Abstract
Resistance of influenza viruses to antiviral drugs can emerge following medication or may result from natural variation. Two classes of anti-influenza virus drugs targeting either the M2 protein (amantadine and rimantadine) or neuraminidase (NA; oseltamivir and zanamivir) are currently licensed. These drugs are expected to be important in controlling the early stages of a potential pandemic. In the present study, we describe how a pyrosequencing method can be used to rapidly detect established molecular markers of resistance to M2 blockers and NA inhibitors in influenza A (H5N1) viruses. The residues L26, V27, A30, S31, and G34 in the M2 protein were targeted for pyrosequencing. The NA residues for pyrosequencing analysis included the established markers of drug resistance (H274 and N294), as well as residues of less certain relevance (V116, I117, Q136, K150, and I222). A single pair of pyro-reverse transcription (RT)-PCR primers was designed to allow amplification of an approximately 600-nucleotide-long amplicon of the NA genes of H5N1 viruses from various clades/subclades associated with infections in humans. The sensitivity of the assay was demonstrated by the successful pyrosequencing of RNA extracted from samples of serially diluted (10−5 to 10−7) virus stocks with initial concentrations ranging from 105 to 108 PFU/ml. The markers of resistance were detected in samples with threshold cycle values ranging from 32 to 37, as determined by real-time RT-PCR. The pyrosequencing approach may provide a valuable tool for rapid detection of markers of drug resistance in H5N1 viruses and facilitate the elucidation of the role of such changes in natural and acquired drug resistance.
Avian influenza A (H5N1) viruses, which first emerged in 1997, have become endemic in Asia and pose a serious threat to agriculture and human health (12, 14, 42). Since their reemergence in 2003, influenza A (H5N1) viruses have continued to infect humans, for whom the mortality rate is high (∼63%) (46). Although vaccines are the primary line of defense for the prophylactic control of influenza virus infections, effective antiviral drugs provide a valuable option for early containment of highly virulent and/or novel strains. Presently, there are two classes of anti-influenza virus drugs licensed by the U.S. Food and Drug Administration that are effective against influenza A viruses, including H5N1 viruses. These drugs are M2 channel blockers, also known as adamantanes (amantadine and rimantadine), and neuraminidase (NA) inhibitors (NAIs) (zanamivir and oseltamivir) (20, 34). Molecular markers of resistance to the older class of drugs, adamantanes, are well established and comprise changes at residues L26, V27, A30, S31, and G34 within the transmembrane domain of the M2 protein (6, 11, 22). Since 2005, a dramatic rise in the resistance of seasonal influenza A viruses to adamantanes has led to changes in CDC recommendations for the use of adamantanes in the control of influenza virus infections (8). In response to a need for rapid monitoring of the susceptibility of seasonal influenza A viruses to adamantanes, the pyrosequencing assay was developed (7). This assay was subsequently adopted by various virus surveillance laboratories for high-throughput screening for adamantane resistance in seasonal influenza A viruses and has proven its usefulness in the timely detection of the emergence and spread of adamantane-resistant H3N2 and H1N1 viruses (3, 4, 9, 17, 39, 40). More recently, pyrosequencing was used to detect quasispecies of the influenza virus in clinical materials as well as in cell cultures of virus isolates (16, 18, 27). In recent years, a significant increase in adamantane resistance has also been reported for some groups of influenza A (H5N1) viruses (11, 24, 25), necessitating close monitoring of adamantane resistance among diverse groups of rapidly evolving influenza A (H5N1) viruses, including viruses circulating in avian populations.
The emergence and spread of adamantane-resistant variants of three antigenic subtypes, H3N2, H1N1, and H5N1, of influenza A viruses influenced the decision to stockpile NAIs to be used for the mitigation of pandemic influenza. The emergence of oseltamivir-resistant viruses in treated individuals has been observed at a low frequency (∼1%) in the adult population (44) and at a higher frequency (up to 30%) in young children (26) and immunocompromised patients. Nevertheless, until recently, resistance to this class of drugs has been very low (<0.5%) in community isolates (23, 25, 32, 33, 36). During the 2007-2008 season, however, a significant rise in resistance to oseltamivir was detected among seasonal influenza A (H1N1) viruses recovered from untreated individuals of different ages and in distant geographic locations (28, 41). The oseltamivir-resistant H1N1 viruses that emerged retained sensitivity to zanamivir. These recent findings emphasize the need for close monitoring of NAI resistance among seasonal influenza viruses as well as among highly virulent H5N1 viruses. In contrast to those for adamantanes, molecular markers of resistance to NAIs are not yet well characterized. Moreover, resistance to NAIs is drug specific and NA type/subtype specific (reviewed in reference 19). Of note, H5N1 viruses of different genetic groups (clades/subclades) exhibit substantial variances in their NA sequences and susceptibilities to NAIs (10, 32, 47). For these reasons, the NA activity inhibition assay is currently used as the primary assay for monitoring resistance to NAIs (21, 35, 41, 43, 45). This method requires virus propagation in cell culture prior to testing and needs to be complemented by sequence analysis of the NA to confirm known markers or to detect novel markers of NAI resistance. Oseltamivir-resistant H5N1 viruses with amino acid changes at two residues, H274Y and N294S, have emerged in humans during oseltamivir treatment (15, 29). The impact of natural drift mutations in the NA on the drug susceptibility of H5N1 viruses has been reported (13, 32, 37) but is not readily predicted. Moreover, when the existing NAIs were designed, the crystal structures of the N1 subtype enzymes, including those of H5N1 viruses, were not available. Recent crystal structure data and conformational studies of the N1 enzyme showed that replacements at residues outside of the conserved active site (e.g., residues Q136 and K150 in the 150 loop of the NA molecule) could affect binding of the existing drugs to the NA by interfering with their induced fit into the active site (10, 30, 38, 47). Although the exact mechanisms by which some changes (e.g., residues V116, I117, Q136, K150, and I222) in the N1 enzyme affect susceptibility to a particular NAI are not well understood, there is a need to monitor their presence in multiple and evolving clades of H5N1 viruses.
