Abstract
Penicillin-binding protein 2x (PBP2x) of Streptococcus pneumoniae represents a primary resistance determinant for beta-lactams, and low-affinity PBP2x variants can easily be selected with cefotaxime. Penicillin-resistant clinical isolates of S. pneumoniae frequently contain in their mosaic PBP2x the mutation T338A adjacent to the active site S337, and T338P as well as T338G substitutions are also known. Site-directed mutagenesis has now documented that a single point mutation at position T338 confers selectable levels of beta-lactam resistance preferentially to oxacillin. Despite the moderate impact on beta-lactam susceptibility, the function of the PBP2x mutants appears to be impaired, as can be documented in the absence of a functional CiaRH regulatory system, resulting in growth defects and morphological changes. The combination of low-affinity PBP2x and PBP1a encoded by mosaic genes is known to result in high cefotaxime resistance. In contrast, introduction of a mosaic pbp1a into the PBP2xT338G mutant did not lead to increased resistance. However, the mosaic PBP1a gene apparently complemented the PBP2xT338G defect, since Cia mutant derivatives grew normally. The data support the view that PBP2x and PBP1a interact with each other on some level and that alterations of both PBPs in resistant clinical isolates have evolved to ensure cooperation between both proteins.
Penicillin resistance in Streptococcus pneumoniae is a complex process which involves multiple amino acid changes in several penicillin-binding proteins (PBPs), the target enzymes for beta-lactam antibiotics (12, 37). PBPs interact with the beta-lactam inhibitor by acylating the active site serine residue located in the first of three conserved motifs within the penicillin-binding domain of all penicillin-interactive enzymes: S*XXK, SXN, and KS/TG (16, 18).
S. pneumoniae contains six PBPs: the three high-molecular-mass (hmm) class A PBPs 1a, 1b, and 2a, the class B hmm PBPs 2x and 2b, and the low-molecular-mass d,d-carboxypeptidase PBP3. All six PBPs have been implicated in resistance (for a review, see reference 12 and references within). However, only the two class B hmm PBPs are primary targets for beta-lactams, i.e., mutations resulting in low-affinity variants can be selected in sensitive strains and mediate resistance, albeit only up to a moderate level. Whereas PBP2b mutations can be selected with penicillins, cefotaxime does not interact with PBP2b (24) and thus selects for PBP2x mutations only (19, 32, 46, 56). Resistance mediated by low-affinity PBP2x in the sensitive background leads to cefotaxime MICs of 0.08 to 0.3 μg/ml, depending on the allele, compared to 0.02 μg/ml in sensitive cells, and a low-affinity PBP2b increases the piperacillin MIC only up to twofold, independent of whether one single point mutation or very-low-affinity variants of highly altered PBP2b from clinical isolates are being investigated (19, 23). High cefotaxime resistance levels in clinical isolates are dependent on a low-affinity PBP2x and require an altered PBP1a as well, resulting in MICs of 1 μg/ml and higher (46).
PBPs are involved in late steps of murein biosynthesis, where the transpeptidation of two muropeptides resulting in cross-linked murein is the critical penicillin-sensitive step. Amino acid alterations in PBPs that confer resistance reduce the binding efficiency for the inhibitor, whereas the actual enzymatic function should not be severely affected. Resistant clinical isolates and transformants containing a low-affinity PBP2x grow perfectly well, at least under laboratory conditions. Therefore, it had been assumed that PBP2x mutations involved in resistance indeed did not affect the protein function. However, it has been shown recently that PBP2x mutations selected in the laboratory require a functional two-component system, CiaRH; the combination of PBP2x mutations with an inactivated Cia system resulted in reduced growth rates, early lysis, and altered cell morphology (38). Depending on the point mutations analyzed, the severity of these effects differed but seemed to be less pronounced with a mosaic PBP2x from a clinical isolate (38).
Resistant clinical isolates contain mosaic pbp2x genes in which parts are replaced by highly divergent sequence blocks as a result of horizontal gene transfer events followed by recombination involving genes of closely related streptococcal species. Mosaic blocks may differ by over 20% from DNA sequences from sensitive PBP genes and from each other, resulting in over 10% amino acid changes, and many different mosaic variants have been identified (12). Due to this variation it is difficult to distinguish PBP mutations involved in resistance development from alterations present due to the distinct origin of the gene. Comparison of a large number of sequences from resistant clinical isolates revealed that at least one of two sites is changed in most resistant PBP2x isolates, both of which are located in the active site cavity of the enzyme: Q552E close to the K547TG triad and T338A(P,G) within the S337TMK motif (3, 7, 14, 43, 44, 47, 54, 57). Curiously, most of the many PBP2x mutations identified in spontaneous cefotaxime-resistant laboratory mutants are distinct from changes seen in clinical isolates, including the T338 mutation (32), suggesting that other selective forces are acting outside the laboratory.
It has been demonstrated that reversion of the mutation at T338 in mosaic PBP2x resulted in a decrease of the resistance level (9, 57), and further confirmation for the importance of this mutation was obtained in kinetic studies with soluble PBP2x derivatives containing a single point mutation at this position (13, 43, 44). However, transformation of the T338 mutation via cefotaxime selection failed (10), and it is therefore unclear to what extent the mutation alone mediates beta-lactam resistance in vivo without the context of a mosaic structure.
Based on observations in our laboratory that T338 mutations can be selected with oxacillin rather than with cefotaxime, the three mutations T338A, T338P, and T338G were introduced into the sensitive S. pneumoniae R6 background and analyzed for their resistance phenotypes. A mosaic pbp1a gene was introduced into the pbp2x mutants under conditions that avoid beta-lactam selection in order to study the impact of such a PBP combination. Moreover, the impact of PBP mutations on cellular growth was investigated in combination with Cia loss-of-function mutants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. pneumoniae R6 is a nonencapsulated beta-lactam-sensitive derivative of the Rockefeller University strain R36A (4). S. pneumoniae 2349 is a member of the high-level penicillin-resistant and multiple antibiotic-resistant Spain23F-1 clone (51). The cefotaxime-resistant laboratory mutant C606, containing four PBP2x mutations, M289T, G422D, G597D, and G601V (32), and R6ciaR::aad9 (39) have been described elsewhere. All other mutants and transformants described here are derivatives of the R6 strain (Tables 1 and 2). Cells were grown at 37°C without aeration in C-medium (31) supplemented with 0.2% yeast extract (Becton, Dickinson and Co., Sparks, MD) or on blood agar plates (D-agar supplemented with 3% defibrinated sheep blood [Oxoid, Wesel, Germany]) (2). Growth in liquid culture was monitored by nephelometry (reported in nephelo units [N]). Escherichia coli strains JM109 and DH5α were used for propagation of plasmids derived from pGEMTeasy (Promega, Madison, WI); they were grown aerobically at 37°C either in LB medium or on LB agar plates. Plasmid pGEMTeasy was selected in E. coli with 100 μg/ml ampicillin. Growth of E. coli was followed by measuring the optical density at 600 nm using a spectrophotometer.
TABLE 1.
Streptococcus pneumoniae strains
TABLE 2.
