Abstract
Enterohemorrhagic Escherichia coli is an emerging pathogen that causes diarrhea and hemolytic uremic syndrome. Much of the genomic information that affects virulence is acquired by horizontal transfer. Genes necessary for attaching and effacing lesions are located in the locus for enterocyte effacement (LEE) pathogenicity island. LEE gene transcription is positively regulated by Ler, which is also encoded by the LEE, and by Pch regulators, which are encoded at other loci. Here we identified genes whose transcription profiles were similar to those of the LEE genes, by comparing the effects of altering ler and pch transcript levels. We assigned these genes into two classes, according to their transcription profiles. By determining the binding profiles for Ler and Pch, we showed that both were involved in regulating one class of genes, but only Pch was involved in regulating the other class. Binding sites were found in the coding region as well as the promoter region of regulated genes, which include genes common to K12 strains as well as 0157-specific genes, suggesting that both act as a global regulator. These results indicate that Ler and Pch orchestrate the transcription of virulence genes, which are captured by horizontal transfer and scattered throughout the chromosome.
Key words: transcriptional regulation, virulence gene, A/E pathogen, DNA-binding protein, captured genome
1. Introduction
Pathogenic bacteria are distinct from related non-pathogenic strains in that they possess and express virulence genes. The major avenue for the acquisition of virulence genes is the horizontal transfer of DNA segments.1 In many pathogenic bacteria, multiple virulence factors function to achieve successful infection and population bursts in niches by protecting bacteria from host defense systems. Multiple virulence genes in different chromosomal loci must often be simultaneously expressed during the various stages of infection. Thus, the integration of the acquired genes, which are scattered throughout the chromosome, into the gene-regulatory systems is important for establishing pathogenic strains. Because the repertoire of virulence traits is often variable among the strains of the same species, bacterial pathogenicity is a dynamic process involving the ongoing addition of novel virulence genes. Therefore, plasticity that allows newly transferred virulence genes to be adopted must also be a characteristic of the virulence-regulatory system. To understand the emergence and evolution of pathogenic bacteria, therefore, the molecular basis of virulence gene regulation must be elucidated.
Enterohemorrhagic Escherichia coli (EHEC) causes hemorrhagic colitis and hemolytic uremic syndrome.2 A comparison of the genomic sequences of EHEC O157:H7 Sakai strain with that of the non-pathogenic E. coli strain K-12 revealed many EHEC O157-specific loci (called S-loops), most of which contain gene clusters.3 One of the virulence-associated loci is the locus for enterocyte effacement (LEE), which encodes a type III secretion system (T3SS), type III secreted effectors, and adhesin. These virulence factors are necessary for the induction of attaching and effacing (A/E) lesions in epithelial cells, which are characterized by a rearrangement of the cytoskeleton and the destruction of microvilli.4,5 In addition to the LEE, gene-encoding effectors for T3SS can be found elsewhere on the chromosome,6 mostly at the end of lambdoid-like prophages, which are scattered throughout the chromosome.7 The mosaic nature of the pathogenic E. coli genome suggests that virulence traits are acquired through the capture of mobile elements such as phages, insertion sequences, and plasmids. In particular, lambdoid phage appears to have a major role in the emergence of EHEC strains, given that a variety of effector genes are encoded by several prophages lysogenized at different loci on the chromosome.7 Therefore, for pathogenicity to be established, the expression kinetics of genes in different chromosomal loci must be appropriately orchestrated.
The transcription of LEE genes is regulated by two positive transcriptional regulators: Pch regulators activate the transcription of the LEE1 operon, which encodes the transcriptional regulator Ler, which in turn regulates other LEE operons and genes in other loci.8 Ler belongs to the H-NS family of nucleoid-associated proteins, exhibiting homology with the carboxy-termini of these proteins corresponding to DNA-binding domain.9 Ler has been shown to activate LEE gene expression by counteracting the repression by H-NS.10 EHEC strains possess five genes that encode homologs of Pch, though only two, pchA and pchB, activate LEE1 transcription.8 Pch is also a small protein as Ler, but it showed no homology with nucleoid-associated protein or transcription regulators. The expression of LEE genes is regulated in response to changes in environmental factors, such as the growth phase, temperature, osmolarity, bicarbonate ion concentration, membrane stress, and nutrient starvation.11–14 Both Pch and Ler are major regulators of LEE gene expression in response to such changes. For example, transient growth arrest caused by insufficient nutrients or by entry into the stationary growth phase induces the expression of LEE genes through increased production of both Pch and Ler, because the transcription of pch genes and the LEE1 operon is positively regulated by starvation-induced signal molecule, ppGpp.13 Furthermore, the activation of the RcsC–RcsD–RcsB phospho-relay system, which is thought to be a signal transduction system for responding membrane stress, affects the expression of LEE genes both positively and negatively through the activation of grvA, encoding a transcription regulator and the repression of pch.12 Thus, environmental factors are evaluated by a sensing system common to all E. coli strains. In EHEC O157:H7, however, the signals from the sensing system also affect the transcription of O157-specific transcription regulators, which results in the coordinated regulation of the virulence genes in response to certain environmental changes.