The ability to rapidly detect markers of antiviral drug resistance is a valuable tool in influenza pandemic preparedness. Especially desirable are rapid, high-throughput methods that are sensitive and that minimize the handling of live viruses. Pyrosequencing has been proven to provide such options for the detection of resistance to adamantanes in seasonal influenza A viruses, and more recently, this method was expanded for use in the detection of resistance to NAIs in seasonal and H5N1 viruses (16, 18, 27).
In the present study, we enhanced the existing pyrosequencing approach by expanding the number of targeted markers (from 2 to 7) in the NAs of H5N1 viruses and provided validation of pyrosequencing assays for both M2 and N1 NAs by testing H5N1 viruses from a variety of genetic groups (clades/subclades). Based on the results of enzyme inhibition assays (2, 5, 25, 27, 32), replacements at some of the targeted residues in the NA (e.g., V116, I117, Q136, K150, and I222) have previously been linked to reduced drug susceptibilities in avian and human viruses carrying N1. However, some of them may yet be proven to have little relevance for clinical resistance. Nevertheless, the pyrosequencing assay developed here provides a robust tool for the detection of influenza virus variants, including those isolated directly from clinical specimens, and thus could be instrumental in identifying new mechanisms of resistance to the newer NAI class of anti-influenza virus drugs.
MATERIALS AND METHODS
Viruses.
Influenza A (H5N1) viruses isolated from humans and birds between 1997 and 2008 and submitted to the WHO Collaborating Center for Surveillance, Epidemiology and Control of Influenza at the CDC, Atlanta, GA, were propagated in allantoic cavities of 9- to 11-day-old embryonated chicken eggs and harvested 24 to 48 h postinoculation. Viruses were handled according to biosafety level 3 containment guidelines with enhancements required by the USDA and the CDC select agents program. Viruses representing different genetic groups (clades and subclades) and associated with confirmed human infections (1) and viruses carrying known markers of resistance to adamantanes and NAIs were chosen for the study. Those viruses included but were not limited to the following: clade 0, A/Hong Kong/482/1997 (I222T); clade 1, A/Vietnam/1203/2004 and A/duck/Vietnam/NCVD-19/2007; clade 2.3.4, A/duck/Vietnam/NCVD100/2007 (I117V) and A/chicken/Laos/NCVD-38/2007 (K150N and I222L); clade 3, A/duck/Hong Kong/380.5/2001 (N294S); and clade 7, A/chicken/Vietnam/NCVD-93/2008 (Table 1). Throughout the manuscript, amino acid residues in the N1 NA are given according to universal N2 subtype numbering.
TABLE 1.
Influenza A (H5N1) viruses recovered from humans and birds and tested in the present study
| Virusa | Clade | Amino acid residues for subtype N1 | Amino acid residues for subtype N2 | Change(s) in M2 sequence(s) |
|---|---|---|---|---|
| A/Hong Kong/482/1997 | 0 | I223T | I222T | None |
| A/Vietnam/1203/2004 | 1 | L26I, S31N | ||
| A/duck/Vietnam/NCVD-19/2007 | 1 | L26I, V27A, S31N | ||
| A/duck/Vietnam/NCVD100/2007 | 2.3.4 | I117V | I117V | None |
| A/chicken/Laos/NCVD-38/2007 | 2.3.4 | K150N, I223L | K150N, I222L | None |
| A/duck/Hong Kong/380.5/2001 | 3 | N295S | N294S | None |
| A/chicken/Vietnam/NCVD-93/2008 | 7 | S31N |
The GenBank accession numbers of the NA sequences of the viruses in the order in which they are listed in the table are AF084272, EF541467, CY030377, FJ538947, CY030487, AY075034, and FJ538949; their M gene sequence accession numbers are AF084282, EF541453, CY030378, FJ538948, CY030488, AY075035, and FJ538950.
Pyrosequencing primer design.
Full-length NA sequences from 1,391 human and avian H5N1 virus isolates collected between 1997 and 2008 were obtained from the CDC sequence database and the Los Alamos influenza sequence database (31) and aligned. Using BioEdit software (version 5.0.6; North Carolina State University), the N1 consensus sequence was generated. Similarly, a consensus sequence based on the alignment of their M gene sequences was generated. These consensus sequences were used to design pyro-reverse transcription (RT)-PCR and sequencing primers with Pyrosequencing assay design software (Biotage). In several instances, primers were modified to accommodate degenerate nucleotides. The primers were synthesized at the CDC Biotechnology Core facility (Table 2).
TABLE 2.
The designed RT-PCR and sequencing primers used to perform pyrosequencing analysis
| Method and primer | Sequence | Target H5N1 gene | NA target residue(s) |
|---|---|---|---|
| RT-PCR | |||
| H5N1-M-F739a | 5′-CAAATGCATCGRTTCAAGTG-3′ | M2 | |
| H5N1-M-R1027-biot | 5′-AGTAGAAACAAGGTAGTTTTTTACTC-3′ | ||
| H5N1-NA-F322C | 5′-ATTGGTTCCARGGGGGATG-3′ | NA | |
| H5N1-NA-R924B-biot | 5′-TTGATTGAARGAYACCCATG-3′ | ||
| Pyrosequencing | |||
| H5N1-NA-F321 | 5′-GATTGGTTCCAAGGGGGATGTG-3′ | V116, I117 | |
| H5N1-NA-F374 | 5′-CCC ACY TGG AAT GCA GAA C-3′ | Q136 | |
| H5N1-NA-F421 | 5′-AATGACAAGCACTCCAAYGGGAC-3′ | K150N | |
| H5N1-NA-F640 | 5′-GACACYATCAAGAGYTGGA-3′ | I222 | |
| H5N1-NA-F796A | 5′-TCAGTCGAATTGAATGCT-3′ | H274 | |
| H5N1-NA-F851 | 5′-ATGCCGGYGAAATCACRTGTGT-3′ | N294 |
The same primer was used at 100 μM for pyrosequencing the region encompassing residues 26, 27, 30, 31, and 34 of the M2 protein.