R6 derivatives constructed in this studya
| Strain | Allele at:
|
MIC (μg/ml)
|
Generation time (min) | Max. cell density (Nb) | Length of stationary phase (h) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| PBP2x | PBP1a | CiaR | Pen | Ctx | Pip | Oxa | ||||
| R6 | WT | WT | WT | 0.02 | 0.020 | 0.04 | 0.08 | 29 | 130 | 5.5 |
| R6pbp 2xT338A | T338A | WT | WT | 0.03 | 0.035 | 0.09 | 0.18 | 31 | 128 | 5.0 |
| R6pbp 2xT338P | T338P | WT | WT | 0.03 | 0.040 | 0.06 | 0.23 | 30 | 122 | 5.0 |
| R6pbp2xT338G | T338G | WT | WT | 0.03 | 0.045 | 0.10 | 0.25 | 32 | 113 | 4.5 |
| R6pbp2x2349 | 2349 | WT | WT | 0.04 | 0.300 | 0.12 | 1.00 | 29 | 122 | 5.0 |
| R6pbp2x2349/pbp1a2349 | 2349 | 2349 | WT | 0.035 | 1.0 | 0.11 | 0.85 | 31 | 120 | 5.0 |
| R6Smrpbp2xT338G/pbp1a2349 | T338G | 2349 | WT | 0.020 | 0.03 | 0.06 | 0.16 | 40 | 102 | 3.0 |
| R6Smrpbp2x2349/pbp1a::Janus | 2349 | Disruption | WT | 0.040 | 0.20 | 0.12 | 0.95 | 39 | 105 | 4.0 |
| R6Smrpbp2xT338G/pbp1a::Janus | T338G | Disruption | WT | 0.020 | 0.03 | 0.09 | 0.21 | 43 | 95 | 2.0 |
| R6ciaR::aad9 | WT | Disruption | Disruption | 0.020 | 0.025 | 0.06 | 0.08 | 32 | 121 | 4.0 |
| R6pbp2xT338A/ciaR::aad9 | T338A | Disruption | Disruption | 0.040 | 0.045 | 0.11 | 0.20 | 33 | 120 | 3.0 |
| R6pbp2xT338P/ciaR::aad9 | T338P | Disruption | Disruption | 0.025 | 0.045 | 0.06 | 0.24 | 35 | 109 | 2.0 |
| R6pbp2xT338G/ciaR::aad9 | T338G | Disruption | Disruption | 0.025 | 0.055 | 0.08 | 0.25 | 38 | 101 | 1.0 |
| R6pbp2x2349/ciaR::aad9 | 2349 | Disruption | Disruption | ND | ND | ND | ND | 30 | 115 | 4.0 |
| R6pbp2x2349/pbp1a2349/ciaR::aad9 | 2349 | 2349 | Disruption | ND | ND | ND | ND | 36 | 107 | 3.0 |
| R6Smrpbp2xT338G/pbp1a2349/ciaR::aad9 | T338G | 2349 | Disruption | ND | ND | ND | ND | 38 | 117 | 3.0 |
| R6Smrpbp2x2349/pbp1a::Janus/ciaR::aad9 | 2349 | Disruption | Disruption | ND | ND | ND | ND | 38 | 104 | 1.5 |
| R6Smrpbp2xT338G/pbp1a::Janus/ciaR::aad9 | T338G | Disruption | Disruption | ND | ND | ND | ND | 50 | 80 | 1.0 |
Data indicate the range of values obtained from at least three independent experiments; the variation of MICs below 0.1 μg/ml differed at most by ±0.02 μg/ml and above 0.1 μg/ml by ±0.05 μg/ml. The generation time varied by ±3 min, nephelometer units by ±10, and the length of stationary phase by ±45 min. Abbreviations: WT, wild type; Pen, benzylpenicillin; Ctx, cefotaxime; Pip, piperacillin; Oxa, oxacillin; ND, not determined.
Nephelo units reached at the end of the exponential growth phase.
MIC determinations.
The MIC for each strain was determined by the agar dilution method on blood agar plates under a natural atmosphere using cells of an exponentially growing culture. A narrow range of antibiotic concentrations was used in order to detect subtle differences in susceptibility. MICs were monitored after 48 h of incubation at 37°C.
Transformation procedure.
Transformation of S. pneumoniae R6 strains was performed according to published procedures (38, 60). The beta-lactam concentrations used for selection are specified in the Results section; streptomycin (Sm) and kanamycin (Km) were used at 200 μg/ml, and spectinomycin was used at 100 μg/ml. Plates were incubated for 24 or 48 h at 37°C. The presence of mutations was verified by DNA sequencing. Transformation of E. coli JM109 was performed using electroporation following published procedures (53).
DNA techniques.
All DNA techniques were performed using standard methods (53). Chromosomal DNA was isolated from S. pneumoniae as described previously (33). Plasmids from E. coli were isolated using the QIAprep Spin Miniprep kit (Qiagen, Hilden, Germany). Genes were amplified from chromosomal DNA by PCR using either GoldStar Taq polymerase (Eurogentec, Seraing, Belgium) or high-fidelity iProof polymerase (Bio-Rad, Hercules, CA) according to the manufacturers's instructions. Alternatively, cells were taken from glycerol stock cultures (approximately 400 μl) and centrifuged, and 1 μl of the pellet was used directly in a PCR. PCR products were purified using the PCR clean-up gel extraction kit (Macherey-Nagel, Düren, Germany). The pbp2x, pbp1a, ciaH, and cpoA genes of the R6 PBP2x mutants were routinely sequenced to verify desired mutations and to confirm that no mutations in the non-PBP genes were present. The DNA oligonucleotides used for PCR and sequencing relevant for this study are listed in Table 3.
TABLE 3.
Oligonucleotides relevant for this study
| Oligonucleotide and use | Sequence (5′→3′) |
|---|---|
| For site-directed mutagenesis | |
| pbp2x_5 | GATTGCTGAGGATGCAACCTCTTATAATGTCTATG |
| pbp2x_3 | GCCTTGAAATTCAAGTTCTATATTGAGCCACTTAGC |
| pbp2x_A-FMa | ATCATCACTTTCATGGCGGAACCTGGCTCATAGTTAC |
| pbp2x_A-RMa | TATGAGCCAGGTTCCGCCATGAAAGTGATGATGTTGG |
| pbp2x_P-FMa | TCATCACTTTCATAGGGGAACCTGGCTCATAGTTAC |
| pbp2x_P-RMa | AACTATGAGCCAGGTTCCCCTATGAAAGTGATGATG |
| pbp2x_G-FMa | ATCATCACTTTCATGCCGGAACCTGGCTCATAGTTAC |
| pbp2x_G-RMa | TATGAGCCAGGTTCCGGCATGAAAGTGATGATGTTG |
| For cia and pbp1a derivatives | |
| CiaR_for | TTGGCAGCAGCTCTTGCATACAGTACAG |
| CiaR_rev | ACCGAAATCGTTGTATCACTATCAAATCC |
| Ja-pbp1a_for | GGCTGGTGCCTTCCCTCAAATTCCTTATC |
| Ja-pbp1a_rev | CAAGTGTTAGCCTAGCCCTATCTGCTCTC |
| Ja-pbp1a-seq_forb | CATAAGGAAAGGGGCCCTAGAACCGCGACTGGGGATCAACTATG |
| Ja-pbp1a-seq_revb | CCATTTCCTCTGGAATAGGCTGTTTCTACTGCTTGGTTAATTCC |
| Ja-Kass_forb | TTAACCAAGCAGTAGAAACAGCCTATTCCAGAGGAAATG |
| Ja-Kass_revb | GTTGATCCCCAGTCGCGGTTCTAGGGCCCCTTTCCTTATGC |
The mutated codon is underlined.
The overlapping termini are shown in italics.
