Abstract
Revolver discovered in the Triticeae plant is a novel class of transposon-like gene and a major component of the large cereal genome. An 89 bp segment of Revolver that is enriched in the genome of rye was isolated by deleting the DNA sequences common to rye and wheat. The entire structure of Revolver was determined by using rye genomic clones, which were screened by the 89 bp probe. Revolver consists of 2929—3041 bp with an inverted repeated sequence on each end and is dispersed through all seven chromosomes of the rye genome. Revolver is transcriptionally active, and the isolated full-length cDNA (726 bp) reveals that Revolver harbors a single gene consisting of three exons (342, 88, and 296 bp) and two introns (750 and 1237 bp), and encodes 139 amino acid residues of protein, which shows similarity to some transcriptional regulators. Revolver variants ranging from 2665 to 4269 bp, in which 5′ regions were destructed, indicate structural diversities around the first exon. Revolver does not share identity with any known class I or class II autonomous transposable elements of any living species. DNA blot analysis of Triticeae plants shows that Revolver has existed since the diploid progenitor of wheat, and has been amplified or lost in several species during the evolution of the Triticeae.
Key words: transposon-like gene, transcript, genomic component, structural and quantitative divergence
1. Introduction
In higher eukaryotes, genes required for cellular function can comprise as little as 20% of the genome1,2 and occur in islands separated by repetitive DNA sequences,3,4 which comprise >70% of the genomes and are often referred to as junk DNA. With the advent of large-scale DNA sequencing, it has become apparent that transposable elements constitute a large proportion of the repetitive DNA component of most eukaryote genomes, e.g. at least 45% of the human genome5 and 50–80% of some grass genomes.6,7
Transposable elements are divided into two major superfamilies, class I and class II, according to their mode of transposition.8 The class I elements can replicate by transcription of genomic copies followed by reverse transcription of an RNA intermediate and integration of the cDNA copy back into the genome.9 A representative class I element is the long-terminal repeat (LTR) retrotransposon, which has both structure and life-cycle similar to those of the retroviruses.2,10,11 LTR retrotransposons are ancient and ubiquitous; they are major components of plant genomes12–16 and are believed to be major contributors to plant genome evolution, as their replication strategy offers the potential for an explosive increase in copy number and for causing insertional genomic change.17,18
A class II transposable element is mobilized by excision from one chromosomal location and reintegration elsewhere in the genome. The class II transposable elements are distinguished by terminal inverted repeats (TIRs) and are divided into three superfamilies—hAT, CACTA and MULU—on the basis of the homology of TIRs and transposase genes carrying out the cut-and-paste process. Transposons are widespread in plants,19,20 but quite low in copy numbers, ranging from tens to hundreds. However, an analysis of the growing database of genomic DNA sequences reveal that plant genomes harbor up to 30000 copies of miniature inverted-repeat terminal elements (MITEs) belonging to class II elements.21 MITEs have made important contributions to plant genome organization as well as LTR retrotransposons.22,23
Widely distributed transposable elements are the most rapidly evolving fraction of the eukaryotic genome,24 because the methylated and heterochromatic state of most highly repetitive elements suffers from sequence change more readily than do genes.25,26 In general, the genomes of higher eukaryotes contain thousands, even millions, of seemingly inactive transposable elements, which have been suggested as a source of interspecific sequence divergence. Species-specific repetitive elements serve as genetic tools for developing DNA markers and polymerase chain reaction (PCR) entry points dispersed throughout the genome.27 As well as the development of molecular markers, active transposable elements, have been significant tools of reverse genetics for functional genomics. Their incorporation into the host genome enables insertional gene mutagenesis and gene tagging. In rice, reverse flow by the RNA of LTR retrotransposon Tos 17 yields mutant lines.28 Ac (hAT) and Mutator (MULU) have been widely used for gene cloning.29
Despite the vast DNA database that exists for higher eukaryotes, most genomic components are invisible and remain to be annotated, except for homologues of known class I and class II elements. We have sought novel active genomic components that might be useful as molecular tools. Among the Triticeae tribe, rye (Secale cereale) has been an important gene source, offering stress resistance useful for wheat (Triticum aestivum L.) and triticale breeding. The rye genome has a 1C DNA content of 3.9 Gb, the highest among the Triticeae, and repetitive sequences comprise 92% of the genome.30 Cot analysis estimates 24% of the rye genome to be rye-specific.1 Rye-specific repetitive sequences have been useful molecular probes for the determination of introgressed genomes and the genomic constitution of wheat–rye hybrids.31,32 However, active transposable elements have not been found in rye, wheat, or wheat relatives. One of the reasons may be that repetitive sequences were isolated mainly from relic DNA that has not suffered from methylation-sensitive restriction enzymes.33–35
In this study, in order to obtain genome-specific elements regardless of the methylation state, we have used genomic deletion of sequences common to wheat and rye. Here, we describe a new class of transposon-like gene, called Revolver, consisting of 2929—3041 bp with TIRs and harboring a transcriptionally active single gene. Revolver exhibits structural diversities and considerable copy number variation through the evolution of the Triticeae, from 2 × 104 in rye to almost rare in wheat.
2. Materials and methods
2.1. Plant materials
The plant materials used in this study included: the rye-inbred pure line IR130 developed at Tottori University (S. cereale L.; 2n = 2x = 14, RR); the wheat variety Chinese Spring (CS) (T. aestivum L.; 2n = 6x = 42); the rye IR130 chromosome 6R addition wheat CS line (6R add. wheat; 2n = 44).
2.2. Genomic deletion
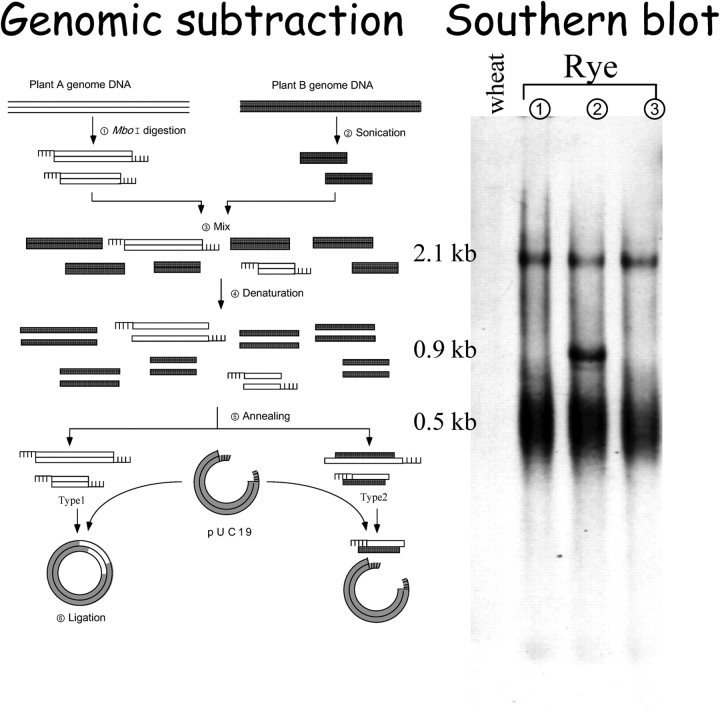
The deletion enrichment scheme36 was used to recover rye-specific DNA sequences from the genome of 6R add. wheat. Genomic DNA was extracted from fresh leaves by the cetyl trimethylammonium bromide (CTAB) method. The genomic DNA of 6R add. wheat was digested completely with the restriction enzyme MboI, while the wheat DNA was sheared randomly to 0.5 kb on average with a sonicator. Then, 13.4 µg of digested 6R add. wheat DNA was mixed with 67.0 µg of the sonicated wheat DNA and denatured by boiling for 10 min and then allowed to anneal at random in 4 mL of PERT reaction mixture (8% phenol, 1.25 M NaClO4, 0.12 M Na2HPO4) in a revolving flask for 72 h at room temperature. The objective was to recover rye-specific repetitive sequences that will anneal by themselves with cohesive ends adjusted to the vector's ends (Fig. 1). The genomic DNA of rye IR130 was digested completely, whereas that of wheat var. CS was sheared randomly with a sonicator. The MboI-digested rye DNA was mixed with the sonicated wheat DNA, denatured by boiling, and then allowed to anneal randomly. Rye-specific sequences that anneal by themselves with cohesive ends of cloning vectors were recovered. After extraction with phenol, the DNA solution was used to ligate with pUC119 linearized by BamHI, followed by transformation of Escherichia coli JM109.