Resistance to adamantanes is associated with mutations occurring at one or more of residues 26, 27, 30, 31, and 34 (7, 11) in the M2 protein. The RT-PCR primers were designed to amplify a region encoding these amino acids (from nucleotide [nt] 739 to nt 1027).
RT-PCR and pyrosequencing.
Viral RNAs were extracted from viral supernatants in lysis buffer. RT-PCR amplifications were performed using the OneStep RT-PCR system (Qiagen, Valencia, CA). Primers were used at 20 μM in a standard 25-μl reaction mixture, and amplification was performed for 45 cycles. Biotinylated amplicons were purified, and the pyrosequencing reactions were carried out as previously described (7), using a PSQ 96MA platform pyrosequencer. Amplification products were washed in a series of buffers, and single-stranded, biotinylated DNA products were hybridized to mutation-specific sequencing primers in a 96-well plate used at final concentrations of 0.45 μM in 40 μl of annealing buffer. Pyrosequencing was performed using Pyro Gold reagents according to Biotage's recommendations. The accuracy of the sequences obtained using pyrosequencing was confirmed by comparison to the results of conventional sequencing.
Assessment of pyrosequencing assay sensitivity.
To determine the sensitivity of the designed pyrosequencing assay in terms of the amount of RNA needed for the detection of markers of resistance in the NAs of H5N1 viruses, we performed the following experiments. The H5N1 viruses grown in eggs had initial concentrations of 105 to 108 PFU/ml. Viral RNA was extracted from the initial stocks and then serially 10-fold diluted in water (10−2 to 10−8). The RNA dilutons were used for amplification of the NA gene fragment, using the RT-PCR primers designed for pyrosequencing, and were also subjected to conventional real-time RT-PCR analysis (the conditions for real-time RT-PCR were consistent with the CDC diagnostic assay for detection and subtyping of influenza viruses currently in use). The primers and probes used in this study are available upon request through a material transfer agreement with the U.S. Centers for Disease Control.
The sensitivity of the pyrosequencing assay was also assessed by using RNA extracted from serially 10-fold-diluted virus stocks. One hundred microliters from each virus dilution was added to an equal volume of lysis buffer. RNAs were extracted and used to perform RT-PCR amplifications as described above, and pyrosequencing was done by using the primer F-796A to analyze codon 274 for the entire panel of viruses (n = 12). This approach allowed determination of the highest dilution of virus stocks that could be used to generate pyrograms of high-quality resolution with the designed pyrosequencing primers. The RNAs from the highest dilution of the virus stock were also subjected to real-time RT-PCR analysis.
RESULTS
Detection of molecular markers of influenza A (H5N1) virus resistance to M2 blockers.
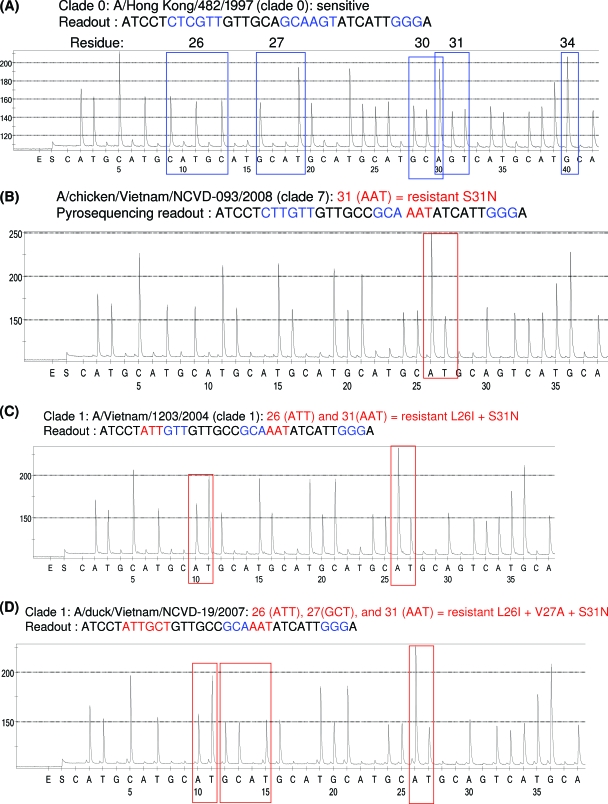
Because of the conserved nature of influenza A virus M2 sequences, it was possible to use the same set of primers to detect adamantane resistance-conferring mutations in two antigenic subtypes, H1N1 and H3N2, of seasonal influenza virus (7). Here, we assessed the ability of the same primers to detect adamantane resistance in the H5N1 subtype. We found that overall the same assay protocol was suitable for the majority of the H5N1 viruses. However, when the modified forward primer (Table 2) was used, it improved the quality of the pyrograms for some H5N1 viruses. A single pair of primers (a modified forward primer and the previously described reverse primer [7]) allowed amplification of all H5N1 viruses tested in the study, and the same forward primer was used to generate sequences from these viruses. The pyrogram (Fig. 1A) for the A/Hong Hong/482/1997 virus shows no markers of resistance to adamantanes, whereas the pyrogram (Fig. 1B) for A/chicken/Vietnam/NCVD-93/2008 clearly indicates a mutation of AGT to AAT (serine to asparagine) at residue 31. The A/Vietnam/1203/2004 virus has mutations CTT to ATT (leucine to isoleucine) at position 26 and AGT to AAT (serine to asparagine) at position 31 (Fig. 1C). This combination of amino acid changes was previously reported for clade 1 H5N1 viruses (11). The M2 protein of the A/duck/Vietnam/NCVD-19/2007 virus contains three mutations: in addition to L26I and S31N (Fig. 1D), it also has V27A (GTT to GCT) at residue 27.