In this study, in order to clarify which genes are involved in the pathogenicity of EHEC and the regulatory networks that govern the LEE and related genes, we isolated genes under the control of the two major virulence regulators, Pch and Ler, by comparing the changes in global transcription caused by altering the pch and ler expression levels. We identified two sets of genes whose transcription profiles were closely correlated with those of LEE genes. Furthermore, we determined the binding sites for PchA and Ler, and found that, although Pch was involved in the regulation of genes in both classes, Ler regulated the expression of only one class of genes.
2. Materials and methods
2.1. Bacterial strains and growth conditions
EHEC O157 Sakai (RIMD 0509952)3 and its derivative strains used in this study are listed in Supplementary Table S1. Deletion mutants of EHEC O157 Sakai were constructed using the method and plasmids of Datsenko and Wanner.15 DNA fragments corresponding to the pchA and ler genes were isolated by polymerase chain reaction (PCR) from the chromosomal DNA of O157 Sakai using specific primers: 5′-CACAGGAATATATCCGTACCC-3′ and 5′-AGTATGTGTCACTGGCCTATACGG-3′ for pchA, and 5′-TTGGCTCACAATACTCATCC-3′ and 5′-GCTTAACTAAATGGAAATGC-3′ for ler. The PCR amplicon was cloned into pGEM-T (Promega) in the direction opposite to that of the lac promoter. The pchA gene of EHEC Sakai ΔpchB ΔpchC was tagged at the 3′-terminus with Strep-tag (IBA), and the ler gene in wild-type O157 Sakai was tagged with 3× FLAG sequence, using an epitope-tagging procedure.16 Bacteria were grown overnight in Luria-Bertani broth (LB), diluted 100-fold, and then cultured in LB or Dulbecco's modified Eagle's medium (DMEM) without fetal calf serum, at 37°C with shaking.
2.2. Transcriptome analysis
Total RNA from EHEC was isolated with TRI reagent (Sigma) using the method recommended by the manufacturer. The RNA was purified again after treatment with RNase-free DNase I (Takara), and the amount and purity of the RNA were determined by measuring A260 and A280. The RNAs from EHEC were used to synthesize cDNAs with Cy-3- or Cy-5-conjugated dUTP. cDNA labeling, microarray hybridization, data capture, and data analysis were performed as described previously.12 We used a DNA microarray with oligo-DNAs specific for each of the CDS of EHEC O157 Sakai.12,17 The expression ratio used here is to indicate the average of the ratios obtained in the two independent experiments in which the signal intensities were greater than the mean ±1 SD of the negative control. The hierarchical clustering of gene expression profiles and conditions were analyzed using GeneSpring GX7.3 (Agilent Technologies). The raw and processed data are available in the GEO of NCBI (in accession no. GSE9752).
2.3. Design of EHEC O157 tiled microarrays
The EHEC O157 Sakai genome sequence and other genetic information were retrieved from the O157 Web site at NAIST (http://genome.naist.jp/bacteria/o157/) and used to design probes, following the Affymetrix guidelines (http://www.affymetrix.com). Two versions of the microarray were used in this study. One contained 25-nt-long probe sequences separated by 103 (±5)-base intervals in the coding region and by intervals of 35 (±2) bases in the intergenic region along both strands of the O157 Sakai chromosome. The other contained 25-nt-long probe sequences separated by 45-base intervals, as possible, from only a single strand of the chromosome. If the sequence was not correct, the probe was offset with sequence within 10 bases. Thus, perfect-match probes and corresponding mismatch probes were synthesized on the chip.