Construction of PBP2x T338 mutants.
S. pneumoniae R6 derivatives containing the PBP2x mutation T338A (R6pbp2xT338A), T338P (R6pbp2xT338P), and T338G (R6pbp2xT338G) were constructed by replacing an internal 1,908-bp fragment of the parental pbp2x with an R6 pbp2x containing a mutated codon to introduce the respective point mutation. Site-directed mutagenesis with R6 pbp2x was performed by overlap extension PCR (26, 27), using chromosomal R6 DNA and the oligonucleotide pairs pbp2x_5 with pbp2x_A-FM and pbp2x_3 with pbp2x_A-RM to introduce T338A, pbp2x_5 with pbp2x_P-FM and pbp2x_3 with pbp2x_P-RM to introduce T338P, and pbp2x_5 with pbp2x_G-FM and pbp2x_3 with pbp2x_G-RM to introduce T338G (Table 3). Mutagenized PCR fragments were subsequently cloned into the vector pGEMTeasy according to the manufacturer's instructions and standard techniques (53). The presence of the mutations was confirmed by double-stranded sequencing. Recombinant vector DNA was used to transform strain R6, followed by selection of transformants with oxacillin as described in the Results section. Introduction of the desired mutation was verified by double-stranded DNA sequencing.
Construction of loss-of-function CiaR derivatives.
Strain R6ciaR::aad9, which contains the spectinomycin resistance gene aad9 from pDL278 (36) within the CiaR gene, has been described previously (39). For construction of ciaR loss-of-function derivatives, the ciaR::aad9 region was amplified by PCR using the oligonucleotides CiaR_for and CiaR_rev (Table 3). The purified PCR fragment was transformed into the R6 PBP mutants using the spectinomycin resistance marker aad9 for selection. Correct integration of the construct was confirmed by DNA sequence analysis.
Introduction of the mosaic PBP1a2349.
The R6Smrpbp2xT338G/pbp1a2349 mutant was constructed via a two-step process using the Janus cassette (58). First, the Janus cassette was introduced into the pbp1a gene of R6Smrpbp2xT338G carrying the AmiA9 streptomycin resistance marker (52). Introduction of the Janus cassette confers a Kmr Sms phenotype. In a second step, the Janus cassette was replaced by the mosaic pbp1a gene of the penicillin-resistant clinical isolate S. pneumoniae 2349, a member of the clone Spain23F-1 (51), resulting in R6Smrpbp2xT338G/pbp1a2349. In detail, two fragments of the PBP1a gene were amplified by PCR using chromosomal R6 DNA and the oligonucleotide pairs Ja-pbp1a_for plus Ja-pbp1a-seq_rev and Ja-pbp1a_rev plus Ja-pbp1a-seq_for (Table 3). These two fragments and the PCR product covering the Janus cassette were combined via another PCR (oligonucleotides Ja-pbp1a_for and Ja-pbp1a_rev), resulting in a product where the Janus cassette was flanked by the two pbp1a fragments, which provided targeting specificity for recombination. This final PCR product was transformed into R6Smrpbp2xT338G followed by screening for kanamycin-resistant, streptomycin-sensitive colonies (R6Smrpbp2xT338G/pbp1a::Janus). These constructs contained a nonfunctional pbp1a, since the transpeptidase domain was disrupted between codons 363 and 366. One of the R6Smrpbp2xT338G/pbp1a::Janus constructs was subjected to further transformation using the plasmid pGEM-1aRes as donor DNA, which contains a 1,920-bp fragment of S. pneumoniae 2349 pbp1a (codon 79 to 719 of mosaic pbp1a covering the entire mosaic block from codon 285 to 667), followed by selection on streptomycin plates. The presence or absence of the PBP1a gene was verified in all constructs by DNA sequence analysis.
Detection of penicillin-binding proteins.
Cells of an exponentially growing culture were centrifuged and resuspended in 20 mM Na-phosphate buffer, pH 7.2. The volume was adjusted so that 5 μl of cell suspension of one sample corresponds to 1 ml of culture at an N of 20, which in turn corresponds to approximately one-sixth of the maximum cell density in wild-type cells. For cell lysis and labeling of PBPs, 20 mM Na-phosphate buffer, pH 7.2, containing 0.2% Triton X-100 and different concentrations of BocillinFL (Invitrogen, Eugene, OR) (63) were added (Fig. 1 and 2). The samples were incubated using different incubation times (30 s to 30 min) and temperatures (0°C to 37°C). PBPs were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 7.5% or 11% polyacrylamide gels with an acrylamide/bisacrylamide ratio of 30:0.8. Bocillin-PBP complexes were visualized with a fluorimager (Molecular Dynamics, Sunnyvale, CA) at 488 nm. Proteins were stained with Coomassie brilliant blue.
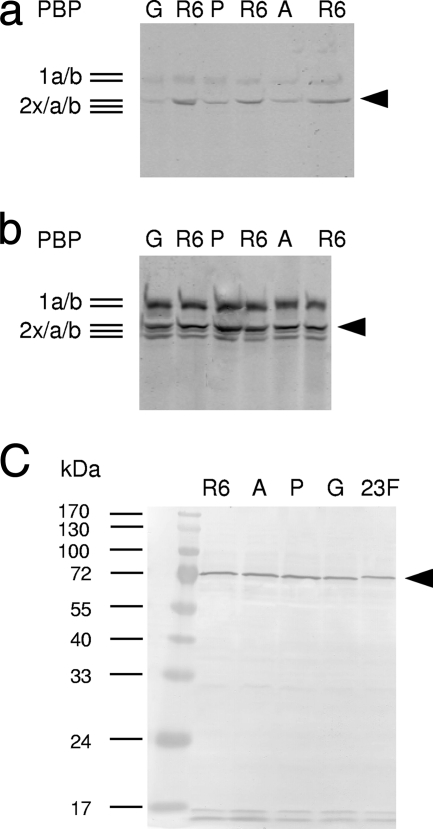
FIG. 1.
PBP profiles of PBP2x mutants. Cell lysates were incubated for 2 min at 20°C. (a and b) BocillinFL at 0.01 μM (a) or 0.3 μM (b) final concentration. Bocillin-PBP complexes were visualized by fluorography after separation on SDS-polyacrylamide gels. (c) Detection of PBP2x after Western blotting of cell lysates and incubation with affinity-purified anti-PBP2x antiserum. Note that the protein encoded by the mosaic gene (2349) has slightly different electrophoretic mobility. R6, control; G, R6pbp2xT338G; P, R6pbp2xT338P; A, R6pbp2xT338A; 23F, R6pbp2x2349. Black arrowhead, PBP2x. The molecular masses of marker proteins are indicated on the left.
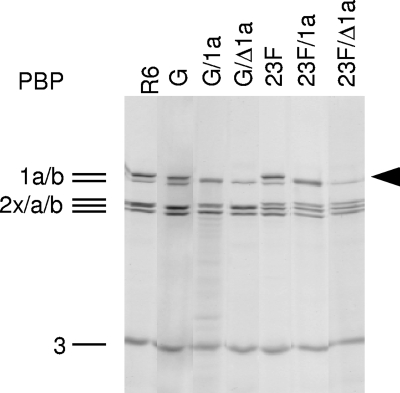
FIG. 2.