FIG. 1.
Detection of established molecular markers of resistance to M2 blockers in H5N1 viruses using pyrosequencing. The sequences generated encompass the region of the M gene which contains the 5-amino-acid residues of M2 protein associated with resistance. (A) A/Hong Kong/482/1997 (sensitive) has no change at any of the 5 codons (blue). (B) A/chicken/Vietnam/NCVD-093/2008 (resistant) contains the mutation S31N (AGT to AAT). (C) A/Vietnam/1203/2004 (resistant) contains two mutations, L26I (CTT to ATT) and S31N (AGT to AAT). (D) A/duck/Vietnam/NCVD-19/2007 (resistant) contains three mutations, L26I (CTT to ATT), V27A (GTT to GCT), and S31N (AGT to AAT).
Detection of molecular markers of influenza A (H5N1) virus resistance to NAIs.
The NA sequence variance of avian H5N1 viruses presents a challenge for designing a universal set of primers for pyrosequencing. The codons of interest include H274, N294, I222, Q136, K150, V116, and I117, and a RT-PCR amplicon must encompass all seven codons. Therefore, primers were designed to allow amplification of the portion of the NA gene from nt 322 to nt 924 (Table 2; also Materials and Methods). Because the codons are distant from each other and because a single pyrosequencing primer can provide reliable sequences only up to 60 nt, an individual primer for sequence reading at each codon was designed and tested (Table 2). The results of pyrosequencing analysis of the representative H5N1 viruses that belong to five different clades/subclades are given below. It is worth noting that the designed assay was also successfully used to analyze sequences at these residues of interest in H5N1 viruses from other subclades (2.1, 2.2, and clade 4; results not shown).
Codon 274.
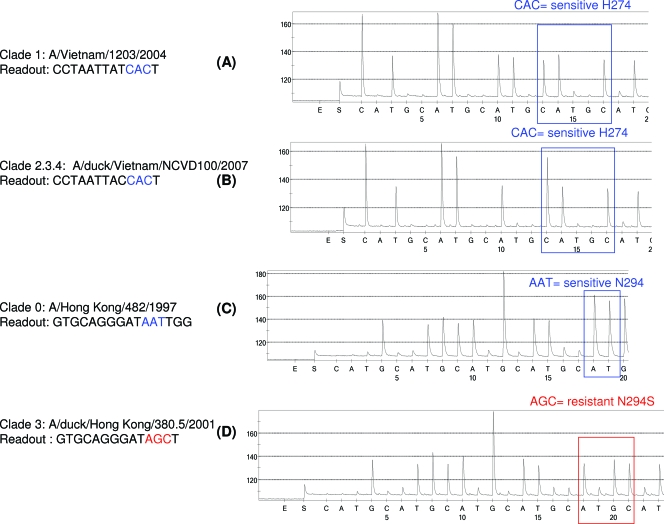
The amino acid change of histidine to tyrosine at residue 274 (CAT to TAT) in the NA of H5N1 viruses has been reported to confer resistance to oseltamivir (15, 29). As anticipated, only a wild-type sequence with CAC (histidine) at codon 274 was detected among the viruses tested. According to the pyrograms and conventional sequencing, A/chicken/Laos/NCVD-38/2007 and A/chicken/Vietnam/NCVD-093/2008 viruses from clades 2.3.4 and 7, respectively, shared identical sequences in the region analyzed (data not shown), whereas sequences of two viruses from clades 1 (A/Vietnam/1203/2004) and 2.3.4 (A/duck/Vietnam/NCVD100/2007) differed by a single nucleotide (Fig. 2A and B).
FIG. 2.
Detection of the most common oseltamivir resistance-conferring mutations in H5N1, using pyrosequencing. One pair of primers (F322C/R924B-biot) was used to generate biotinylated PCR products from the set of H5N1 viruses used in the study. The pyrograms show the NA sequences at codon 274. (A and B) Wild-type viruses A/Vietnam/1203/2004 (A) and A/duck/Vietnam/NCVD100/2007 (B), from clades 1 and 2.3.4, respectively, contain CAC (H274). (C and D) Oseltamivir-sensitive A/Hong Kong/482/1997 contains AAT (N294) (C), and oseltamivir-resistant A/duck/Hong Kong/380.5/2001 contains AGC (N294S) (D).
Codon 294.
The effect of the amino acid change N294S on the NAI susceptibilities of H5N1 viruses in vitro has previously been reported (29, 48). Pyrograms confirmed the presence of a wild-type NA sequence in A/Hong Kong/482/1997 (Fig. 2C) and an AAT to AGC mutation (asparagine to serine) at residue 294 in the A/duck/Hong Kong/380.5/2001 virus from clade 3 (Fig. 2D). The latter virus is oseltamivir resistant, according to NAI assays (data not shown).
Codon 222.
Substitutions at a conserved residue, I222, have been shown to alter susceptibility to NAIs (2, 19, 41). As expected, wild-type sequence ATA (isoleucine) at codon 222 was detected in A/Vietnam/1203/2004 (Fig. 3A), while pyrograms show mutations at codon 222 in A/Hong Kong/482/1997 (threonine, ACA) and A/chicken/Laos/NCVD-38/2007 (leucine, TTA) (Fig. 3B and C).
FIG. 3.
