maeda, K., H. Nakata, Y. Koh, T. Miyakawa, H. Ogata, Y. Takaoka, S. Shibayama, K. Sagawa, D. Fukushima, J. Moravek, Y. Koyanagi, and H. Mitsuya. 2004. Spirodiketopiperazine-based CCR5 inhibitor which preserves CC-chemokine/CCR5 interactions and exerts potent activity against R5 human immunodeficiency virus type 1 in vitro. J. Virol. 78:8654-8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nei, M. 1987. DNA polymorphism within and between populations, p. 254-286. In Molecular evolutionary genetics. Columbia University Press, New York, NY.
- 23.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14. [Google Scholar]
- 24.Nichols, W. G., H. M. Steel, T. Bonny, K. Adkison, L. Curtis, J. Millard, K. Kabeya, and N. Clumeck. 2008. Hepatotoxicity observed in clinical trials of aplaviroc (GW873140). Antimicrob. Agents Chemother. 52:858-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petropoulos, C., W. Huang, J. Toma, S. Fransen, S. Bonhoeffer, and J. Whitcomb. 2004. Resistance to HIV-1 entry inhibitors may occur by multiple molecular mechanisms. Antivir. Ther. 9:S25. [Google Scholar]
- 26.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 28.Pugach, P., A. J. Marozsan, T. J. Ketas, E. L. Landes, J. P. Moore, and S. E. Kuhmann. 2007. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology 361:212-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richman, D. D., S. C. Morton, T. Wrin, N. Hellmann, S. Berry, M. F. Shapiro, and S. A. Bozzette. 2004. The prevalence of antiretroviral drug resistance in the United States. AIDS 18:1393-1401. [DOI] [PubMed] [Google Scholar]
- 30.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 31.Strizki, J. M., L. Wojcik, A. J. Marozsan, S. Kuhmann, W. Huang, J. Whitcomb, C. Petropoulos, and J. P Moore. 2005. Properties of in vitro generated HIV-1 variants resistant to the CCR5 antagonist SCH 351125 and SCH 417690. Antivir. Ther. 10:S66. [Google Scholar]
- 32.Swofford, D. L. 1998. PAUP*, phylogenetic analysis using parsimony (* and other methods) (4.0b10). Sinauer Associates, Sunderland, MA.
- 33.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walmsley, S., B. Bernstein, M. King, J. Arribas, G. Beall, P. Ruane, M. Johnson, D. Johnson, R. Lalonde, A. Japour, S. Brun, and E. Sun. 2002. Lopinavir-ritonavir versus nelfinavir for the initial treatment of HIV infection. N. Engl. J. Med. 346:2039-2046. [DOI] [PubMed] [Google Scholar]
- 36.Watson, C., S. Jenkinson, W. Kazmierski, and T. Kenakin. 2005. The CCR5 receptor-based mechanism of action of 873140, a potent allosteric noncompetitive HIV entry inhibitor. Mol. Pharmacol. 67:1268-1282. [DOI] [PubMed] [Google Scholar]
- 37.Westby, M., M. Lewis, J. Whitcomb, M. Youle, A. L. Pozniak, I. T. James, T. M. Jenkins, M. Perros, and E. van der Ryst. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80:4909-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westby, M., C. Smith-Burchnell, J. Mori, M. Lewis, M. Mosley, M. Stockdale, P. Dorr, G. Ciaramella, and M. Perros. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 81:2359-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitcomb, J. M., W. Huang, S. Fransen, K. Limoli, J. Toma, T. Wrin, C. Chappey, L. D. B. Kiss, E. E. Paxinos, and C. J. Petropoulos. 2007. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob. Agents Chemother. 51:566-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeni, P., A. LaMarca, D. Berger, P. Cimoch, A. Lazzarin, P. Salvato, F. M. Smaill, E. Teofilo, S. J. Madison, W. G. Nichols, K. K. Adkison, T. Bonny, J. Millard, D. McCarty, and the EPIC (CCR100136) Study Team. 2009. Antiviral activity and safety of aplaviroc, a CCR5 antagonist, in combination with lopinavir/ritonavir in HIV-infected, therapy-naive patients: results of EPIC study (CCR100136). HIV Med. 10:116-124. [DOI] [PubMed] [Google Scholar]