PBP profiles of S. pneumoniae R6 derivatives containing different pbp2x and pbp1a alleles. Lanes: R6 (control); G, R6pbp2xT338G; G/1a, R6Smrpbp2xT338G/pbp1a2349; G/Δ1a, R6Smrpbp2xT338G/pbp1a::Janus; 23F, R6pbp2x2349; 23F/1a, R6pbp2x2349/pbp1a2349; 23F/Δ1a, R6Smrpbp2x2349/pbp1a::Janus. Black arrowhead, PBP1a. Cell lysates were incubated for 30 min at 37°C using 6.7 μM BocillinFL (final concentration). Proteins were separated by SDS-PAGE, and Bocillin-PBPcomplexes were visualized by fluorography. PBP2x2349 has a slower electrophoretic mobility than PBP2xR6 and derivatives, and PBP2x and PBP2a are not resolved in the first four lanes.
Western blotting.
After separation on SDS-PAGE, proteins were transferred onto a polyvinylidene difluoride membrane. PBPs were visualized after incubation with affinity-purified PBP2x antibodies (1:10,000 in phosphate-buffered saline) (40) or PBP1a antibodies (1:3,000 in phosphate-buffered saline) (21) followed by incubation with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Sigma-Aldrich, St. Louis, MO) and staining with 4-nitrobluetetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate.
Microscopic techniques.
Cell morphology and vitality of S. pneumoniae were monitored during exponential growth in C-medium containing 2% choline chloride, which inhibits cell separation (8) due to the inhibition of the LytB glucosaminidase implicated in this process (17); therefore, morphological changes are more visible. A Nikon Eclipse E600 microscope equipped with a 40×/numerical aperture 0.75 and a 100×/numerical aperture 1.4 oil objective was used for phase contrast and fluorescence microscopy. Photos were taken with a Nikon CCD-1300B camera (Nikon, Düsseldorf, Germany). Vitality of cells was determined using the LIVE/DEAD BacLight bacterial viability kit L13152 (Invitrogen, Eugene, OR) according to the manufacturer's instructions. Cells were centrifuged, washed, and resuspended in H2O, labeled with SYTO9 and propidium iodide fluorescent stains, and immobilized on slides with poly-l-lysine hydrobromide (Sigma-Aldrich, St. Louis, MO). For parallel observation of both stains, a B-2A green long-pass filter (EX 450 to 490, DM 505, BA 520; Nikon) was used.
RESULTS
T338 mutations in PBP2x confer selectable resistance to beta-lactam antibiotics.
To assess the importance of mutations at Thr338 for beta-lactam resistance in PBP2x, three mutants were constructed in S. pneumoniae R6 containing an Ala, Pro, or Gly substitution at this position. For site-directed mutagenesis the codons that appear most frequently in resistant streptococci were chosen (Table 3). Transformants of the R6 strain were isolated using the cloned pbp2x variants as donor DNA followed by plating on 0.11 to 0.15 μg/ml oxacillin, concentrations which are slightly above the MIC of R6 (0.08 μg/ml). Under these conditions, no colonies appeared on the R6 control plates.
Mutations in ciaH occur frequently in such transformants, especially at higher concentrations of the selective beta-lactam, and mutations in cpoA encoding a glycosyltransferase have been identified in piperacillin-resistant mutants (15, 22). Therefore, only transformants from the lowest selective concentration (0.11 μg/ml) were used for further analysis. At this concentration, resistant colonies were obtained with a frequency of approximately 10−5 (mutants A and G) and 10−2 (mutant P). The absence of mutations in ciaH and cpoA as well as the presence of the respective PBP2x point mutation were confirmed by DNA sequencing.
Determination of MICs for benzylpenicillin, cefotaxime, piperacillin, and oxacillin of the R6pbp2xT338 mutants revealed slightly different resistance levels, depending on the particular point mutation in PBP2x (Table 2). Although the absolute values differed by less than twofold, differences between the three mutations were highly reproducible as determined using narrow concentration ranges of 0.01-μg/ml steps for the MIC determination. In general, the highest MICs were obtained for oxacillin (up to three times the MIC of the parental strain R6), whereas the effects on benzylpenicillin susceptibility were less pronounced (1.5 times the MIC). With all three antibiotics, the T338G mutation had the greatest effect.
For comparison we used R6pbp2x2349, which contains the mosaic PBP2x gene of the clinical isolate S. pneumoniae 2349, a member of the high-level penicillin-resistant Spain23F-1 clone (41), which includes the T338A change within its mosaic block (34, 51). This construct showed considerably higher oxacillin and cefotaxime MICs compared to those observed with the single PBP2x mutations, whereas piperacillin and benzylpenicillin MICs increased only slightly (Table 2).
The effect of the point mutations on the interaction with beta-lactams was tested using Bocillin FL and visualization of the PBP-antibiotic complexes after SDS-PAGE and fluorometry (Fig. 1a and b). In all cases decreased labeling of the mutated PBP2x was observed using low concentrations and a short incubation time (Fig. 1a). This was most pronounced in the R6pbp2xT338G mutant, in agreement with the reduced acylation efficiency reported for the soluble PBP2x derivatives (43). No effect on the protein amount was detected on Western blots using affinity-purified PBP2x antibodies (Fig. 1c). The growth of the R6pbp2xT338 mutants was only marginally affected in liquid medium. The generation time was slightly longer (30 to 32 min) compared to the R6 strain (29 min), and the cultures grew to a lower cell density (N = 113 to 128) compared to R6 (N = 130). Again, R6pbp2xT338G showed the greatest effect (Table 2).
Introduction of the mosaic pbp1a2349 into PBP2x mutants.
In order to investigate the effect of a combination of a pbp2x allele containing a single point mutation with the mosaic pbp1a, we introduced pbp1a2349 via the Janus cassette, a bicistronic cassette conferring a Kmr Sms phenotype which enables the replacement of genes through negative selection (58). Insertion of the Janus cassette in the first step results in the disruption of the gene. Therefore, this technique can only be applied for nonessential genes, such as pbp1a (29, 49). The R6pbp2xT338G mutant was chosen as recipient after introduction of the Smr allele, since we expected that any effect on resistance would be more visible in this strain compared to the other single-point PBP2x mutants. Using this strategy the mosaic pbp1a from strain 2349 was then successfully introduced into R6Smrpbp2xT338G (Table 2; Fig. 2).
The effect of PBP1a2349 differed markedly, depending on the PBP2x allele present. With the cognate PBP2x2349, the cefotaxime MIC increased to 1 μg/ml, compared to 0.3 μg/ml with the PBP2x allele alone, whereas a slight but reproducible decrease in resistance for the other three beta-lactams was observed (Table 2). In contrast, in the PBP2xT338G background, the MIC decreased for all four beta-lactams tested (Table 2). The presence of PBP1a2349 had no effect on the amount of PBP2x or PBP1a as determined on Western blots with affinity-purified anti-PBP2x antibodies or PBP1a antiserum (not shown).
The impact of a disrupted PBP1a was then tested in combination with the two PBP2x alleles. With PBP2x2349, a decrease in the cefotaxime MIC was observed, as expected, and in the case of PBP2xT338G a slight decrease in MICs for all beta-lactams was apparent (Table 2). In all cases, the cells grew significantly slower with inactivated PBP1a (Table 2).
A functional CiaRH system is required in R6pbp2xT338 mutants.