Figure 1.
The scheme for cloning genome-specific elements by subtracting sequences common to wheat and rye. Genomic subtraction recovered a repetitive sequence, which is enriched in rye, but absent from wheat. Southern blot analysis of the EcoO109I digests confirmed that the rye 89 bp fragment (AB304276) hybridized well to the genome of rye ( Secale cereale,
Secale cereale,  S.vavillovi and
S.vavillovi and  S.montanum), but did not hybridize to the wheat genomic DNA. The full structure of the element was determined by sequencing positive genomic clones screened by the initial rye-specific probe.
S.montanum), but did not hybridize to the wheat genomic DNA. The full structure of the element was determined by sequencing positive genomic clones screened by the initial rye-specific probe.
A total of 222 recombinant plasmids were isolated and 1 µg of each plasmid DNA was spotted onto two sheets of nylon membranes, which were hybridized for 3 h in 5× SSC, 2% blocking reagent (Roche), 0.1% N-lauroylsarcosine, 0.02% SDS, and 1 µg/mL sheared pUC119 DNA. One membrane was probed with 5 ng of rye IR130 genomic DNA and the other was probed with 10 ng of wheat CS DNA, each labeled with digoxigenin-11-dUTP, for 14 h at 68°C, washed twice in 2× SSC, 0.1% SDS at room temperature for 5 min, and twice in 0.1× SSC, 0.1% SDS at 68°C for 15 min. The hybridization signals were detected on X-ray films using as the chemiluminescent substrate a 25 mM solution of disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo [3.3.1.13,7] decan}-4-yl) phenyl phosphate (CSPD, Boehringer Mannheim Biochemica). From the differential blotting, 14 plasmid clones that hybridized strongly to rye DNA, but not wheat DNA, were screened. In this study, a single 89 bp sequence was used for further characterization.
2.3. Phage library screening and DNA sequencing
In order to determine the full structure of the element harboring the 89 bp segment, approximately 5 × 104 plaques of a lambda FIX II library constructed from rye genomic DNA were screened with the cloned 89 bp fragment labeled with digoxigenin-dUTP. Positive plaques were detected with anti-dig-AP and CDP-star (Roche). Positive six lambda clones were chosen at random and restriction sites of BamHI and SacI were mapped on the six insertions. A 21.6 kb insertion of the three lambda clones carrying the 89 bp segment was subcloned into pBluescript and fully sequenced on both strands using the ALFexpress sequencer (Pharmacia Biotech). Overlapping fragments were assembled with the ALFexpress assembler software. Completed sequences were compared with the non-redundant GenBank+ EMBL + DDBJ database using BLASTN and BLASTX homology search software.
2.4. RNA gel blot analysis and cDNA library construction
Total RNA was extracted from young leaves of rye according to a modified guanidine isothiocyanate procedure and then treated with DNase I. Twenty micrograms of total RNA was electrophoresed in a 1.1% agarose/18% formamide gel and transferred to a nylon membrane. The membranes were prehybridized at 65°C for 4 h in hybridization solution containing 5 × Denhardt's, 10% dextran sulfate, 6× SSC and 0.2% SDS. A Revolver 1.5 kb probe (25 ng) excised from a genomic clone of lambda-1 and labeled with (α)32P-dCTP (Amersham) was added to the pre-hybridization solution and allowed to hybridize at 65°C for 14 h. Blots were washed in 2× SSC, 0.1% SDS at 65°C for 1 min and three times in 2× SSC, 0.1% SDS at 65°C for 20 min, then exposed to Fuji RX-U film. Polyadenylated mRNA was prepared from the rye leaf total RNA by affinity chromatography using an oligo(dT) column, and the first and second cDNA strands were synthesized for cloning into bacteriophage lambda ZAPII.
2.5. Reverse transcriptase–polymerase chain reaction (RT–PCR)
Plaque lifts from a rye leaf cDNA library constructed in lambda ZAPII were screened by a Revolver 1.5 kb probe excised from a genomic clone of lambda-1. The nucleotide sequence of a positive cDNA clone (726 bp) was determined by using its phagemid DNA as a cycle sequencing template. Total RNA for Reverse transcriptase–polymerase chain reaction (RT–PCR) extracted from seedlings was treated with DNase I. Single-strand cDNAs were synthesized by Avian Myeloblastosis virus (AMV) reverse transcriptase (Life Science) using an oligo(dT) primer. The primers for amplification of Revolver cDNA were designed from both ends of a positive cDNA clone. Reaction mixtures contained 10 ng of template cDNA, 50 pmol of each primer (5′-GGCACGAGGGTACGAGTCCGAG-3′, 5′-GGCACAACTCATGTAAAAGAGGG-3′), 0.4 mM dNTPs, 1 × LA PCR buffer II, 2.5 mM MgCl2, and 0.5 U of LA Taq polymerase (Takara) in a volume of 25 µl. The PCR reaction program consisted of 30 cycles of 30 s at 95°C, 30 s at 63°C, 1 min at 72°C. The RT–PCR products were purified, ligated to the pGEM-T vector (Promega), and sequenced.
CapFishingTM technology (Seegene), which employs the CapFishingTM adapter and the oligo(dT) primer, was used to obtain a full-length cDNA of Revolver. The single-strand cDNAs were synthesized from CapFishingTM adapter-added total RNA by SuperScript III reverse transcriptase (Invitrogen) using an oligo(dT) primer extended with 23 bp of 3′ Revolver sequence (5′-TTTTTTTTTTTTTTGGCACAACTCATGTAAAAGAGGG-3′). Because the CapFishingTM adapter (patent pending) binds to the CAP structure of the full-length mRNA, the 3′ end of the first-strand full-length cDNA was extended using the CapFishingTM adapter as template. Then, the PCR reaction for amplifying the Revolver-related full-length cDNA was carried out using the 5′ CapFishingTM adapter primer (5′-GTCTACCAGGCATTCGCTTCAT-3′) and the 3′ Revolver primer (5′-GGCACAACTCATGTAAAAGAGGG-3′) in a 30 µl mixture including 5 µl of the single-strand cDNA solution, 0.3 µM of each primer, 0.4 mM dNTPs, 1 × LA PCR buffer II, 2.5 mM MgCl2 and 0.5 U of LA Taq polymerase (Takara). The cycling profile was: 35 cycles at 94°C for 30 s, 60°C for 30 s, 72°C for 2 min. The double-strand cDNA products were TA-cloned and sequenced.
2.6. PCR
The primers for amplification of Revolver genomic DNA were designed from both ends of Revolver-2 in lambda-2. Reaction mixtures contained 10 ng of template genomic DNA, 50 pmol of each primer (5′-GCCTTTCGGCCTTCCTCTCAGGCGG-3′, 5′-GTAGTCGTCAGGAGTCCTCACCA-3′), 0.4 mM dNTPs, 1 × LA PCR buffer II, 2.5 mM MgCl2 and 0.5 U of LA Taq polymerase (Takara) in a volume of 50 µl. The PCR reaction program consisted of 30 cycles of 30 s at 95°C, 1 min at 70°C, 3 min at 72°C. The PCR products were purified, ligated to the pGEM-T vector (Promega), and sequenced.
2.7. DNA gel blot analysis
Genomic DNA (10 µg) of Triticum aestivum (AABBDD), Triticum dicoccum (AABB), Triticum monococcum (AA), Aegilops speltoides (SS), Aegilops tauschii (DD), Dasypyrum villosum (VV), Secale cereale, Secale vavillovi, Secale silvestre, Secale montanum, Secale fragile (RR) and X Triticosecale (AABBRR) were digested to completion with restriction enzymes, subjected to electrophoresis in a 1% agarose gel, then transferred, and crosslinked to nylon membranes. The membranes were pre-hybridized at 65°C for 3 h in hybridization solution containing 5× SSC, 2% blocking reagent (Roche), 0.1% N-lauroylsarcosine and 0.02% SDS. A Revolver cDNA probe (pSc5, 693 bp, 25 ng) labeled with digoxigenin-11-dUTP was added to the pre-hybridization solution and allowed to hybridize at 65°C for 14 h. Blots were washed twice in 2× SSC, 0.1% SDS at room temperature for 5 min and twice in 0.1× SSC, 0.1% SDS at 65°C for 15 min, then exposed to Kodak X-OMAT film using CDP-star.