2.4. ChIP-on-chip
Chromosome immunoprecipitation and DNA microarray analysis were performed as described by Oshima et al.18 with slight modifications. EHEC O157 Sakai strain SKI1172 or SKI1282 was grown in DMEM to logarithmic phase (OD600 = 0.7 − 0.8) at 37°C. Formaldehyde (final concentration of 1%) was added to 10 mL of the culture, and the mixture was incubated at room temperature for 20 min. To terminate the crosslinking reaction, glycine (final concentration of 0.45 M) was added, and the mixture was incubated at room temperature for 5 min. Bacteria were collected by centrifugation and washed first with Tris buffered saline, then with washing buffer [50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 0.01% Tween-20] twice, suspended in 1 mL of lysis buffer [50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 1% Tween-20, 20 mg/mL lysozyme], and stored at −20°C until use. Bacteria were lysed by incubation at 37°C for 30 min and diluted in 4 mL of washing buffer containing phenylmethanesulphonylfluoride (final concentration of 1 mg/mL). The solution was then sonicated 15 times for 30 s at 30 s intervals at 4°C using a Bioruptor UCD-200 (Cosmo Bio Co., Ltd.). Cell debris was removed by centrifugation at 15 000 rpm for 30 min. An 800 µL aliquot of the supernatant was mixed with magnetic beads bound to an anti-FLAG antibody (M2 monoclonal antibody, Sigma-Aldrich) or MagStrep (IBA) antibody and incubated at 4°C overnight. The beads were washed three times with washing buffer and then with TE (10 mM Tris-HCL (pH8.0), 1 mM EDTA). The protein–DNA complex bound to the beads was released in 100 mL of elution buffer [50 mM Tris–HCl (pH 7.5), 10 mM EDTA, 1% SDS] by heating at 65°C for 20 min for Ler-FLAG, or in Biotin buffer [50 mM Tris (pH 7.8), 150 mM NaCl, 5 mM EDTA, 10 mM Biotin] incubated at room temperature for 10 min for PchA-Strep. DNA fragments crosslinked to proteins were released by incubating the eluate with proteinase K (1 mg/mL) at 42°C for 2 h, then 65°C for 6 h, and purified using the Qiagen PCR Clean-up kit (Qiagen) in 200 µL of TE. Recovered DNA fragments were amplified, terminally labeled, and hybridized with the EHEC O157 tiled microarray as described previously.18 The raw intensity data (CEL files) were processed using the In Silico Molecular Cloning Array Edition software (In Silico Biology) for analysis and visualization. Signal intensities were obtained by subtracting the signal intensities of the mismatch probes from those of the perfect-match probes, and probes with negative values were excluded from further analysis. To compensate for differences among the probes caused by different probabilities of DNA disruption and hybridization efficiency, signals were normalized by dividing them by the signal intensities from DNA samples taken before the beads were added. The binding sites for PchA-Strep and Ler-FLAG were assessed manually. The reproducibility and specificity of binding of PchA or Ler to several novel sites were confirmed by ChIP-PCR for each site.
3. Results and discussion
3.1. Identification of Pch- and/or Ler-regulated genes
The genes in the LEE pathogenicity island are organized mainly into five operons, LEE1 to LEE5, and several cistrons. The transcription of the LEE genes is positively regulated by the Ler protein, which is encoded by the first gene of the LEE1 operon. Transcription of the LEE1 operon is regulated by several proteins conserved in the non-pathogenic E. coli K12 strain, including IHF, QseA, and H-NS, and by the O157-specific regulator, Pch.8,14,19,20 EHEC O157:H7 Sakai strain possesses three intact pch genes, which encode proteins that differ by only one or two amino acids, and are located in different prophage-like elements.
To elucidate the network created by O157-specific regulators, the effect of the overproduction or deletion of Ler or Pch on global transcription levels was determined using DNA microarray hybridization. We determined differences in the transcript levels for all the genes in EHEC O157 Sakai strain in 13 comparisons (Fig. 1): ler null mutant against wild-type EHEC, a pchA-pchB-pchC triple mutant against wild type, EHEC harboring a high copy number of the ler or pchA gene against EHEC harboring a control vector, the ler mutant harboring a high copy number of the pchA gene against the ler mutant harboring the control vector, the pchA-pchB-pchC triple mutant harboring a high copy number of the ler gene against the pch triple-mutant harboring the control vector, and the pchA-pchB double mutant harboring a high copy number of ler against the double mutant harboring the control vector. Since the expression level of LEE genes in EHEC O157, when grown in tissue culture medium DMEM, is much higher than that in EHEC grown in LB, we included the differences in broth media as other factors affecting expression of EHEC genes. Except for the last case (grown in LB), all comparisons were carried out for bacteria grown in LB and DMEM.