It was shown recently that disruption of the two-component signal-transducing system CiaRH in combination with particular PBP2x mutations selected in the laboratory results in dramatic growth defects and morphological changes, indicating that these mutations somehow impair the function of the PBP and that these defects can be complemented by an active Cia system (38). In order to assess the impact of the T338 mutation on PBP2x function, the CiaR gene encoding the response regulator was disrupted in each of the three R6pbp2xT338 mutants and the R6pbp2x2349 construct, and cellular growth and morphology were followed during incubation in liquid medium. Due to the operon structure of ciaRH, a disruption of ciaR affects both genes, i.e., neither CiaH nor CiaR are produced (62).
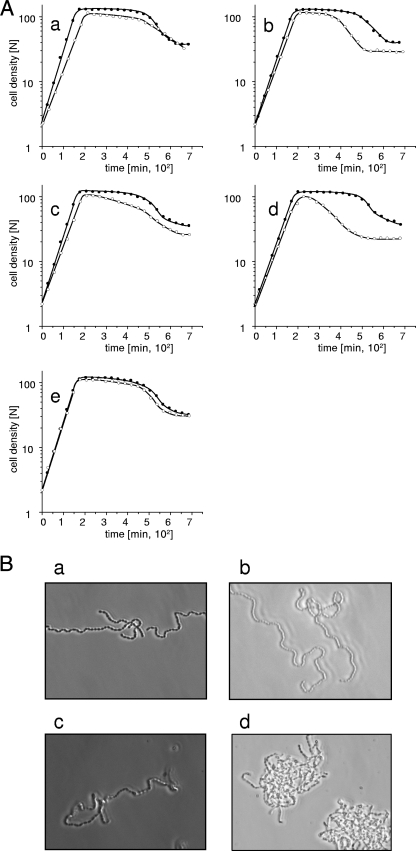
In all three PBP2x mutants, disruption of ciaR resulted in a longer generation time (Table 2). Also, growth ceased at a lower cell density, and an abbreviated or no stationary phase was followed by early lysis (Fig. 3A, graphs a to e). Again, the R6pbp2xT338G/ciaR::aad9 mutant displayed the most dramatic effect compared to the parental R6pbp2xT338G, with a generation time of 38 min compared to 32 min, a maximal cell density of 100 N compared to 120 N, and an extremely reduced stationary phase (Fig. 3A, graph d), whereas growth of R6pbp2xT338G/ciaR::aad9 and R6pbp2xT338P/ciaR::aad9 was less impaired (Fig. 3A, graphs b and c; Table 2). R6pbp2x2349/ciaR::aad9 containing the mosaic pbp2x grew almost normally, and stationary lysis was only slightly enhanced (Fig. 3A, graph e), as shown previously (38), suggesting the presence of compensatory mutations within the PBP2x gene of the clinical isolate.
FIG. 3.
Growth and cell morphology of PBP2x mutants with and without a functional CiaRH system. (A) Cells of an exponentially growing culture were diluted 1:20 in prewarmed C-medium. The cell density is given in nephelometer units (N). Closed circles, PBP2x mutants; open circles, ciaR::aad9 derivatives. (a) R6; (b) R6pbp2xT338A; (c) R6pbp2xT338P; (d) R6pbp2xT338G; (e) R6pbp2x2349. (B) Cells were grown in C-medium containing 2% choline. Samples were taken during the mid-exponential growth phase (N ≈ 60), and cells were examined using phase-contrast microscopy as described in Materials and Methods. (a) R6; (B) R6ciaR::aad9; (c) R6pbp2xT338G; (d) R6pbp2xT338G/ciaR::aad9.
Cell morphology and division were monitored using the LIVE/DEAD staining procedure during growth in medium containing 2% choline, which induces chain formation (8), therefore allowing alterations to be detected more easily. The R6pbp2xT338/ciaR::aad9 constructs grew in twisted chains of misshaped cells which accumulated in cell clumps (examples are shown in Fig. 3B) and contained a considerable proportion of nonviable cells (not shown). These effects were most dramatic in the R6pbp2xT338G/ciaR::aad9 mutant (Fig. 3B, panel d). Disruption of CiaR did not change the MICs and also had no effect on the amount of PBP2x produced in the cells (Table 2).
PBP1a and the Cia system.
We then used the same strategy, i.e., constructing ciaR::aad9 derivatives, to investigate the role of PBP1a in pbp2x mutants. First, the two PBP2x mutants with the mosaic pbp1a R6Smrpbp2xT338G/pbp1a2349 and R6pbp2x2349/pbp1a2349 and, in a second set of experiments the pbp1a deletion mutants R6Smrpbp2xT338G/pbp1a::Janus and R6Smrpbp2x2349/pbp1a::Janus, were examined in combination with Cia loss-of-function mutations (Fig. 4).
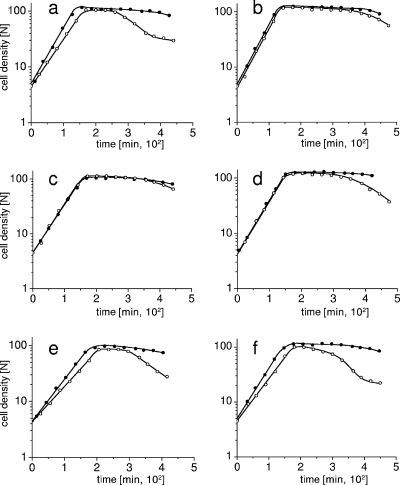
FIG. 4.
Effects of ciaR disruption in PBP2x/PBP1a double mutants. Cells of an exponentially growing culture were diluted 1:20 in prewarmed C-medium. The cell density is given in nephelometer units (N). Closed circles, wild-type Cia system; open circles, ciaR::aad9 derivatives. (a and b) Parental strains R6pbp2xT338G (a) and R6pbp2x2349 (b). (c and d) PBP1a2349 derivatives R6Smrpbp2xT338G/pbp1a2349 (c) and R6pbp2x2349/pbp1a2349 (d). (e and f) Δpbp1a derivatives R6Smrpbp2xT338G/pbp1a::Janus (e) and R6Smrpbp2x2349/pbp1a::Janus (f).
Surprisingly, the presence of the mosaic pbp1a allowed normal growth of the cells in the absence of a functional Cia system in the PBP2xT338G mutant (R6Smrpbp2xT338G/pbp1a2349/ciaR::aad9), and only a slight tendency for early lysis after reaching stationary phase was observed (Fig. 4c). In contrast, the deletion of PBP1a in combination with the PBP2x and Cia mutations (R6Smrpbp2xT338G/pbp1a::Janus/ciaR::aad9) enhanced the growth defects already seen in the R6pbp2xT338G/ciaR::aad9 mutant (Fig. 4e). This strongly indicates that the mosaic PBP1a somehow compensates the PBP2xG338 mutation and this compensation becomes visible in the absence of the Cia system.
In the case of the mosaic PBP2x2349, disruption of CiaR resulted only in a slightly enhanced stationary-phase lysis, independent of the presence or absence of the mosaic PBP1a (Fig. 4b and d). However, deletion of PBP1a in combination with a nonfunctional Cia system also resulted in severe growth deficiencies (Fig. 4f), indicating that CiaRH is required under these conditions.
DISCUSSION
PBP2x mutations at T338 and beta-lactam resistance.
Mutations at T338 in the S. pneumoniae PBP2x are among the best-characterized changes relevant to beta-lactam resistance in clinical isolates. The threonine residue adjacent to the active site S337 is frequently replaced by A or G, or occasionally by P or S (3, 7, 14, 43, 44, 47, 54, 57). Such mutations occur in strains of a whole range of resistance levels, with G338 having been observed especially in context with high-level cefotaxime resistance in S. pneumoniae (43) and in commensal streptococci as well (5).