Pyrosequencing analysis of codons 222, 136, 150, and 116 and 117 in the NAs of H5N1 viruses. Viruses from clades 0, 1, and 2.3.4 were tested. (A) Wild-type sequence ATA (I222) in A/Vietnam/1203/2004. (B and C) Mutated sequences ACA (I222T) and TTA (I222L) from A/Hong Kong/482/1997 (B) and A/chicken/Laos/NCVD-38/2007 (C), respectively. (D and E) Wild-type viruses A/Hong Kong/482/1997 (D) and A/duck/Vietnam/NCVD100/2007 (E) with CAA and CAG, respectively, at residue 136. (F and G) Wild-type A/chicken/Vietnam/NCVD-093/2008 (F) with AAA (lysine) at position 150 and mutant A/chicken Laos/NCVD-38/2007 (G) with AAC (asparagine) at position 150. (H and I) Wild-type A/Vietnam/1203/2004 (H) with V116 (GTT) and ATA (I117) and variant A/duck/Vietnam/NCVD100/2207 (I), from clade 2.3.4, with GTT corresponding to the mutated sequence I117V.
Codons 136 and 150.
The analysis of the crystal structures of N1 NAs from H1N1 and H5N1 viruses (10, 38) revealed the importance of the residues forming the 150 loop and glutamine at residue 136 for binding of the drug to the NAs. Moreover, amino acid mutation Q136K was reported to cause zanamivir resistance in seasonal influenza viruses carrying the enzyme of the N1 subtype (A. C. Hurt, personal communication; also unpublished CDC data). The results (Fig. 3D and E) show the presence of either CAA or CAG at codon 136, which corresponds to wild-type Q136.
The ability of the assay to detect changes at codon 150 was tested, using viruses from three clades: 1, 2.3.4, and 7. These viruses, A/Vietnam/1203/2004, A/duck/Vietnam/NCVD100/2007 (clade 2.3.4), and A/chicken/Vietnam/NCVD-93/2008 (clade 7), shared the identical wild-type sequence AAA (lysine) at position 150 (Fig. 3F). In contrast, the pyrogram from A/chicken/Laos/NCVD-38/2007 (clade 2.3.4) showed the presence of the drug resistance-conferring mutation AAA to AAC (lysine to asparagine (Fig. 3G).
Codons 116 and 117.
Amino acid residue V116 appears to be conserved among NA types and subtypes, and the replacement of valine with alanine at this position in H5N1 viruses resulted in reduced susceptibility to NAIs (25). Furthermore, mutation I117V was reported to be associated with reduced susceptibility to NAIs in both H5N1 (25, 32) and seasonal H1N1 (27. In our tests, A/Hong Kong/482/1997, A/Vietnam/1203/2004, and A/duck/Hong Kong/380.5/2001, from clades 0, 1, and 3, respectively, shared the identical target sequences with GTT (valine) at position 116 and ATA (isoleucine) at position 117 (Fig. 3H). In accordance with conventional sequence data, pyrograms (Fig. 3I) showed the presence of wild-type sequence GTT (valine) at position 116 and mutant sequence GTT (valine) at residue 117 of the NA of A/duck/Vietnam/NCVD100/2007 from clade 2.3.4.
To evaluate the sensitivity of the pyrosequencing assay, real-time RT-PCR analysis was conducted on RNA dilutions ranging from 10−5 to 10−7, which yielded quality pyrograms following pyrosequencing (see Materials and Methods). Results from the real-time RT-PCR amplification analysis confirmed that the pyrosequencing assay was successful in reliably identifying each of the targeted NA mutations at all 7 codons in samples with threshold cycle (CT) values ranging from 31 to 35 (Fig. 4). In addition, the sensitivity of the pyrosequencing assay was tested by analyzing the pyrograms obtained directly from the highest dilutions of the H5N1 virus stocks. Similar to that for the RNA dilutions, the sensitivity of the pyrosequencing assay was found to range from 10−5 to 10−7 virus dilutions of the initial virus stocks (105 to 108 PFU/ml). Real-time-RT-PCR performed on the highest virus dilutions had CT values ranging from 32 to 37. This level of sensitivity was within the range of detection of the established CDC influenza virus diagnostic real-time RT-PCR assay.
FIG. 4.
Evaluation of the sensitivity of the pyrosequencing assay designed for detection of markers of resistance to NAIs in H5N1 viruses. RNAs were prepared from the 7 viruses shown in Table 1 and from 5 additional viruses that belong to clades/subclades 1, 2.1, and 2.2. Viral RNA was extracted, and 10-fold (10−2 to 10−8) dilutions of viral RNA were prepared for each virus. Pyrosequencing analysis was performed on each RNA dilution. The highest level of dilution at which sequences were obtained was used in conventional real-time RT-PCR amplification to determine the CT values. Shown are amplification curves from the 12 RNA samples. The CT values ranged from 31 to 35. dRn, delta raw fluorescence normalizing.
DISCUSSION
Monitoring of drug resistance among H5N1 viruses is a challenging task due to their rapid evolution and the need for enhanced biosafety level 3 containment when working with live viruses. In this study, we demonstrated the development of a rapid and high-throughput pyrosequencing method that could be utilized for reliable detection of known molecular markers of influenza viruses resistant to NAIs or adamantanes from a variety of influenza A (H5N1) viruses. It is worth noting that because our assay was designed to encompass all seven currently known markers associated with drug resistance in the influenza virus NA, it differs markedly from other described methods (18, 27), which were designed to target only one or two markers. Moreover, recent work that elucidated the structure of N1 NA showed that previous work on the design of NA inhibitors may not have been optimal for targeting the NA of the N1 subtype, and natural resistance to such designed drugs can be conferred by changes in residues outside of the immediate active site of N1 (10, 30, 38, 47, 49). Indeed, drug susceptibility testing using the NA inhibition assay showed that naturally occurring amino acid replacements outside of the NA active site (e.g., V116 and I117) can alter the susceptibility of avian H5N1 viruses to oseltamivir and/or zanamivir (25, 32). Although clinical resistance to NAIs associated with each of these genetic markers has not been conclusively established, it is prudent to have reliable tools for rapid and high-throughput detection of such variants in the event of an epidemic or pandemic outbreak of influenza. It is noteworthy that the pyrosequencing assay developed and reported here can be easily performed with small amounts of available RNA, which might be limited in clinical specimens, and generates short gene sequences, making the product less affected by RNA quality in virus samples. For seasonal influenza viruses, we and others have demonstrated a reproducible ability to detect molecular markers associated with drug resistance from a variety of clinical specimens (16, 18, 27).