2.8. Slot blot analysis
For slot blot analysis, genomic DNA from wheat and wheat relatives was blotted onto hybond N+ membranes (Amersham). The non-radioactive chemiluminescence method (Gene Images, Amersham) was used for probe labeling, hybridization, and detection of hybridization sites. The Revolver cDNA sequences (pSc5, 693 bp) were used as a probe. The membranes were hybridized at 60°C for 30 min in hybridization buffer and then hybridized with labeled probes at 60°C overnight. The membranes were washed in 1× SSC, 0.1% SDS at 60°C for 15 min, then in 0.5× SSC, 0.1% SDS for 15 min, followed by incubation for 60 min at room temperature in 10% (W/V) blocking reagent in antibody wash buffer. Then the membrane was incubated in the presence of anti-fluorescein antibody-alkaline phosphatase (AP) conjugate. Unbound conjugate was removed by three washes in 0.3% (V/V) Tween 20 in antibody wash buffer at room temperature. Hybridization sites were detected using the CDP-star detection reagent. Decomposition of the stabilized dioxetane was catalyzed by probe-bound AP, and the light emitter was exposed on X-ray film and then recorded by a fluoro-image analyzer (FUJIFILM FLA-2000). The copy number of the Revolver probe was determined by quantitative slot blot hybridization to a series of measured amounts of Revolver cDNA.
2.9. DNA:DNA in situ hybridization
Rye chromosomes were prepared from root tip meristems of germinating seeds, which were cut off and placed into distilled water at 0°C for 24 h. After fixation in ethanol:glacial acetic acid (3:1, V/V) for 24 h and three washes with water, the root tips were digested for 30 min in an enzyme solution consisting of 1% pectolyase and 2% cellulase Onozuka RS in 75 mM KCl, 7.5 mM EDTA at pH 4.5. The root tips were washed with water and squashed in 45% acetic acid on glass microscope slides. After removal of the coverslip, the slides were frozen on dry ice, treated with RNase A, dehydrated through an ethanol series, and dried. Revolver cDNA was labeled with biotin-16-dUTP by PCR. The subtelomeric repetitive DNA probe was labeled with digoxigenin-11-dUTP (Boehringer Mannheim). Fluorescence in situ hybridization (FISH) and signal detection were performed as described by Heslop-Harrison et al.37
A 15 µM portion of the hybridization mixture containing 50% (V/V) formamide in 2× SSC, 1 µg/mL probe DNA, and 100 µg/mL salmon sperm DNA was covered with a 22 × 30 mm2 coverslip, sealed by a rubber solution, and put on a thermal cycler (PHC-3 Techne) programmed for denaturing DNA at 82°C for 8 min; and in situ hybridization at 37°C for 18 h. The preparations were then washed four times at 40°C for 10 min each time, three times in 2× SSC and once in 4× SSC, then blocked with 5% (W/V) BSA in 0.05% (V/V) Tween 20, 0.1 M NaHCO3 at 37°C for 5 min. The probes were detected with 10% (V/V) avidin-FITC and 10% (V/V) anti-dig-rhodamine (Boehringer Mannheim) in a humid chamber at 38°C for 1 h. After three washes in 0.05% (V/V) Tween 20, 0.1 M NaHCO3 and one wash in 2× SSC at 40°C (10 min each wash), the preparations were counter-stained with a mixture of 3 µg/mL DAPI (Boehringer Mannheim) and 10% (V/V) antifade (Oncor) in glycerol. The preparations were observed with a light microscope (OLYMPUS BX4), and the fluorochrome signals were visualized.
3. Results and discussion
3.1. Structure of dispersed transposon-like element Revolver recovered by deletion
A rye genome-specific DNA library was constructed by the deletion of sequences common to wheat and rye (Fig. 1). From this library, 222 plasmid DNAs were screened by differential dot hybridization, and 14 clones hybridized strongly to rye DNA, but did not hybridize to wheat DNA. Among them, seven clones belong to the tandem 350 bp family33,34 and one clone belongs to the dispersed R173 family,38,39 both of which are rye-specific repetitive sequences. On the other hand, a single clone contained an 89 bp unknown sequence (AB304276). These results indicate that deletion of sequences common to wheat and rye was effective, and the 89 bp sequence was used for further characterization.
Southern blot analysis confirmed that the rye 89 bp fragment hybridized well to the genome of S. cereale, S. vavillovi, and S. montanum, but did not hybridize to the wheat genomic DNA (Fig. 1). Therefore, the 89 bp segment is derived from a repetitive sequence family dispersed specifically in the R genome of the genus Secale. A strongly hybridized 1.5 kb band was observed on the SacI digest(data not shown), indicating that the repetitive sequences commonly contain a 1.5 kb sequence, and that the full structure is more than 1.5 kb long. In order to determine the full sequence of the repetitive element harboring the 89 bp fragment, a rye genomic library constructed in lambda FIX (Stratagene) was screened by the 89 bp probe. The plaque hybridization analysis found approximately 800 positive plaques for the membranes of 5 × 104 plaques of the rye genomic DNA library. Six positive lambda clones were chosen at random, and the restriction sites of BamHI and SacI were mapped differently on the six insertions, which are, therefore, derived from different areas of the rye genome.
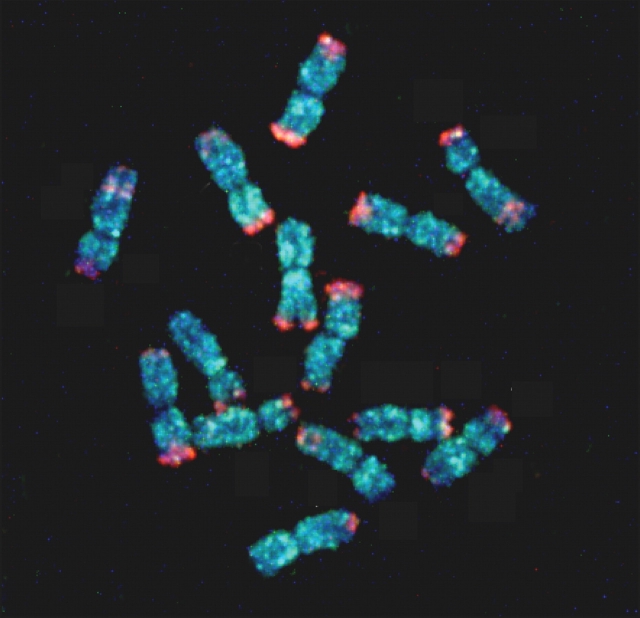
Three lambda clones were sequenced in full. Two lambda clones, which are 92% identical, included the full structure of the repetitive element harboring the 89 bp segment, the one in lambda-1 is 3041 bp long and the one in lambda-2 is 2929 bp long (in accession nos. AB124639–124640). The regions flanking the 3 kb elements do not show any homology between the two lambda clones. The consensus sequence contains 20 bp of incomplete TIRs on both ends (5′-TGTgAcGCCCgaGAccGACg-3′, 5′-TGTaAtGCCCagGAtggGAC-3′) and subterminal short repeat sequences on the 5′ end (5′-TCCAGAAGAT-3′). The transposable element includes TIRs and often also sequence motifs in the subterminal regions. The 3 kb elements are insertions flanked by 2 bp repeats, TC in lambda-1, and AG in lambda-2, which may be terminal sequence duplications. As shown in Fig. 2, a FISH experiment probed by the 3041 bp sequence in lambda-1 showed that the 3 kb insertion is dispersed on the seven chromosomes of rye. These findings indicate that a transposon-like 3 kb element is dispersed throughout the rye genome.
Figure 2.
The transposon-like gene Revolver is dispersed on all seven chromosomes of rye. A FISH experiment was conducted using dual probes of Revolver labeled by biotin-16-dUTP and the subtelomeric 350 bp family labeled by digoxigenin-11-dUTP. Each probe was visualized by avidin-FITC (blue color) and anti-dig-rhodamine (red color), respectively.