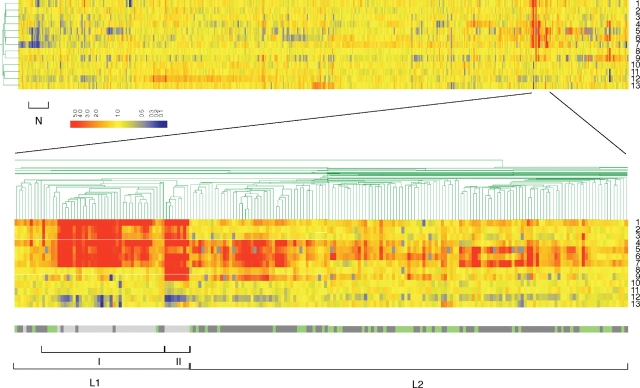
Figure 1.
Hierarchical cluster analysis of gene expression changes in response to changes in Pch or Ler levels. Gene expression data from 13 microarray experiments (lines) and whole EHEC O157 Sakai 5454 genes (columns) are indicated. The microarray experiments involved 13 comparisons between EHEC O157 Sakai and derivative strains: (1) Wild type (WT) harboring pGEM-ler to WT harboring pGEM vector grown in LB. (2) The pchApchB mutant harboring pGEM-ler to the pchApchB mutant harboring pGEM-vector grown in LB. (3) The pchApchBpchC mutant harboring pGEM-ler to the pchApchBpchC mutant harboring pGEM-vector grown in LB. (4) WT harboring pGEM-pchA to WT harboring pGEM-vector grown in LB. (5) WT harboring pGEM-ler to WT harboring pGEM-vector grown in DMEM. (6) WT harboring pGEM-pchA to WT harboring pGEM-vector grown in DMEM. (7) The pchApchBpchC mutant harboring pGEM-ler to the pchApchBpchC mutant harboring pGEM-vector grown in DMEM. (8) The ler mutant harboring pGEM-pchA to the ler mutant harboring pGEM vector grown in LB. (9) The ler mutant harboring pGEM-pchA to the ler mutant harboring pGEM vector grown in DMEM. (10) The ler mutant to WT grown in LB. (11) The pchApchBpchC mutant to WT grown in LB. (12) The pchApchBpchC mutant to WT grown in DMEM. (13) The ler mutant to WT grown in DMEM. The lower panel shows gene clusters in and around the LEE genes along with a color bar indicating the gene context (green: common to K12; dark grey: EHEC O157 specific; light grey: LEE). The scale bar indicates color coding of the RNA levels. Class L1 with subclasses I and II, class L2, and class N are indicated (see text).
The EHEC genes were subjected to clustering analysis by standard correlation to determine their transcript expression patterns (Fig. 1). As expected, genes belonging to the LEE showed similar expression profiles, and we classified them as part of a single group, class L1, which also included genes that were not encoded within the LEE (Fig. 2). Within the LEE-encoded genes of class L1, there were two distinct subgroups. One subgroup contained all the LEE1 operon genes except ler, and the other contained most of the other LEE genes (Supplementary Table S2). We found that the transcript levels of the LEE1 genes were increased by PchA overexpression in both wild-type EHEC and the ler deletion mutant, but the transcript levels of the other LEE genes were enhanced by PchA overexpression only in the presence of the ler gene. Because the transcript data were obtained from ler null mutant or from strains harboring multiple copies of ler, the ler gene itself could not be classified in this experiment.