Soluble PBP2x derivatives containing A338, -G, or -P have been studied in vitro, revealing that the acylation efficiency for cefotaxime is reduced between two- and eightfold, given a k2/K ratio of 209,000 M−1 s−1 for the wild-type PBP2x (9, 10, 43, 44). The T338A mutation in the active site interferes with the local hydrogen bonding network, which involves a crucial water molecule, thereby possibly affecting the recognition of substrate muropeptides (13, 44). In agreement with these findings, reversion of A338T in the context of a mosaic PBP2x resulted in an increased acylation efficiency, and the cefotaxime MIC of an R6 transformant containing the revertant of the mosaic PBP2xA338T plus the cognate mosaic PBP1a dropped from 1 to 0.75 μg/ml (9). Similarly, reversion of T338P resulted in four- and eightfold reductions of the MICs for cefotaxime and benzylpenicillin, respectively, in a strain harboring the cognate altered PBP1a, PBP2b, and MurM genes (57).
Whereas cefotaxime has generally been used to select PBP2x mutants and transformants (19, 32, 56), we have now shown that oxacillin can efficiently select T338 mutations and that depending on the amino acid substitution (A, G, or P), the oxacillin MIC increases between two- and threefold in the R6 background. Fine-tuning of the MICs revealed subtle differences mediated by a single point mutation, with T338G appearing the most effective one and which might have larger effects in the context of other mutations in PBP2x and other PBPs as well. The cefotaxime MIC increased only by approximately twofold, whereas the mutation G601V has been shown to result in a fourfold increase (40) and mutations at T550 result in MICs of 0.2 to 0.3 μg/ml (11, 19).
PBPs and the Cia system.
Despite the apparently minor impact on MICs, the PBP2x mutants showed some growth defects when compared to the wild-type strain (Table 2). When the PBP2x mutations were combined with a nonfunctional CiaRH regulatory system, these effects became more pronounced: the generation time decreased further and many dead cells were observed, and the stationary phase was much shorter or not existent followed by early cell lysis. Again, the severity of these phenotypes was most apparent in the T338G mutant. Similar phenotypes have been described in other laboratory PBP2x mutants (38).
In contrast to the PBP2x point mutations, the combination of a highly altered mosaic PBP2x of clinical isolates with a nonfunctional CiaRH system appears to be less critical (Fig. 3) (38), strongly suggesting the presence of compensatory mutations in the mosaic PBP2x. In this context it would be interesting to compare the structure of PBP2x laboratory mutants with the known structures of mosaic PBP2x (13, 50).
The role of the CiaRH regulatory system in this context is not clear. Mutants in the Cia system have a pleiotropic phenotype, affecting cellular lysis, genetic competence, and resistance to a variety of different lytic agents (38). Although the Cia regulon has been described in detail, the molecular basis for these phenotypes is not well understood (25). So far, no mutations in ciaH have been observed in clinical isolates, whereas all cefotaxime-resistant and some piperacillin-resistant laboratory mutants contain CiaH mutations (61, 62) (our unpublished data). Cia mutants show a reduced virulence when tested in mouse models (35, 55, 59), and thus selective forces to maintain a functional Cia system in vivo seem obvious. The experiments described here confirm that Cia mutants can be used to detect functional defects in PBP2x of resistant strains.
PBP2x and PBP1a.
A third property of the PBP2x mutants relates to their interaction with PBP1a variants. Whereas the combination of a low-affinity mosaic PBP2x with the cognate mosaic PBP1a leads to high-level cefotaxime resistance (11, 46), introduction of a mosaic PBP1a into the R6pbp2xT338G mutant resulted in a slight drop in resistance, similar to the disruption of PBP1a in the PBP2x mutant. Failure to observe a resistance increase with PBP1a must be due to the nature of the PBP2x alterations, since also with another PBP2x mutant (PBP2xC606), which displays an unusually low acylation efficiency for cefotaxime (k2/K value below 50 M−1 s−1) (40), no resistant transformants were obtained with the mosaic pbp1a.
On the other hand, the presence of PBP1a2349 appeared to compensate the growth defect mediated by the disrupted Cia system in the PBP2xT338G mutant background (Fig. 4b), whereas disruption of PBP1a led to slower growth and early lysis in combination with PBP2xT338G as well as PBP2x1a2349 (Fig. 4e and f). These data strongly suggest that PBP2x and PBP1a interact on some level. In E. coli, interactions between the class A hmm PBP1b and the class B PBP3 have been documented (6). Both murein-synthesizing enzymes also interact with FtsN (45), and all these proteins are localized at the division septum (1, 6), where they might be part of a larger murein-synthesizing multienzyme complex (28). Also in S. pneumoniae, PBP2x and PBP1a appear to be located in the cell equator, together with FtsZ (42). The aberrant morphology of cell chains observed in PBP2x mutants, especially in the absence of a functional Cia system, indicates that their division plane cannot be synthesized properly (Fig. 3B). It is thus feasible that the PBP2x mutations have an impact on the interaction with other components of the cell division machinery, such as PBP1a, and that in clinical isolates compensatory mutations have occurred. Alternatively, the PBP mutations might affect the enzymatic function of the proteins; an in vitro assay system with the actual substrate of the enzyme would be desirable to solve these questions.
Two issues are noteworthy in this context. One is the fact that no PBP1a mutations occurred in cefotaxime- or piperacillin-resistant laboratory mutants, although resistance levels of 1 μg/ml and higher were obtained (23). Instead, PBP2a and PBP3 mutations were found in addition to PBP2x or PBP2b (30) (our unpublished results). Similarly, no mutations in PBP1a, the homologue of S. pneumoniae PBP1a, were detected in penicillin-resistant Streptococcus gordonii mutants with up to a 500-fold increase of the MIC, whereas again, mutations in PBP2x, PBP2b, and non-PBP genes were selected (20). In that study, deletion of PBP1A (and PBP2a) profoundly slowed resistance development but only moderately affected resistance in already highly resistant mutants. Those authors suggested that the transglycosylation activity of class A PBPs might facilitate early development of resistance by stabilizing penicillin-altered peptidoglycan via transglycosylation, whereas they might be less indispensable in highly resistant mutants which have reestablished a penicillin-insensitive cell wall-building machinery.
Second, most of the PBP2x mutations selected with cefotaxime in the laboratory have not been found in clinical isolates, with the exceptions of Q552E and T550A (19, 56). T550A occurs rarely and has been found mainly in unusually high-level cefotaxime-resistant isolates (3, 11, 48, 54). These data clearly show that the selective forces used in the laboratory are distinct from those acting on clinical isolates, and the data also indicate that the mosaic genes of clinical isolates have evolved by selection not only for resistance but for maintenance of other functions as well to ensure functionality of the cell wall synthesis apparatus and the cell division complex. Given the fact that many distinct PBP2x mosaic variants have been described, the identification of mutations relevant for resistance and the distinctions from alterations required for in vivo function of this protein will be difficult.
Taken together, the data show that a single point mutation in PBP2x affects cellular growth on several levels, although its impact on resistance in a beta-lactam-sensitive background is only minor. Cell division appears to be impaired in the PBP2x mutants, and it remains to be seen to what degree the cell wall structure is affected as well. Whether this is the consequence of an altered enzymatic activity, due to disturbance of the interaction with a partner protein such as PBP1a, or related to the activity of the Cia system remains to be clarified.