Despite extensive characterization of the repetitive elements in the rye genome, the 3 kb element does not show similarity to any known rye repetitive element: the 350 bp family,33,34 the 120 bp family,33,40 the 5.3H3 family,35 the R173 family,38,39 or pSc250.41 The entire structure of the 3 kb transposon-like element does not share whole identity with class I or class II transposable elements. We have named the novel transposon-like element Revolver.
3.2. Identification of Revolver mRNA
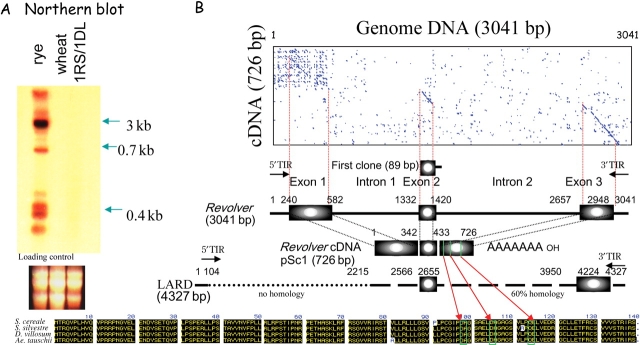
Northern analysis showed that Revolver is expressed extensively in rye, but there is no transcript in wheat (Fig. 3A). Weak transcripts are observed in a rye-chromosome-translocated wheat line. A Revolver cDNA clone was identified from a leaf cDNA library by the 3 kb Revolver probe. A positive rye cDNA, entirely 80% identical, is 726 bp long (AB124665). By using cap site-specific cloning method, a single size of cDNA (0.7 kb) was amplified. The obtained full-length cDNA of Revolver, 728 bp long (AB124666), shows 94% identity with the positive cDNA. Nucleotide sequence comparison between the 3 kb Revolver genomic DNAs (Revolver-1 in lambda-1 and Revolver-2 in lambda-2) and the cDNAs revealed that the Revolver element consists of three exons of 342, 88, and 292 bp, and two introns of 750 and 1237 bp (Fig. 3B). Splice acceptor and donor sites certainly exist in the exon–intron junctions. A putative TATA box is located at base 221, with a cap site at base 261 and a possible polyadenylation signal AATAAA at base 2918. Therefore, Revolver harbors a single gene consisting of three exons and the initial 89 bp clone obtained by genomic subtraction is located around the second exon (Fig. 3B).
Figure 3.
Identification of Revolver mRNA. (A) Multiple sizes of transcripts homologous to Revolver were found in rye but no transcript was found in wheat. (B) A full-length Revolver cDNA clone (AB124665–124666), 726 bp long, was identified. Nucleotide sequence comparison between Revolver and the cDNA revealed that Revolver includes a single gene consisting of three exons and two introns. The predicted 139 amino acid sequence is highly conserved in Revolver cDNAs (AB124645, AB124666, AB304271–304275), which are recovered from S. cereale, S. silvestre, D. villosum, and Ae. tauschii, indicating activity in all of these species. The ORF shows similarity to some DNA-binding proteins. The Revolver ORF includes a DDE motif. The initial 89 bp clone (AB304276) obtained by genomic subtraction corresponds to the second exon. Revolver (AB124639, AB124640) shows 60% homology to parts of both ends (5′ side 104 bp, 3′ side 2213 bp) of LARD (4327 bp), however, in which the region extending from exon 1 to the half of intron 1 of Revolver is replaced by a different sequence and the coding region is not present.
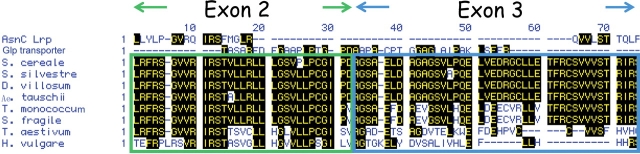
Revolver cDNA was recovered from S. silvestre, D. villosum, T. monococcum, and Ae. tauschii after isolation by RT–PCR using 22 mer primers of both ends. These Revolver cDNAs (AB124645, AB124666, AB304271–AB304275) contain a single open reading frame (ORF) encoding 139 amino acid residues of protein (Fig. 3). The ORF does not show homology to known transposases; however, the predicted Revolver product shows similarity to a transcriptional regulator family of AsnC/Lrp or a glycerol-3-phosphate transporter and includes a DDE motif (Fig. 3 and 4). Because of the presence of terminal inverted repeats and encoding a DNA-binding-like protein, Revolver is considered to be a class II transposable element. The inverted repeated sequences determining the family identity of a transposon and the encoding gene are unique sequences that are quite different from known class II transposons. Moreover, the entire structure of Revolver does not share whole identity with either class I or class II autonomous transposable elements.
Figure 4.
Revolver cDNAs isolated by RT–PCR from S. cereale, S. silvestre, S. fragile, D. villosum, Ae. tauschii, and T. monococcum (AB124645, AB304271–304275). Wheat and barley ESTs showing partial homology to Revolver were found in the Triticeae EST database. The predicted Revolver ORF shows similarity to some transcriptional regulators with DNA-binding ability. The ORF is highly conserved among the top four species, indicating that Revolver is active in these species. In wheat and barley, however, only partial sequences encoded in exon 2 are retained.
The Ty1-copia group retrotransposon is a major component of plant genomes.12,27 BARE-1, one of the Ty1-copia retrotransposons,42,43 is dispersed and transcribed in the genome of barley Hordeum vulgare.44,45 Revolver is quite different from retrotransposons, although it shows partial homology (60%) to both end regions (5′ end 123 bp, 3′ end 777–2112 bp) of LARD LTR (3130–4960 bp), one of which is found as a nested insertion in the 3′ LTR of BARE-142 and is regarded as solo LTRs of the non-autonomous retorotransposon elements named LARD (large retrotransposon derivative) in barley.46 The two sequences of 5 kb were found to be similar to this insertion in a 66 kb stretch of the barley genome and were described as an LTR of LARD.46,47 The LARD LTRs include the region homologous to both 5′ and 3′ ends of the Revolver element (Fig. 3B). At the 5′ terminus, LARD LTRs have the region of 123–149 bp homologous to upstream of the transcription initiation site of Revolver. At the 3′ terminus, LARD LTR-1 (4960 bp) has a region of 2112 bp showing 60% homology to Revolver, from the middle of the first intron to the 3′ terminus of Revolver. Another LARD LTR nested in BARE-1 has a region of 777 bp showing 60% homology to the middle of the second intron to downstream of the third exon. Both 3′ termini coincide with the 3′ terminus of the untranslated region downstream of the third exon of Revolver. However, no homology to Revolver was observed in 631–2176 bp of LARD LTR-1 or 598–2353 bp of LARD LTR nested in BARE-1. As to the region of about 2 kb between these end regions, LARD LTRs lack the region from the first exon to the middle of the first intron. Moreover, the central regions of LARD LTRs are highly variable, except for the 5′ and 3′ ends,46 but Revolver is entirely conserved. LARD LTRs have sequences not present in Revolver; instead by the first exons of Revolver and result in non-coding sequences. Revolver and LARD LTR may be evolutionally related, but LARD LTR is a structural part of an LTR retrotransposon, and on the contrary, Revolver is a single gene consisting of the exon–intron structure. Gene-encoding LTR has never been found. Therefore, Revolver is quite distinguished from LARD LTRs, in which the exons 1 and 2 of Revolver are replaced by different sequences and the coding region is not present, and whose autonomous element has never been reported. The presence of a Revolver-like element in barley suggests a wide prevalence of Revolver among the Triticeae.