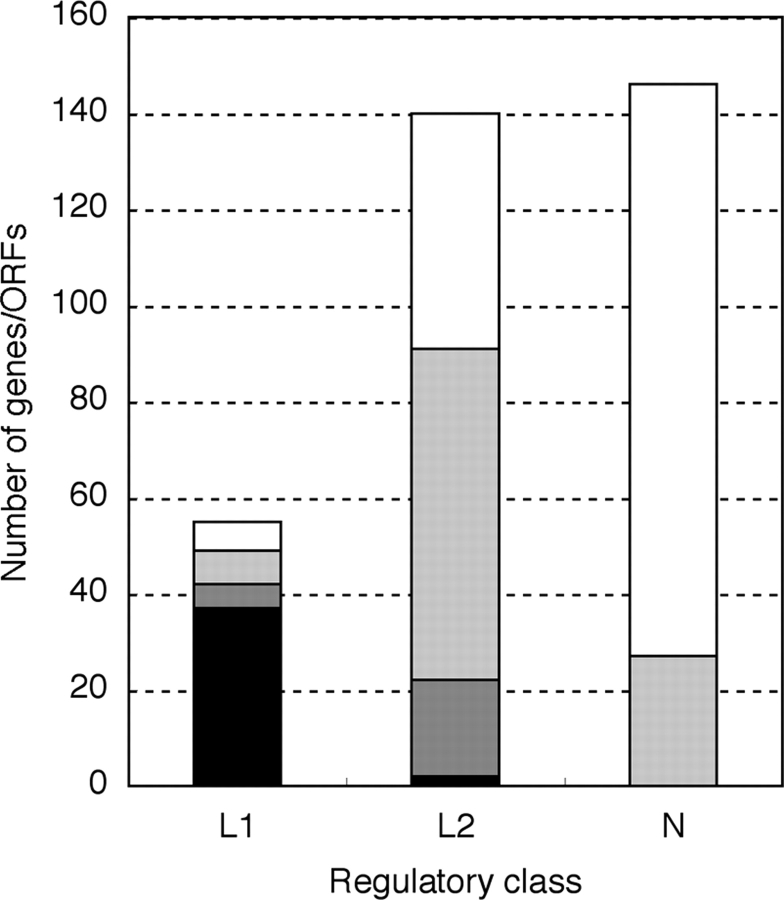
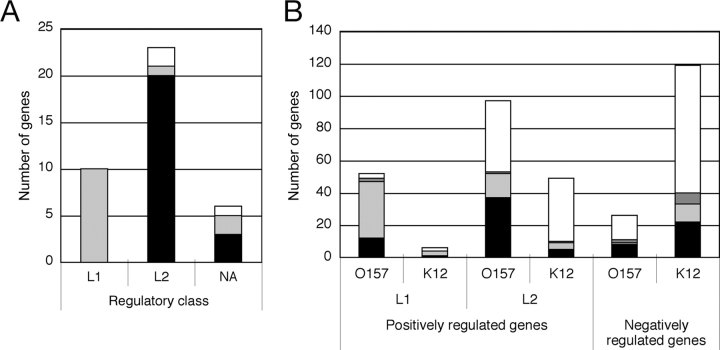
Figure 2.
Characteristics of genes in each cluster. Number of genes in three gene classes, L1, L2 and N, which were isolated by transcriptomic analysis, was shown. Genes were sorted by chromosomal locus and function as follows: black, LEE genes; grey, non-LEE virulence-associated genes; light grey, other genes in S-loop; white, genes on K12-common loci.
The three class L1 effector genes that lie outside the LEE were nleA, espF-U/tccP, and espJ. The nleA/espI gene is a single cistron on prophage Sp9 and is required for Citrobacter rodentium, an A/E mouse pathogen, to colonize the mouse colon.21 EspF-U/TccP is necessary for the recruitment of actin to Tir beneath attached bacteria, which is essential for A/E lesion formation.22,23 Two genes, espF-U/tccP and espJ, are located on lambdoid-like prophage Sp14 and constitute an operon.24,25 Since the espF-M gene of EHEC O157 Sakai is a pseudogene,23 it is likely that espF-M was detected because of cross-hybridization with espF-U/tccP. Finally, class L1 included three unknown O157-specific genes in an S-loop, ECs0355, ECs0356, ECs0814, and the plasmid-born gene tagA. In addition, the transcription levels of three genes common to EHEC O157:H7 and K-12 in the E. coli backbone chromosome were regulated in close correlation with the LEE genes. These genes, ECs0512 (ylaD), ECs0513 (hha), and ECs0514 (ybaJ), are likely to comprise an operon. The hha gene encodes a transcription regulator for ler.26
The expression profile of the class L1 cluster was correlated with that of another cluster of genes. We designated these genes as class L2 (Fig. 1). Class L2 contained 140 CDS/genes, consisting of 89 genes in an S-loop and 51 genes in the backbone or K-12 common chromosome (Fig. 2 and Supplementary Table S3). Although the transcript levels of the class L2 genes increased with increased Pch levels, like the genes in class L1, their response to increased levels of Ler was different, in that the L2 transcript levels increased only slightly with the overexpression of Ler when grown in LB. In contrast, the genes in L1 showed a marked increase in transcript level with high levels of Ler, even in LB.