Acknowledgments
This work was supported by the EU (LSHB-CT-2005-512061 and LSHM-CT-2004-512138) and the Stiftung für Innovation Rheinland-Pfalz.
We thank Sonja Schröck from the Nano+Bio Center from the University of Kaiserslautern for technical assistance in DNA sequencing and Patrick Maurer and Bernhard Henrich for helpful discussions.
Footnotes
Published ahead of print on 15 December 2008.
REFERENCES
- 1.Addinall, S. G., and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25:303-309. [DOI] [PubMed] [Google Scholar]
- 2.Alloing, G., C. Granadel, D. A. Morrison, and J.-P. Claverys. 1996. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol. Microbiol. 21:471-478. [DOI] [PubMed] [Google Scholar]
- 3.Asahi, Y., Y. Takeuchi, and K. Ubukata. 1999. Diversity of substitutions within or adjacent to conserved amino acid motifs of penicillin-binding protein 2x in cephalosporin-resistant Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 43:1252-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79:137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann, C. 2003. Epidemiologie eines Mosaikgens: pbp2x in β-Lactam-sensitiven und -resistanten oralen Streptokokken aus Spanien. Ph.D. dissertation. Universität Kaiserslautern, Kaiserslauten, Germany.
- 6.Bertsche, U., T. Kast, B. Wolf, C. Fraipont, M. Aarsman, K. Kanneberg, M. von Rechenberg, M. Nguyen-Distèche, T. Den Blaauwen, J.-V. Höltje, and W. Vollmer. 2006. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol. Microbiol. 61:675-690. [DOI] [PubMed] [Google Scholar]
- 7.Bicmen, M., Z. Gulay, S. V. Ramaswamy, D. M. Musher, and D. Gur. 2006. Analysis of mutations in the pbp genes of penicillin-non-susceptible pneumococci from Turkey. Clin. Microbiol. Infect. 12:150-155. [DOI] [PubMed] [Google Scholar]
- 8.Briese, T., and R. Hakenbeck. 1983. Interaction between choline and the N-acetyl-muramyl-L-alanine-amidase of Streptococcus pneumoniae, p. 173-178. In R. Hakenbeck, J.-V. Höltje, and H. Labischinski (ed.), The target of penicillin. Walter de Gruyter & Co., Berlin, Germany.
- 9.Carapito, R., L. Chesnel, T. Vernet, and A. Zapun. 2006. Pneumococcal beta-lactam resistance due to a conformational change in penicillin-binding protein 2x. J. Biol. Chem. 281:1771-1777. [DOI] [PubMed] [Google Scholar]
- 10.Chesnel, L., L. Pernot, D. Lemaire, D. Champelovier, J. Croize, O. Dideberg, T. Vernet, and A. Zapun. 2003. The structural modifications induced by the M339F substitution in PBP2x from Streptococcus pneumoniae further decreases the susceptibility to beta-lactams of resistant strains. J. Biol. Chem. 278:44448-44456. [DOI] [PubMed] [Google Scholar]
- 11.Coffey, T. J., M. Daniels, L. K. McDougal, C. G. Dowson, F. C. Tenover, and B. G. Spratt. 1995. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 39:1306-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denapaite, D., F. Chi, P. Maurer, O. Nolte, and R. Hakenbeck. 2007. Mechanism of penicillin resistance in Streptococcus pneumoniae: targets, gene transfer, and mutations, p. 290-303. In R. Hakenbeck and G. S. Chhatwal (ed.), Molecular biology of streptococci. Horizon Bioscience, Wymondham, Norfolk, United Kingdom.
- 13.Dessen, A., N. Mouz, E. Gordon, J. Hopkins, and O. Dideberg. 2001. Crystal structure of PBP2x from a highly penicillin-resistant Streptococcus pneumoniae clinical isolate: a mosaic framework containing 83 mutations. J. Biol. Chem. 276:45105-45112. [DOI] [PubMed] [Google Scholar]
- 14.du Plessis, M., E. Bingen, and K. P. Klugman. 2002. Analysis of penicillin-binding protein genes of clinical isolates of Streptococcus pneumoniae with reduced susceptibility to amoxicillin. Antimicrob. Agents Chemother. 46:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edman, M., S. Berg, P. Storm, M. Wikström, S. Vikström, A. Öhmann, and A. Wieslander. 2003. Structural features of glycosyltransferases synthesizing major bilayer and nonbilayer-prone membrane lipids in Acholeplasma laidlawii and Streptococcus pneumoniae. J. Biol. Chem. 278:8420-8428. [DOI] [PubMed] [Google Scholar]
- 16.Frère, J.-M., C. Duez, J.-M. Ghuysen, and J. Vanderkerkhove. 1976. Occurrence of a serine residue in the penicillin-binding site of the exocellular DD-carboxy-peptidase-transpeptidase from Streptomyces R61. FEBS Lett. 70:257-260. [DOI] [PubMed] [Google Scholar]
- 17.Garcia, P., M. P. Gonzalez, E. Garcia, R. Lopez, and J. L. Garcia. 1999. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 31:1275-1277. [DOI] [PubMed] [Google Scholar]
- 18.Goffin, C., and J.-M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grebe, T., and R. Hakenbeck. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob. Agents Chemother. 40:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haenni, M., and P. Moreillon. 2006. Mutations in penicillin-binding protein (PBP) genes and in non-PBP genes during selection of penicillin-resistant Streptococcus gordonii. Antimicrob. Agents Chemother. 50:4053-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakenbeck, R., T. Briese, L. Chalkley, H. Ellerbrok, R. Kalliokoski, C. Latorre, M. Leinonen, and C. Martin. 1991. Variability of penicillin-binding proteins from penicillin-sensitive Streptococcus pneumoniae. J. Infect. Dis. 164:307-312. [DOI] [PubMed] [Google Scholar]
- 22.Hakenbeck, R., T. Grebe, D. Zähner, and J. B. Stock. 1999. β-Lactam resistance in Streptococcus pneumoniae: penicillin-binding proteins and non penicillin-binding proteins. Mol. Microbiol. 33:673-678. [DOI] [PubMed] [Google Scholar]
- 23.Hakenbeck, R., C. Martin, C. Dowson, and T. Grebe. 1994. Penicillin-binding protein 2b of Streptococcus pneumoniae in piperacillin-resistant laboratory mutants. J. Bacteriol. 176:5574-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hakenbeck, R., S. Tornette, and N. F. Adkinson. 1987. Interaction of non-lytic β-lactams with penicillin-binding proteins in Streptococcus pneumoniae. J. Gen. Microbiol. 133:755-760. [DOI] [PubMed] [Google Scholar]
- 25.Halfmann, A., M. Kovacs, R. Hakenbeck, and R. Brückner. 2007. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of fifteen promoters drive expression of small noncoding RNAs. Mol. Microbiol. 66:110-126. [DOI] [PubMed] [Google Scholar]
- 26.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interaction. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 28.Höltje, J.-V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoskins, J., P. Matsushima, D. L. Mullen, J. Tang, G. Zhao, T. I. Meier, T. I. Nicas, and S. R. Jaskunas. 1999. Gene disruption studies of penicillin-binding proteins 1a, 1b and 2a in Streptococcus pneumoniae. J. Bacteriol. 181:6552-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krauß, J., and R. Hakenbeck. 1997. A mutation in the D,D-carboxypeptidase penicillin-binding protein 3 of Streptococcus pneumoniae contributes to cefotaxime resistance of the laboratory mutant C604. Antimicrob. Agents Chemother. 41:936-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacks, S., and R. D. Hotchkiss. 1960. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim. Biophys. Acta 39:508-517. [DOI] [PubMed] [Google Scholar]