The novel high-copy element Revolver is transcriptionally active in rye. Some of transposon-like elements exist in high copy numbers in the genomes of most eukaryotes, but the great majority of them are inactive, and only a small portion of them retain the ability to transpose.48,49 Very few transposons have been shown to be transcriptionally active. A copia-like retroelement BARE-1 dispersed in the barley genome43 is transcribed in somatic tissues.44 Some LTR retrotransposons, such as tobacco Tnt1, Tto1, and OARE-1 that are largely inactive, can be transcriptionally activated under the conditions of biotic and abiotic stress, including wounding, oxidative stress, and pathogen infection.50,51 After stress-induced transcription, the rice LTR retrotransposon Tos17 increased the genomic copy number.52 In maize, a survey of more than 4 × 105 expressed sequence tag (EST) sequences identified only 56 retrotransposon cDNAs, supporting the notion that most retrotransposons are inactive.7 Furthermore, most of these sequences are derived from the low-to-middle repetitive LTR retrotransposons, and not from the very high copy number elements that have been responsible for doubling the size of the maize genome in the past 5–6 million years. In humans, only 30–60 L1 elements out of 5 × 105 comprising 45% of the genome are thought to be active.53 In contrast to the low-level activity of high-copy retrotransposons, highly repetitive Revolver is transcribed strongly and may retain mobility and mutagenetic potential.
A transcriptionally active Revolver gene is well conserved among the Triticeae members. The methylated and heterochromatic state of most transposons can cause them to change sequence more rapidly than the genes.25,26 For example, regulation at any stage of the replication cycle for retrotransposons (transcription, translation, reverse transcription, and integration of element cDNA) can limit the transposition. Furthermore, the paucity of maize retrotransposon-derived ESTs indicates that some epigenetic mechanisms might have been repressing the transcription of their large fraction in the genome. In contrast to these silenced retrotransposons, Revolver is transcriptionally active, and short RNA homologues causing RNAi silencing54–56 were not observed on Northern blots (Fig. 3A). Revolver has not suffered from epigenetical silencing systems and might has retained strong transcriptional activity during the long evolution of the Triticeae.
3.3. Structural diversities of Revolver in the rye genome
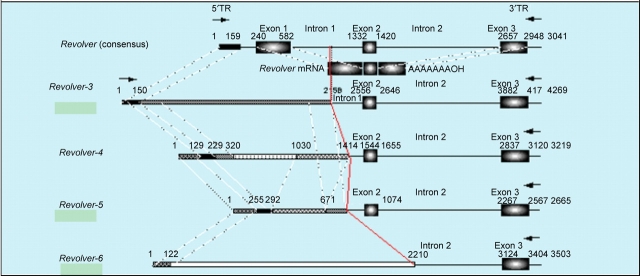
Polymerase chain reaction was performed using the primer derived from one clone of Revolver (Revolver-2). Several sizes of Revolver variants ranging from 2665 to 4269 bp were amplified from the rye genome, but nothing was amplified from the wheat genome. All of the lengthy variants may be non-autonomous elements of Revolver, because they have the downstream region of second intron of Revolver, but they have structural mutation occurred at the 5′-side, in which the coding regions, especially in the first exon, are considerably destructed (Fig. 5).
Figure 5.
Structural diversities of Revolver in the rye genome. Several sizes of Revolver variants ranging from 2665 to 4269 bp (Revolver-3–6, AB124641–12464) were obtained from the rye genome. All of the lengthy variants may be non-autonomous elements of Revolver, because they have the downstream region of second intron of Revolver, but they have structural mutation occurred at the 5′ side, in which the coding region especially in the first exon is considerably destructed. Among the Revolver family showing extremely structural diversities, Revolver-1 (3041 bp) in lambda-1 and Revolver-2 (2929 bp) in lambda-2, which indicates the highest coincidence with a full-length cDNA of 728 bp, should have basic structure of Revolver.
First, Revolver-3 (AB124641) comprises total length of 4269 bp (Fig. 5), and at the 3′ side, it has the region of 2093 bp from the middle of the first intron of Revolver through the third exon and reaching to the 3′ terminal. At the 5′ side, Revolver-3 has the homologous region of 150 bp including the inverted repeat sequence. However, as to the region of about 2 kb between these sequences, Revolver-3 lacks the region from the first exon to the middle of the first intron of Revolver. In the region from 631 to 2176 bp at the 5′ side of Revolver-3, there exist short repetitive sequences occurring repeatedly, but no homology was observed with Revolver. Referring to the homology with Revolver of each region, Revolver-3 exhibited homology from 77 to 93%.
Next, the total length of Revolver-4 (AB124642) consists of 3219 bp (Fig. 5), and at the 3′ side, it has the region of 1806 bp from the second exon to the 3′ terminal of Revolver. However, in the 1413 bp at the 5′ side, the region homologous to Revolver is limited to only 101 bp at the 5′ terminal. Further, Revolver-5 (AB124643) has a total length of 2665 bp (Fig. 5), and at the 3′ side, it has the region of 1826 bp from the second exon to the 3′ terminal of Revolver. At its 5′ side, the region homologous to Revolver is limited to only 37 bp at the terminal region, but the region of about 670 bp is homologous to Revolver-4. Finally, Revolver-6 (AB124644) has a total length of 3503 bp (Fig. 5), and at the 3′ side it has the region of 1294 bp from the middle of the second intron to the 3′ terminal of Revolver; however, at the 5′ side there is not a region homologous to Revolver, and 121 bp at the 5′ terminal is homologous to Revolver-4 and Revolver-5.
Revolver shows extremely structural diversities, especially around the first exon. Among them since Revolver-1 in lambda-1 and Revolver-2 in lambda-2 show the highest correspondence with a full-length cDNA of 728 bp, they should be basic structure of Revolver, despite the not so high homology of the first exon. Multiple transcripts longer than 0.7 kb RNA fragment (Fig. 3A) may be unspliced versions of several lengthy variants or a read-through transcript from an external promoter. The major 3 kb transcripts may be unspliced immature transcripts, which are caused by structural degeneration in 5′ region around the first exon. It suggests that such variants are prevalent than the normal spliced transcripts.
3.4. Amplification and elimination of Revolver in the Triticeae genome
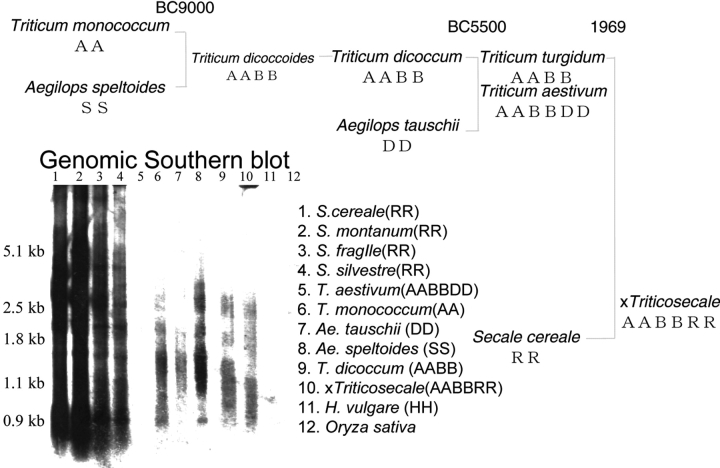
The distribution of Revolver in the Triticeae was analyzed by Southern blotting. As shown in Fig. 6, the Revolver cDNA probe hybridized strongly to gel blots of the wheat relatives Secale sp. (RR), D. villosum (VV), and moderately to wheat ancestral species T. monococcum (AA), Ae. speltoides (SS), T. dicoccum (AABB), and Ae. tauschii (DD). These cereals may share a large superfamily of Revolver stemming from their common progenitor. On the contrary, Revolver cDNA did not show hybridization to T. aestivum genomic DNA (Fig. 6). DNA slot blot analysis also shows that Revolver elements are abundant in the genomes of Secale sp. (RR) and D. villosum (VV) (over 104 copies), and Revolver is present with moderate repeats in the wheat ancestral species T. monococcum (AA), T. dicoccum (AABB) and Ae. tauschii (DD) (3 × 103 − 8 × 103 copies), but very rare in bread wheat (103 copies). While these facts indicate that Revolver has existed since the diploid progenitor of wheat and been amplified in rye and some other species, it has been lost from bread wheat through evolution accompanied by hexaploidization. Revolver exhibits quantitative variation during evolution of the Triticeae.
Figure 6.