In the present study, we conducted an assessment of the pyrosequencing assay using two approaches. Viral RNA was extracted from virus stocks, and then serially diluted RNA preparations were subjected to RT-PCR and pyrosequencing. Alternatively, serial dilutions were made from the virus stocks, and then viral RNA was extracted and used for RT-PCR amplification followed by pyrosequencing. The second approach may provide a better estimation of the assay's sensitivity. In both instances, RNA was used to determine the CT values of the highest dilutions that generated good-quality pyrograms (sequences) in the real-time RT-PCR assay. The CT values (ranging from 31 to 35 versus 32 to 37) obtained with either approach yielded results indicative of a high level of sensitivity of the pyrosequencing assay designed in the present study. It is noteworthy that the latter approach more accurately reflects the content of viral RNA available in a specimen.
Unlike that to NAIs, resistance to amantadine and rimantadine is relatively high among clade 1 and 2.1 H5N1 viruses (11, 24, 25, 49). Using pyrosequencing methodology, we were able to analyze the presence of the 5 molecular markers of resistance to M2 blockers in a wide variety of H5N1 viruses. We have shown here only examples of the detection of mutations I26L (CTT to ATT), V27A (GTT to GCT), and S31N (AGT to AAT) in H5N1 viruses from clades 0, 1, and 7; however, it is important to emphasize that the assay also successfully allowed testing of viruses from subclades 2.1 and 2.2 and clade 4 (data not shown). In conclusion, the sensitivity of the pyrosequencing assay was optimized to allow analyses of viral samples with small quantities of RNA. This could permit detection of drug-resistant H5N1 viruses in clinical specimens, thus reducing the time needed to complete antiviral susceptibility testing.
The assay developed in this study lends itself for use in rapid and reliable detection of known molecular markers of resistance to anti-influenza virus drugs. The results obtained can be utilized to guide decisions regarding antiviral use in prophylaxis and treatment in the event of local outbreaks or potential pandemics.
Acknowledgments
We are grateful to Nancy Cox, Tim Uyeki, Michael Shaw, and Charles Davis from the Influenza Division, CDC, for their valuable contributions to the project. We thank David Swayne, South-East Poultry Research Laboratory, USDA, and the National Centre for Veterinary Diagnostics, Vietnam, for sharing the H5N1 viruses.
The findings and conclusions of this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print on 5 January 2009.
REFERENCES
- 1.Abdel-Ghafar, A. N., T. Chotpitayasunondh, Z. Gao, F. G. Hayden, D. H. Nguyen, M. de Jong, A. Naghdaliyev, J. S. Peiris, N. Shindo, S. Soeroso, and T. M. Uyeki. 2008. Update on avian influenza A (H5N1) virus infection in humans. N. Engl. J. Med. 358:261-273. [DOI] [PubMed] [Google Scholar]
- 2.Abed, Y., B. Nehme, M. Baz, and G. Boivin. 2008. Activity of the neuraminidase inhibitor A-315675 against oseltamivir-resistant influenza neuraminidases of N1 and N2 subtypes. Antivir. Res. 77:163-166. [DOI] [PubMed] [Google Scholar]
- 3.Barr, I. G., A. C. Hurt, N. Deed, P. Iannello, C. Tomasov, and N. Komadina. 2007. The emergence of adamantane resistance in influenza A(H1) viruses in Australia and regionally in 2006. Antivir. Res. 75:173-176. [DOI] [PubMed] [Google Scholar]
- 4.Barr, I. G., A. C. Hurt, P. Iannello, C. Tomasov, N. Deed, and N. Komadina. 2006. Increased adamantane resistance in influenza A(H3) viruses in Australia and neighbouring countries in 2005. Antivir. Res. 73:112-117. [DOI] [PubMed] [Google Scholar]
- 5.Baz, M., Y. Abed, J. McDonald, and G. Boivin. 2006. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin. Infect. Dis. 43:1555-1561. [DOI] [PubMed] [Google Scholar]
- 6.Boivin, G., N. Goyette, and H. Bernatchez. 2002. Prolonged excretion of amantadine-resistant influenza A virus quasi species after cessation of antiviral therapy in an immunocompromised patient. Clin. Infect. Dis. 34:E23-E25. [DOI] [PubMed] [Google Scholar]
- 7.Bright, R. A., M. J. Medina, X. Xu, G. Perez-Oronoz, T. R. Wallis, X. M. Davis, L. Povinelli, N. J. Cox, and A. I. Klimov. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 366:1175-1181. [DOI] [PubMed] [Google Scholar]
- 8.Bright, R. A., D. Shay, J. Bresee, A. Klimov, N. Cox, and J. R. Ortiz. 2006. High levels of adamantane resistance among influenza A (H3N2) viruses and interim guidelines for use of antiviral agents—United States, 2005-2006 influenza season. MMWR Morb. Mortal. Wkly. Rep. 55:44-46. [PubMed] [Google Scholar]