- 32.Laible, G., and R. Hakenbeck. 1991. Five independent combinations of mutations can result in low-affinity penicillin-binding protein 2x of Streptococcus pneumoniae. J. Bacteriol. 173:6986-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laible, G., R. Hakenbeck, M. A. Sicard, B. Joris, and J.-M. Ghuysen. 1989. Nucleotide sequences of the pbpX genes encoding the penicillin-binding protein 2x from Streptococcus pneumoniae R6 and a cefotaxime-resistant mutant, C506. Mol. Microbiol. 3:1337-1348. [DOI] [PubMed] [Google Scholar]
- 34.Laible, G., B. G. Spratt, and R. Hakenbeck. 1991. Inter-species recombinational events during the evolution of altered PBP2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 5:1993-2002. [DOI] [PubMed] [Google Scholar]
- 35.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 36.LeBlanc, D. J., L. N. Lee, and J. M. Inamine. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1804-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macheboeuf, P., C. Contreras-Martel, V. Job, O. Dideberg, and A. Dessen. 2006. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30:673-691. [DOI] [PubMed] [Google Scholar]
- 38.Mascher, T., M. Heintz, D. Zähner, M. Merai, and R. Hakenbeck. 2006. The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and mutations in pbp2x involved in beta-lactam resistance. J. Bacteriol. 188:1959-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mascher, T., M. Merai, N. Balmelle, A. de Saizieu, and R. Hakenbeck. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J. Bacteriol. 185:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maurer, P., B. Koch, I. Zerfaß, J. Krauß, M. van der Linden, J.-M. Frère, C. Contreras-Martel, and R. Hakenbeck. 2008. Penicillin-binding protein 2x of Streptococcus pneumoniae: three new mutational pathways for remodelling an essential enzyme into a resistance determinant. J. Mol. Biol. 376:1403-1416. [DOI] [PubMed] [Google Scholar]
- 41.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiological Network (PMEN). J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morlot, C., A. Zapun, O. Dideberg, and T. Vernet. 2003. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol. Microbiol. 50:845-855. [DOI] [PubMed] [Google Scholar]
- 43.Mouz, N., A. M. Di Guilmi, E. Gordon, R. Hakenbeck, O. Dideberg, and T. Vernet. 1999. Mutations in the active site of penicillin-binding protein PBP2x from Streptococcus pneumoniae. Role in the specificity for β-lactam antibiotics. J. Biol. Chem. 274:19175-19180. [DOI] [PubMed] [Google Scholar]
- 44.Mouz, N., E. Gordon, D.-M. Di Guilmi, I. Petit, Y. Petillot, Y. Dupont, R. Hakenbeck, T. Vernet, and O. Dideberg. 1998. Identification of a structural determinant for resistance to β-lactam antibiotics in gram-positive bacteria. Proc. Natl. Acad. Sci. USA 95:13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller, P., C. Ewers, U. Bertsche, M. Anstett, T. Kallis, E. Breukink, C. Fraipont, M. Terrak, M. Nguyen-Distèche, and W. Vollmer. 2007. The essential cell division proteins FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J. Biol. Chem. 282:36394-36402. [DOI] [PubMed] [Google Scholar]
- 46.Muñóz, R., C. G. Dowson, M. Daniels, T. J. Coffey, C. Martin, R. Hakenbeck, and B. G. Spratt. 1992. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 6:2461-2465. [DOI] [PubMed] [Google Scholar]
- 47.Nagai, K., T. A. Davies, M. R. Jacobs, and P. C. Appelbaum. 2002. Effects of amino acid alterations in penicillin-binding proteins (PBPs) 1a, 2b, and 2x on PBP affinities of penicillin, ampicillin, amoxicillin, cefditoren, cefuroxime, cefprozil, and cefaclor in 18 clinical isolates of penicillin-susceptible, -intermediate, and -resistant pneumococci. Antimicrob. Agents Chemother. 46:1273-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Negri, M. C., M. I. Morosini, M. R. Baquero, R. R. Campo, J. Blázquez, and F. Baquero. 2002. Very low cefotaxime concentrations select for hypermutable Streptococcus pneumoniae populations. Antimicrob. Agents Chemother. 46:528-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paik, J., I. Kern, R. Lurz, and R. Hakenbeck. 1999. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J. Bacteriol. 181:3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pernot, L., L. Chesnel, A. Le Gouellec, J. Croizé, T. Vernet, O. Dideberg, and A. Dessen. 2004. A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics. J. Biol. Chem. 279:16463-16470. [DOI] [PubMed] [Google Scholar]
- 51.Reichmann, P., A. König, J. Liñares, F. Alcaide, F. C. Tenover, L. McDougal, S. Swidsinski, and R. Hakenbeck. 1997. A global gene pool for high-level cephalosporin resistance in commensal Streptococcus spp. and Streptococcus pneumoniae. J. Infect. Dis. 176:1001-1012. [DOI] [PubMed] [Google Scholar]
- 52.Salles, C., L. Creancier, J. P. Claverys, and V. Méjean. 1992. The high level streptomycin resistance gene from Streptococcus pneumoniae is a homologue of the ribosomal protein S12 gene from Escherichia coli. Nucleic Acids Res. 20:6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 54.Sanbongi, Y., T. Ida, M. Ishikawa, Y. Osaki, H. Kataoka, T. Suzuki, K. Kondo, F. Ohsawa, and M. Yonezawa. 2004. Complete sequences of six penicillin-binding protein genes from 40 Streptococcus pneumoniae clinical isolates collected in Japan. Antimicrob. Agents Chemother. 48:2244-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sebert, M. E., L. M. Palmer, M. Rosenberg, and J. N. Weiser. 2002. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sifaoui, F., M.-D. Kitzis, and L. Gutmann. 1996. In vitro selection of one-step mutants of Streptococcus pneumoniae resistant to different oral β-lactam antibiotics is associated with alterations of PBP2x. Antimicrob. Agents Chemother. 40:152-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, A. M., and K. P. Klugman. 2005. Amino acid mutations essential to production of an altered PBP2X conferring high-level beta-lactam resistance in a clinical isolate of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 49:4622-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sung, C. K., H. Li, J. P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and K. R. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 60.Tiraby, J.-G., and M. S. Fox. 1974. Marker discrimination and mutagen-induced alterations in pneumococcal transformation. Genetics 77:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zähner, D., T. Grebe, E. Guenzi, J. Krauß, M. van der Linden, K. Terhune, J. B. Stock, and R. Hakenbeck. 1996. Resistance determinants for β-lactam antibiotics in laboratory mutants of Streptococcus pneumoniae that are involved in genetic competence. Microb. Drug Resist. 2:187-191. [DOI] [PubMed] [Google Scholar]
- 62.Zähner, D., K. Kaminski, M. van der Linden, T. Mascher, M. Merai, and R. Hakenbeck. 2002. The ciaR/ciaH regulatory network of Streptococcus pneumoniae. J. Mol. Microbiol. Biotechnol. 4:211-216. [PubMed] [Google Scholar]
- 63.Zhao, G., T. I. Meir, S. D. Kahl, K. R. Gee, and L. C. Blaszczak. 1999. Bocillin FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 43:1124-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]