Distribution of Revolver in plant genomes as revealed by genomic Southern blotting. Revolver cDNA hybridized strongly to Secale species and moderately to T. monococcum (AA), Ae. speltoides (SS), Ae. tauschii (DD), and T. dicoccum (AABB). In contrast, Revolver was not detected in the common wheat genome. These facts indicate that Revolver has existed since the diploid progenitor of wheat (3000–8000 copies), and it has been amplified in rye (20 000 copies), but it has been lost from bread wheat after the allopolyploidy event.
The rye haploid genome contains 3.9 Gb of DNA and is the largest among the Triticeae genomes. Highly repetitive Revolver was visualized over the entire length of rye chromosomes by a FISH experiment (Fig. 2). The copy number of Revolver was calculated by slot blot hybridization with a Revolver probe as 2 × 104 per rye genome, but only 103 copies in the wheat genome. Generally, angiosperm genomes vary tremendously in size; some species have less than 50 Mb of DNA per haploid genome and others have more than 85 000 Mb.57 The lack of correlation between the complexity of the organism and the size of the genome has long been recognized as a C value paradox.58 There is great variability in genome size among the grass family Poaceae: 450 Mb for rice, 2500 Mb for maize, 5000 Mb for barley, and 16 000 Mb for hexaploid wheat.59 Most of the variation in genome size is attributable mainly to the amount of repetitive DNA, which comprises 70% of cereal genomes.1,3 Cot analysis estimated that repetitive sequences comprise 92% of the rye genome,30 and 24% of them were differentiated in a rye-specific array.1 The considerable accumulation of Revolver may be a reason why the rye genome became so enlarged when compared to other members of the Triticeae. If Revolver is a mobile element, it is reasonable that as many as 2 × 104 copies of the family have been generated and spread throughout the rye genome since the evolutionary event that separated rye and wheat. The considerable variation of Revolver copy number among the wheat-related species indicates their propagation activity during evolution of the Triticeae tribe. The name of the novel transposon-like gene Revolver means a dynamic factor to construct genomes through evolution of the Triticeae.
Among the repetitive DNA, especially the LTR retrotransposons, the copy number has increased,60,61 and repetitive DNA comprises 60% or more of many large plant genomes, such as those of maize (Zea mays), wheat, and barley,6,7,45,62 which has contributed greatly to the expansion of the genome.5,17,63,64 Over a broad range of organisms, retrotransposon copy number appears to be correlated with genome size. The small genome of the yeast Saccharomyces cerevisiae (13 × 106 bp) contains 51 full-length retrotransposons.65 In the large genome of maize (2500 Mb), some retrotransposons have copy numbers exceeding 2 × 104 per haploid genome6,62,63,66 and all of the retrotransposons sequenced were shown to have inserted within the last six million years, leading to a doubling of the size of the maize genome.26 Revolver is quite different from LTR retrotransposons and features a DNA-type transposable element. The copy number of the DNA-type transposable element is generally less than those of the highly repetitive retrotransposons, but recent analysis of the growing genomic database revealed that plant genomes harbor a huge copy of DNA-type transposable elements termed MITEs.21,23 The plant genome could also be expanded by the class II element MITE.67,68 The DNA-type transposon-like gene Revolver is a novel member of a major genomic component, as well as LTR retrotransposons and MITEs, which might have contributed to plant genome construction and evolution.
Revolver is less frequent in the genome of hexaploid bread wheat (T. aestivum; 2n = 42, AABBDD), which is an allohexaploid carrying three different subgenomes, A, B, and D69 resulting from two independent hybridization events (Fig. 6). The first combined the A genome of the wild diploid wheat T. monococcum (AA) and the B genome, which has the greatest genetic similarity to the S genome of Ae. speltoides.70–72 This resulted in the tetraploid ancestor of modern Triticum species T. dicoccum and Ae. tauschii, the diploid donor of the D genome hybridized over 8000 years ago, resulting in hexaploid wheat.73 Revolver existed in the wheat ancestral species, T. monococcum (AA), T. dicoccum (AABB), and Ae. tauschii (DD) (3–8 × 103 copies). However, it is considerably less frequent in T. aestivum (103 copies). The polyploidy that arise either by duplication of a single genome or by the acquisition of a few genomes from related species (allopolyploidy) is a major force in the evolution of plants; 50–70% of angiosperms have experienced at least one episode of polyploidy in their history.74,75 The combination of A, B and, D genomes into a single nucleus may generate more incompatibility than harmony for the propagation of some genes.76–78 Revolver contains a single ORF encoding a 139 amino acid residue protein, which features a transcriptional regulator able to bind DNA, suggesting that the Revolver element can transpose in a cut-and-paste fashion, like class II elements. In wheat, no transcript of the Revolver gene is observed on the Northern blot (Fig. 3A) and no Revolver cDNA is recovered by RT–PCR. A EST homologue is highly degenerated from the obtained cDNAs of other wheat-related species (Fig. 4). Revolver may have been eliminated from the wheat genome because of losing the gene activity needed to reintegrate into the host wheat genome after that polyploidy induced incompatibility among the three genomes.
As well as a history of genome expansion, flowering plants appear to have undergone genome contraction.79–81 One possible mechanism that reduces genome size is unequal intrastrand recombination between the two tandem repeats in direct orientation on the same chromatid. The outcome of the event can yield the net deletion of one repeat and the sequences between the repeats. Unequal intrastrand recombination between adjacent LTR retrotransposons might lead to a decrease in the size of a genome, a phenomenon that is supported by the abundance of solo LTRs and the general absence of the LTR–internal-LTR structure.79 Deletions of retrotransposons are more common in insects.82 Highly saturated Revolver in the wheat polyploid genome can offer unequal intrastrand recombination, and recombination between adjacent Revolver elements might have reduced the copy number in the wheat genome. The considerably destructive variants of Revolver (Fig. 5) may be stemmed from unequal recombination.
Revolver is enriched in the genomes of wild wheat relatives but rare in the common wheat genome. A cultivated rye (S. cereale L.) has been used as a gene source for wheat and triticale breeding by interspecific chromosome introgression and rearrangement or by translocation or substitution. A representative achievement of this type of manipulation is the introduction of a stem rust resistance gene into wheat. The rye genome still has a gene resource potential for future improvement of wheat. Rye-specific repeated sequences have been useful as probes to determine alien chromatins and chromosome constitutions in wheat–rye cross-breeding.31,32 Revolver is an effective tool for developing molecular tags for transferring useful germplasm of wheat relatives, rye, and Dasypyrum into the wheat genome. Revolver offer FISH, Southern probe for genotyping, and dispersed PCR entry points to amplify rye-specific multiple genomic fragments.83 Revolver is attractive as an index of genomic evolution and as a landmark of chromosomes useful for evaluating evolutionary relationships among the tribe Triticeae.
In conclusion, genomic deletion cloning led to the discovery of Revolver, a novel member of the major genomic component, as well as LTR retrotransposons and MITEs, which show drastically different copy number between bread wheat and rye, a useful genetic resource of wheat. Revolver consists of a single gene encoding a DNA-binding-like protein, which is transcriptionally active and does not suffer from epigenetic silencing. Revolver elements feature an autonomous transposon, which may complement in trans the propagation of related elements, including LARD, that might have contributed to plant genome expansion, rearrangement, and evolution.
Funding
The authors acknowledge Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT) for the Grant-in-Aid for Scientific Research Nos. 04760006 and 1360006 that supported this work.