- 9.Bright, R. A., D. K. Shay, B. Shu, N. J. Cox, and A. I. Klimov. 2006. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA 295:891-894. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, L. S., R. E. Amaro, D. Xu, W. W. Li, P. W. Arzberger, and J. A. McCammon. 2008. Ensemble-based virtual screening reveals potential novel antiviral compounds for avian influenza neuraminidase. J. Med. Chem. 51:3878-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, C. L., J. M. Rayner, G. J. Smith, P. Wang, T. S. Naipospos, J. Zhang, K. Y. Yuen, R. G. Webster, J. S. Peiris, Y. Guan, and H. Chen. 2006. Distribution of amantadine-resistant H5N1 avian influenza variants in Asia. J. Infect. Dis. 193:1626-1629. [DOI] [PubMed] [Google Scholar]
- 12.Claas, E. C., J. C. de Jong, R. van Beek, G. F. Rimmelzwaan, and A. D. Osterhaus. 1998. Human influenza virus A/HongKong/156/97 (H5N1) infection. Vaccine 16:977-978. [DOI] [PubMed] [Google Scholar]
- 13.Collins, P. J., L. F. Haire, Y. P. Lin, J. Liu, R. J. Russell, P. A. Walker, J. J. Skehel, S. R. Martin, A. J. Hay, and S. J. Gamblin. 2008. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 453:1258-1261. [DOI] [PubMed] [Google Scholar]
- 14.de Jong, J. C., E. C. Claas, A. D. Osterhaus, R. G. Webster, and W. L. Lim. 1997. A pandemic warning? Nature 389:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Jong, M. D., T. T. Tran, H. K. Truong, M. H. Vo, G. J. Smith, V. C. Nguyen, V. C. Bach, T. Q. Phan, Q. H. Do, Y. Guan, J. S. Peiris, T. H. Tran, and J. Farrar. 2005. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 353:2667-2672. [DOI] [PubMed] [Google Scholar]
- 16.Deyde, V. M., M. Okomo-Adhiambo, T. G. Sheu, T. R. Wallis, A. Fry, N. Dharan, A. I. Klimov, and L. V. Gubareva. 2009. Pyrosequencing as a tool to detect molecular markers of resistance to neuraminidase inhibitors in seasonal influenza A viruses. Antivir. Res. 81:16-24. [DOI] [PubMed] [Google Scholar]
- 17.Deyde, V. M., X. Xu, R. A. Bright, M. Shaw, C. B. Smith, Y. Zhang, Y. Shu, L. V. Gubareva, N. J. Cox, and A. I. Klimov. 2007. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis. 196:249-257. [DOI] [PubMed] [Google Scholar]
- 18.Duwe, S., and B. Schweiger. 2008. A new and rapid genotypic assay for the detection of neuraminidase inhibitor resistant influenza A viruses of subtype H1N1, H3N2, and H5N1. J. Virol. Methods 153:134-141. [DOI] [PubMed] [Google Scholar]
- 19.Ferraris, O., and B. Lina. 2008. Mutations of neuraminidase implicated in neuraminidase inhibitors resistance. J. Clin. Virol. 41:13-19. [DOI] [PubMed] [Google Scholar]
- 20.Gubareva, L. V., L. Kaiser, and F. G. Hayden. 2000. Influenza virus neuraminidase inhibitors. Lancet 355:827-835. [DOI] [PubMed] [Google Scholar]
- 21.Gubareva, L. V., R. G. Webster, and F. G. Hayden. 2002. Detection of influenza virus resistance to neuraminidase inhibitors by an enzyme inhibition assay. Antivir. Res. 53:47-61. [DOI] [PubMed] [Google Scholar]
- 22.Hay, A. J., M. C. Zambon, A. J. Wolstenholme, J. J. Skehel, and M. H. Smith. 1986. Molecular basis of resistance of influenza A viruses to amantadine. J. Antimicrob. Chemother. 18(Suppl. B):19-29. [DOI] [PubMed] [Google Scholar]
- 23.Hayden, F. G. 2006. Antivirals for influenza: historical perspectives and lessons learned. Antivir. Res. 71:372-378. [DOI] [PubMed] [Google Scholar]
- 24.He, G., J. Qiao, C. Dong, C. He, L. Zhao, and Y. Tian. 2008. Amantadine-resistance among H5N1 avian influenza viruses isolated in Northern China. Antivir. Res. 77:72-76. [DOI] [PubMed] [Google Scholar]
- 25.Hurt, A. C., P. Selleck, N. Komadina, R. Shaw, L. Brown, and I. G. Barr. 2007. Susceptibility of highly pathogenic A(H5N1) avian influenza viruses to the neuraminidase inhibitors and adamantanes. Antivir. Res. 73:228-231. [DOI] [PubMed] [Google Scholar]
- 26.Kiso, M., K. Mitamura, Y. Sakai-Tagawa, K. Shiraishi, C. Kawakami, K. Kimura, F. G. Hayden, N. Sugaya, and Y. Kawaoka. 2004. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364:759-765. [DOI] [PubMed] [Google Scholar]
- 27.Lackenby, A., J. Democratis, M. M. Siqueira, and M. C. Zambon. 2008a. Rapid quantitation of neuraminidase inhibitor drug resistance in influenza virus quasispecies. Antivir. Ther. 13:809-820. [PubMed] [Google Scholar]
- 28.Lackenby, A., O. Hungnes, S. G. Dudman, A. Meijer, W. J. Paget, A. J. Hay, and M. C. Zambon. 31 January 2008. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Eurosurveillance 13(5):pii=8026. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=8026. [DOI] [PubMed] [Google Scholar]
- 29.Le, Q. M., M. Kiso, K. Someya, Y. T. Sakai, T. H. Nguyen, K. H. Nguyen, N. D. Pham, H. H. Ngyen, S. Yamada, Y. Muramoto, T. Horimoto, A. Takada, H. Goto, T. Suzuki, Y. Suzuki, and Y. Kawaoka. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437:1108. [DOI] [PubMed] [Google Scholar]
- 30.Luo, M. 2006. Structural biology: antiviral drugs fit for a purpose. Nature 443:37-38. [DOI] [PubMed] [Google Scholar]