Class L2 contained 20 non-LEE effector genes that encode T3SS-secreted proteins. Most of these effector genes are located on exchangeable effector loci (EEL) of lambda-like prophages,7 including Sp3, Sp6, Sp9, Sp10, Sp11, Sp17, and SpLE3 (Fig. 2). This observation of the expression and T3SS-dependent secretion of these effectors when the EHEC harbored a high copy number of the pchA gene is consistent with our previous report.7 All the effectors, except EspM2 (encoded by ECs3485), that were found in the culture supernatant of EHEC were regulated coordinately with the LEE genes. The expression of another effector gene, ECs4653, which encodes EspY4, was also upregulated by Ler and Pch, just like the other non-LEE effector genes.
Some genes were regulated in conjunction with their neighbors because of members of the same operon. One cluster of these genes, ECs0240–ECs0245, lies in the rhsI element, which is one of nine rhs elements in EHEC O157 Sakai. ECs0815 and ECs0816 seem to belong to the same operon and are transcribed in the opposite direction from ECs0814, which belongs to class L1. Another cluster of genes, from ECs3512 to ECs3508, seems to comprise an operon that lies at the end of prophage-like element Sp17. In addition, class L2 included genes for type-II secretion machinery (etpC, etpD, etpE, etpH, and etpK), toxB, and hemolysin (hlyD) on the virulence plasmid pO157 (Supplementary Table S3).
One cluster of genes showed a negative correlation in their transcription with changes in Ler or Pch expression (shown as class N in Fig. 1 and Supplementary Table S4). Most drastically affected were genes associated with an acid-resistant phenotype, which are closely clustered on the chromosome. These genes include hdeA, hdeB, hdeD, gadE (yhiE), gadX, and gadW. One hundred and nineteen genes conserved between K12 and EHEC O157 and 27 O157-specific genes on S-loops formed clusters (Fig. 2). These genes showed very similar changes in transcript levels in response to changes in Ler/Pch levels. Several genes in this class, in addition to gadE, gadX, and gadW, regulate glutamate-dependent acid resistance. The EvgS–EvgA two-component regulatory system controls the expression of gadE, and transcription of the gadXW operon is dependent on alternative sigma factor RpoS, whose gene was also included in this class. Several of these factors, including GadE (YhiE), EvgS–EvgA, and RpoS, negatively affect the expression of LEE genes or bacterial adherence.27–29 Although the ClpXP protease has been shown to contribute indirectly to the expression of the LEE genes,28 the transcription of the clpX–clpP genes was negatively regulated by Ler and Pch.
3.2. Identification of binding sites for Pch and Ler
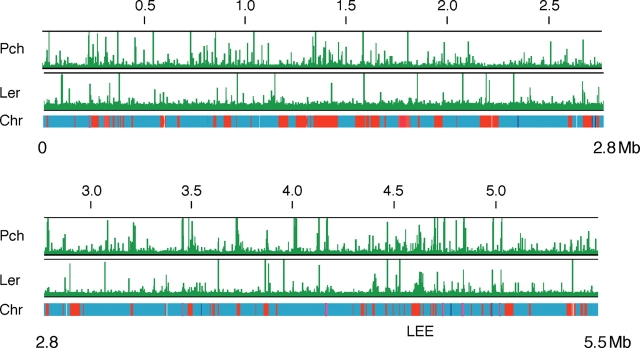
Although transcriptomic analyses revealed that Pch and/or Ler affect the transcription of many genes on S-loops and in the backbone or K-12 common chromosome, the transcript levels of some of them might be regulated indirectly, through proteins from genes that are directly regulated by Pch and/or Ler. To distinguish directly regulated genes from indirectly regulated genes, we determined the binding sites of PchA and Ler on the EHEC O157 Sakai chromosome by chromosome immunoprecipitation (ChIP) or chromosome affinity precipitation, in combination with DNA microarray hybridization. A derivative strain of EHEC O157 Sakai that expresses Strep-tagged PchA (PchA-Strep) or FLAG-tagged Ler (Ler-FLAG) was grown in DMEM to the logarithmic growth phase at 37°C, which is the optimal condition for expression of the LEE genes, and treated with formalin to crosslink the DNA with bound proteins. DNA fragments that bound to PchA-Strep or Ler-FLAG were separated from the bacterial lysate with Strep-Tactin-coated magnetic beads (MagStrep) or anti-FLAG-conjugated magnetic beads. After decrosslinking and purification, the DNA fragments that bound to PchA or Ler were mapped by hybridization with a high-density tiled microarray. We found 259 loci for Pch and 59 for Ler on the chromosome (Fig. 3 and Supplementary Tables S5 and S6). Of these, 43.1% of the Pch-bound loci and 39.0% of the Ler-bound loci were in S-loops or junctions with S-loops and in the backbone or K12-common chromosome. Considering that only 25.5% of the chromosome was unique to the EHEC O157 Sakai strain compared with E. coli K12 strain MG1655, it was clear that Pch and Ler preferentially bound to S-loop sequences rather than to the backbone or K12-common chromosome.