References
- 1.Flavell R. B., Rimpau J. R., Smith D. B. Repeated sequence DNA relationships in four cereal genomes. Chromosoma. 1977;63:205–222. [Google Scholar]
- 2.Grandbastien M. A. Retroelements in higher plants. Trends Genet. 1992;8:103–108. doi: 10.1016/0168-9525(92)90198-d. [DOI] [PubMed] [Google Scholar]
- 3.Barakat A., Carels N., Bernardi G. The distribution of genes in the genomes of Gramineae. Proc. Natl. Acad. Sci. USA. 1997;94:6857–6861. doi: 10.1073/pnas.94.13.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panstruga R., Buschges R., Piffanelli P., Schulze-Lefert P. A contiguous 60 kb genomic stretch from barley reveals molecular evidence for gene islands in a monocot genome. Nucleic Acids Res. 1998;26:1056–1062. doi: 10.1093/nar/26.4.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park E. T., Kazazian H. H., Jr Mobile elements and the human genome. Nat. Rev. Genet. 2000;1:134–144. doi: 10.1038/35038572. [DOI] [PubMed] [Google Scholar]
- 6.SanMiguel P., Bennetzen J. L. Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 1998;82:37–44. [Google Scholar]
- 7.Myers B. C., Tingey S.V., Morgante M. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 2001;11:1660–1676. doi: 10.1101/gr.188201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finnegan D. J. Transposable elements. Curr. Opin. Genet. Dev. 1992;2:861–867. doi: 10.1016/s0959-437x(05)80108-x. [DOI] [PubMed] [Google Scholar]
- 9.Boeke J. D., Chapman K. B. Retrotransposition mechanisms. Cuur. Opin. Cell Biol. 1991;3:502–507. doi: 10.1016/0955-0674(91)90079-e. [DOI] [PubMed] [Google Scholar]
- 10.Adams S. E., Mellor J., Gull K., Sim R. B., Tuite M. F., Kingsman S. M., Kingsman A. J. The function and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell. 1987;49:111–119. doi: 10.1016/0092-8674(87)90761-6. [DOI] [PubMed] [Google Scholar]
- 11.Doolittle R. F., Feng D.-F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Quant. Rev. Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 12.Flavell A. J., Dunbar E., Anderson R., Pearce S. R., Hartley R., Kumar A. Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res. 1992;20:3639–3644. doi: 10.1093/nar/20.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voytas D. F., Cummings M. P., Konieczny A. K., Ausubel F. M., Rodermel S. R. Copia-like retrotransposons are ubiquitous among plant. Proc. Natl. Acad. Sci. USA. 1992;89:7124–7128. doi: 10.1073/pnas.89.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennetzen J. L. The Mutator transposable element system of maize. Curr. Top. Microbiol. Immunol. 1996;204:195–229. doi: 10.1007/978-3-642-79795-8_9. [DOI] [PubMed] [Google Scholar]
- 15.Suoniemi A., Tanskanen J., Schulman A. H. Gypsy-like retrotransposons are widespread in the plant kingdom. Plant J. 1998;13:699–705. doi: 10.1046/j.1365-313x.1998.00071.x. [DOI] [PubMed] [Google Scholar]
- 16.Vicient C. M., Kalendar R., Schulman A. H. Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res. 2001;11:2041–2049. doi: 10.1101/gr.193301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennetzen J. L. Transposable element contribution to plant gene and genome evolution. Plant Mol. Biol. 2000;42:251–269. [PubMed] [Google Scholar]
- 18.Bennetzen J. L., Ma J., Devos K. M. Mechanisms of recent genome size variation in flowering plants. Annals Botany. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flavell A. J., Pearce S. R., Kumar A. Plant transposable elements and the genome. Curr. Opin. Genet. Dev. 1994;4:838–844. doi: 10.1016/0959-437x(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 20.Kunze R., Well C. F. The hAT and CACTA superfamilies of plant transposons, In. In: Craig N. L., Craigie R., Gellert M., Lambowitz M., editors. Mobile DNA II. Washington, DC: American Society of Microbiology; 2002. pp. 565–610. [Google Scholar]
- 21.Zhang X., Jiang N., Feschotte C., Wessler S. R. PIF- and Pong-like transposable elements: distribution, evolution and relationship with Tourist-like miniature inverted-repeat transposable elements. Genetics. 2004;166:971–986. doi: 10.1534/genetics.166.2.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wessler S. R., Bureau T. E., White S.E. LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 23.Feschotte C., Wessler S. R. Mariner-like transposases are widespread and diverse in flowering plants. Proc. Natl. Acad. Sci. USA. 2002;99:280–285. doi: 10.1073/pnas.022626699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Von Strenberg R. M., Novick G. E., Gao G-P., Herrera R. J. Genome canalization: the coevolution of transposable and interspersed repititive elements with single copy DNA, In. In: McDonald J. F., editor. Transposable Elements and Evolution. Dordrecht: Kluwer Academic Publishers; 1993. pp. 108–139. [DOI] [PubMed] [Google Scholar]
- 25.Marillonnet S., Wessler S. R. Extreme structural heterogeneity among the members of a maize. Genetics. 1998;150:1245–1246. doi: 10.1093/genetics/150.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.SanMiguel P., Gaut B. S., Tikhonov A., Nakajima Y., Bennetzen J. L. The paleontology of intergene retrotransposons of maize. Nature Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 27.Kumar A., Pearce S. R., McLean K., Harrison G., Heslop-Harrison J. S., Waugh R., Flavell A. J. The Ty1-copia group of retrotransposons in plants: genomic organisation, evolution, and use as molecular markers. Genetica. 1997;100:205–217. [PubMed] [Google Scholar]
- 28.Hirochika H., Sugimoto K., Otsuki Y., Tsugawa H., Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennetzen J. L. The contribution of retroelements to plant genome organization, function and evolution. Trends Microbiol. 1996;4:347–353. doi: 10.1016/0966-842x(96)10042-1. [DOI] [PubMed] [Google Scholar]
- 30.Flavell R. B., Bennett M. D., Smith J. B., Smith D. B. Genome size and the proportion of repeated sequence DNA in plants. Biochem. Genet. 1974;12:257–269. doi: 10.1007/BF00485947. [DOI] [PubMed] [Google Scholar]
- 31.Guidet F., Rogowsky P., Taylor C., Song W., Langridge P. Cloning and characterization of a new rye-specific repeated sequence. Genome. 1991;34:81–87. [Google Scholar]
- 32.Cuadrado A., Vitellozzi F., Jouve N., Ceoloni C. Fluorescence in situ hybridization with multiple repeated DNA probes applied to the analysis of wheat-rye chromosome pairing. Theor. Appl. Genet. 1997;94:347–355. [Google Scholar]
- 33.Bedbrook J. R., Jones J., O'Dell M., Thompson R. D., Flavell R. B. A molecular description of telomeric heterochromatin in Secale species. Cell. 1980;19:545–560. doi: 10.1016/0092-8674(80)90529-2. [DOI] [PubMed] [Google Scholar]
- 34.Appels R., Dennis E. S., Smyth D. R., Peacock W. J. Two repeated DNA sequences from the heterochromatic regions of rye (Secale cereale) chromosomes. Chromosoma. 1981;84:265–277. [Google Scholar]
- 35.Appels R., Moran L. B., Gustafson J. P. Rye heterochromatin. I. Studies on clusters of the major repeating sequence and the identification of a new dispersed repetitive sequence element. Can. J. Genet. Cytol. 1986;28:645–657. [Google Scholar]
- 36.Lamar E. E., Palmer E. Y-encoded, species-specific DNA in mice: evidence that the Y chromosome exists in two polymorphic forms in inbred strains. Cell. 1984;37:171–177. doi: 10.1016/0092-8674(84)90312-x. [DOI] [PubMed] [Google Scholar]
- 37.Heslop-Harrison J.S, Brandes A., Taketa S., Schmidt T., Vershinin A. V., Alkhimova E. G., Kamm A., Doudrick R., Schwarzacher T., Katsiotis A., et al. The chromosomal distribution of Ty1-copia group retrotransposable elements in higher plants and their implication for genome evolution. Genetica. 1997;100:197–204. [PubMed] [Google Scholar]
- 38.Rogowsky P. M., Manning S., Liu J. Y., Langridge P. The R173 family of rye-specific repetitive DNA sequences: a structural analysis. Genome. 1991;34:88–95. doi: 10.1007/BF00029152. [DOI] [PubMed] [Google Scholar]
- 39.Rogowsky P. M., Liu J. Y., Manning S., Taylor C., Langridge P. Structural heterogeneity in the R173 family of rye-specific repetitive DNA sequences. Plant Mol. Biol. 1992;20:95–102. doi: 10.1007/BF00029152. [DOI] [PubMed] [Google Scholar]