- 31.Macken, C., H. Lu, J. Goodman, and L. Boykin. 2001. The value of a database in surveillance and vaccine selection, p. 103-106. In A. D. M. E. Osterhaus, N. Cox, and A. W. Hampson (ed.), Options for the control of influenza, vol. IV. Elsevier Science, Amsterdam, The Netherlands. [Google Scholar]
- 32.McKimm-Breschkin, J. L., P. W. Selleck, T. B. Usman, and M. A. Johnson. 2007. Reduced sensitivity of influenza A (H5N1) to oseltamivir. Emerg. Infect. Dis. 13:1354-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monto, A. S., J. Rotthoff, E. Teich, M. L. Herlocher, R. Truscon, H. L. Yen, S. Elias, and S. E. Ohmit. 2004. Detection and control of influenza outbreaks in well-vaccinated nursing home populations. Clin. Infect. Dis. 39:459-464. [DOI] [PubMed] [Google Scholar]
- 34.Moscona, A. 2005. Neuraminidase inhibitors for influenza. N. Engl. J. Med. 353:1363-1373. [DOI] [PubMed] [Google Scholar]
- 35.Mungall, B. A., X. Xu, and A. Klimov. 2003. Assaying susceptibility of avian and other influenza A viruses to zanamivir: comparison of fluorescent and chemiluminescent neuraminidase assays. Avian Dis. 47:1141-1144. [DOI] [PubMed] [Google Scholar]
- 36.Mungall, B. A., X. Xu, and A. Klimov. 2004. Surveillance of influenza isolates for susceptibility to neuraminidase inhibitors during the 2000-2002 influenza seasons. Virus Res. 103:195-197. [DOI] [PubMed] [Google Scholar]
- 37.Rameix-Welti, M. A., F. Agou, P. Buchy, S. Mardy, J. T. Aubin, M. Véron, S. van der Werf, and N. Naffakh. 2006. Natural variation can significantly alter the sensitivity of influenza A (H5N1) viruses to oseltamivir. Antimicrob. Agents Chemother. 50:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell, R. J., L. F. Haire, D. J. Stevens, P. J. Collins, Y. P. Lin, G. M. Blackburn, A. J. Hay, S. J. Gamblin, and J. J. Skehel. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45-49. [DOI] [PubMed] [Google Scholar]
- 39.Saito, R., D. Li, and H. Suzuki. 2007. Amantadine-resistant influenza A (H3N2) virus in Japan, 2005-2006. N. Engl. J. Med. 356:312-313. [DOI] [PubMed] [Google Scholar]
- 40.Saito, R., D. Li, Y. Suzuki, I. Sato, H. Masaki, H. Nishimura, T. Kawashima, Y. Shirahige, C. Shimomura, N. Asoh, S. Degawa, H. Ishikawa, M. Sato, Y. Shobugawa, and H. Suzuki. 2007. High prevalence of amantadine-resistance influenza A (H3N2) in six prefectures, Japan, in the 2005-2006 season. J. Med. Virol. 79:1569-1576. [DOI] [PubMed] [Google Scholar]
- 41.Sheu, T., V. Deyde, M. Okomo-Adhiambo, R. Garten, X. Xu, R. A. Bright, T. R. Wallis, N. Cox, A. Klimov, and L. V. Gubareva. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob. Agents Chemother. 52:3284-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subbarao, K., A. Klimov, J. Katz, H. Regnery, W. Lim, H. Hall, M. Perdue, D. Swayne, C. Bender, J. Huang, M. Hemphill, T. Rowe, M. Shaw, X. Xu, K. Fukuda, and N. Cox. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 43.Tisdale, M. 2000. Monitoring of viral susceptibility: new challenges with the development of influenza NA inhibitors. Rev. Med. Virol. 10:45-55. [DOI] [PubMed] [Google Scholar]
- 44.Ward, P., I. Small, J. Smith, P. Suter, and R. Dutkowski. 2005. Oseltamivir (Tamiflu) and its potential for use in the event of an influenza pandemic. J. Antimicrob. Chemother. 55(Suppl. 1):i5-i21. [DOI] [PubMed] [Google Scholar]
- 45.Wetherall, N. T., T. Trivedi, J. Zeller, C. Hodges-Savola, J. L. Kimm-Breschkin, M. Zambon, and F. G. Hayden. 2003. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: report of the neuraminidase inhibitor susceptibility network. J. Clin. Microbiol. 41:742-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.World Health Organization. 2009. Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO. http://www.who.int/csr/disease/avian_influenza/country/cases_table_2009_01_19/en/index.html.
- 47.Xu, X., X. Zhu, R. A. Dwek, J. Stevens, and I. A. Wilson. 2008. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J. Virol. 82:10493-10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yen, H. L., N. A. Ilyushina, R. Salomon, E. Hoffmann, R. G. Webster, and E. A. Govorkova. 2007. Neuraminidase inhibitor-resistant recombinant A/Vietnam/1203/04 (H5N1) influenza viruses retain their replication efficiency and pathogenicity in vitro and in vivo. J. Virol. 81:12418-12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Q., J. Yang, K. Liang, L. Feng, S. Li, J. Wan, X. Xu, G. Yang, D. Liu, and S. Yang. 2008. Binding interaction analysis of the active site and its inhibitors for neuraminidase (N1 subtype) of human influenza virus by the integration of molecular docking, FMO calculation and 3D-QSAR CoMFA modeling. J. Chem. Inf. Model. 48:1802-1812. [DOI] [PubMed] [Google Scholar]