- 40.McIntyre C. L., Pereira S., Moran L. B., Appels R. New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome. 1990;33:635–640. doi: 10.1139/g90-094. [DOI] [PubMed] [Google Scholar]
- 41.Vershinin A. V., Schwarzacher T., Heslop-Harrison J. S. The large-scale genomic organization of repetitive DNA families at the telomeres of rye chromosomes. Plant Cell. 1995;7:1823–1833. doi: 10.1105/tpc.7.11.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manninen I., Schulman A. H. BARE-1, a copia-like retroelement in barley (Hordeum vulgare L.) Plant Mol. Biol. 1993;22:829–846. doi: 10.1007/BF00027369. [DOI] [PubMed] [Google Scholar]
- 43.Suoniemi A., Anamthawat-Jonsson K., Arna T., Schulman A. H. Retrotransposon BARE-1 is a major, dispersed component of the barley (Hordeum vulgare L.) genome. Plant Mol. Biol. 1996;30:1321–1329. doi: 10.1007/BF00019563. [DOI] [PubMed] [Google Scholar]
- 44.Suoniemi A., Narvanto A., Schulman A. H. The BARE-1 retrotransposon is transcribed in barley from an LTR promoter active in transient assays. Plant Mol. Biol. 1996;31:295–306. doi: 10.1007/BF00021791. [DOI] [PubMed] [Google Scholar]
- 45.Vicient C. M., Suoniemi A., Anamthawat-Jonsson K., Tanskanen J., Beharav A., Nevo E., Schulman A. H. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell. 1999;11:1769–1784. doi: 10.1105/tpc.11.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalendar R., Vicient C. M., Peleg O., Anamthawat-Jonsson K., Bolshoy A., Schulman A. H. Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics. 2004;166:1437–1450. doi: 10.1534/genetics.166.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shirasu K., Schulman A. H., Lahaye T., Schuze-Lefert P. A contiguous 66 kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 2000;10:908–915. doi: 10.1101/gr.10.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grandbastien M. A. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998;3:181–187. [Google Scholar]
- 49.Kumar A., Bennetzen J. L. Plant retrotransposons. Annu. Rev. Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 50.Vernhettes S., Grandbastien M.-A., Casacuberta J. M. In vivo characterization of transcriptional regulatory sequences involved in the defence-associated expression of the tobacco retrotransposon Tnt1. Plant Mol. Biol. 1997;35:673–679. doi: 10.1023/a:1005826605598. [DOI] [PubMed] [Google Scholar]
- 51.Takeda S., Sugimoto K., Otsuki H., Hirochika H. Transcriptional activation of the tobacco retrotransposon Tto1 by wounding and methyl jasmonate. Plant Mol. Biol. 1998;36:365–376. doi: 10.1023/a:1005911413528. [DOI] [PubMed] [Google Scholar]
- 52.Hirochika H. Activation of tobacco retrotransposons during tissue culture. EMBO J. 1993;12:2521–2528. doi: 10.1002/j.1460-2075.1993.tb05907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ostertag E. M., Kazazian H. H., Jr Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 54.Hamilton A., Voinnet O., Chappell L., Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Llave C., Kasschau K. D., Rector M. A., Carrington J. C. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sunker R., Girke T., Zhu J-K. Identification and characterization of endogenous small interfering RNAs from rice. Nucleic Acids Res. 2005;33:4443–4454. doi: 10.1093/nar/gki758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennett M. D., Leitch I. J. Nuclear DNA amounts in angiosperms. Ann. Bot. 1995;76:113–176. [Google Scholar]
- 58.Thomas C. A. The genetic organization of chromosomes. Annu. Rev. Genet. 1971;5:237–256. doi: 10.1146/annurev.ge.05.120171.001321. [DOI] [PubMed] [Google Scholar]
- 59.Arumuganathan K., Earle E. D. Nuclear DNA content of some important plant species. Plant Mol. Biol. Reporter. 1991;9:208–218. [Google Scholar]
- 60.Doolittle W. F., Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 61.Orgel L. E., Crick F. H. C. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 62.Wicker T., Stein N., Albar L., Feuillet C., Schlagenhauf E., Keller B. Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J. 2001;26:307–316. doi: 10.1046/j.1365-313x.2001.01028.x. [DOI] [PubMed] [Google Scholar]
- 63.SanMiguel P., Tikhonov A., Jin Y. K., Motchoulskaia N., Zakharov D., Melake-Berhan A., Springer P. S., Edwards K. J., Lee M., Avramova Z., et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 64.Tikhonov A. P., SanMiguel P. J., Nakajima Y., Gorensten N. M., Bennetzen J. L., Avramova Z. Colinearity and its exceptions on orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim J. M., Vanguri S., Boeke J. D., Gabriel A., Voytas D. F. Transpossble elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 66.Ramakrishna W., Dubcovsky J., Park Y.-J., Busso C., Emberton J., SanMiguel P., Bennetzen J.L. Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics. 2002;162:1389–1400. doi: 10.1093/genetics/162.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wicker T., Guyot R., Yahiaoui N., Keller B. CACTA transposons in Triticeae: a diverse family of high-copy repetitive elements. Plant Physiol. 2003;132:52–63. doi: 10.1104/pp.102.015743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langdon T., Jenkins G., Hasterok R., Jones R. N., King I. P. A high-copy-number CACTA family transposon in temperate grasses and cereal. Genetics. 2003;163:1097–1108. doi: 10.1093/genetics/163.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dvorak J., McGuire P. E., Cassidy B. Apparent sources of the A genomes of wheats inferred from polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome. 1988;30:680–689. [Google Scholar]
- 70.Dvorak J., Zhang H. B. Reconstruction of the phylogeny of the genus Triticum from variation in repeated nucleotide sequences. Theor. Appl. Genet. 1992;84:419–429. doi: 10.1007/BF00229502. [DOI] [PubMed] [Google Scholar]
- 71.Friebe B., Gill B. S. Chromosome banding and genome analysis in diploid and cultivated polyploid wheats, In. In: Jauhar P. P., editor. Methods of Genome Analysis in Plants. Boca Raton, FL, NY: CRC Press; 1996. pp. 39–60. [Google Scholar]
- 72.Tsunewaki K. Plasmon analysis as the counterpart of genome analysis, In. In: Jauhar P. P., editor. Methods of Genome Analysis in Plants. Boca Raton, FL, NY: CRC Press; 1996. pp. 271–299. [Google Scholar]
- 73.Feldman M., Lupton F. G. H., Miller T. E. Wheats, In. In: Smartt J., Simmonds N. W, editors. Evolution of Crops. 2nd Ed. London: Longman Scientific; 1995. pp. 184–192. [Google Scholar]
- 74.Leitch I. J., Bennett M. D. Polyploidy in angiosperms. Trends Plant Sci. 1997;2:470–476. [Google Scholar]
- 75.Wendel J. F. Genome evolution in ployploids. Plant Mol. Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- 76.Feldman M., Liu B., Segal G., Abbo S., Levy A. A., Vega J. M. Rapid elimination of low-copy DNA sequence in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics. 1997;147:1381–1387. doi: 10.1093/genetics/147.3.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ozkan H., Levy A. A., Feldman M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell. 2001;13:1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.SanMiguel P., Remakrishna W., Bennetzen J. L., Busso C., Dubcovsky J. Transposable elements, genes and recombination in a 215 kb contig from wheat chromosome 5Am. Funct. Integr. Genomics. 2002;2:70–80. doi: 10.1007/s10142-002-0056-4. [DOI] [PubMed] [Google Scholar]
- 79.Bennetzen J. L., Kellogg E. A. Do plants have a one-way ticket to genomic obesity? Plant Cell. 1997;9:1509–1514. doi: 10.1105/tpc.9.9.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kellogg E. A. Relationships of cereal crops and other grasses. Proc. Natl. Acad. Sci. USA. 1998;95:2005–2010. doi: 10.1073/pnas.95.5.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leitch I. J., Chase M. W., Bennett M. D. Phylogenetic analysis of DNA C-value provides evidence for a small ancestral genome size in flowering plants. Annal. Bot. 1998;82:85–94. [Google Scholar]
- 82.Petrov D. A., Sangster T. A., Johnston J. S., Hartl D. L., Shaw K. L. Evidence for DNA loss as a determinant of genome size. Science. 2000;287:1060–1062. doi: 10.1126/science.287.5455.1060. [DOI] [PubMed] [Google Scholar]
- 83.Tomita M. Revolver-2: a novel transposon-like element from rye. 2005 United States Patent and Trademark Office, Published Application No. 20050091710.