Figure 3.
Distribution of PchA and Ler binding over the EHEC O157 Sakai chromosome. Graphs show PchA and Ler binding to the chromosome of the EHEC O157 Sakai strain (horizontal bar). Red and blue segments indicate S-loops (O157:H7 strain-specific regions) and backbone (regions conserved with E. coli K12), respectively. Vertical bars indicate the relative hybridization intensity of the precipitated DNA compared with DNA in the supernatant.
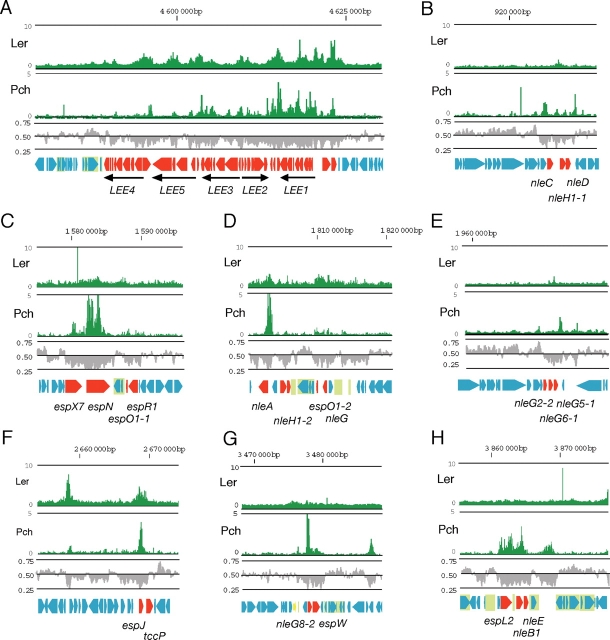
The expression of the LEE genes is positively regulated by a molecular cascade: Pch is a positive transcriptional regulator for the LEE1 promoter.8 Activation of the LEE1 operon leads to the expression of Ler, which activates the other LEE genes in operons LEE2 to LEE5 and several cistrons. As expected, Pch bound to the LEE1 promoter region (Fig. 4A). In addition, Pch bound to sites throughout the LEE sequence, including the coding region of the LEE1 operon. Furthermore, although the binding was weaker than in the LEE1 operon, Pch bound to other parts of the LEE island: the upstream region of the grlR–grlA operon, the 5′ portions of the LEE2 and LEE3 operons, the downstream portion of the LEE3 operon, the upstream portion of tir, the coding region of cesT, and the downstream portion of eae. Ler bound at similar levels to several regions in the LEE island, mostly in the 5′ regions of operons or genes (Fig. 4A): the intergenic regions between LEE1 and espG and between grlR and ECs4579 (rorf3), the 5′ portions of the LEE2 and LEE3 operons, the intergenic region between cesF and map, and the 5′ regions of escD and sepL.
Figure 4.
Distribution of PchA and Ler binding in the LEE and EELs. Graphs show PchA and Ler binding as in Fig. 2. Upper line indicates the position from the replication origin of the chromosome. Middle row indicates G + C contents. (A) Chromosomal region around the LEE. Genes in the LEE are represented by red arrows. The five main LEE operons, LEE1 to LEE5, are represented by arrows below the open reading frames map. (B) EEL on prophage-like element Sp3. Genes encoding effector proteins are indicated by red arrows. (C) EEL on prophage-like element Sp6. (D) EEL on prophage-like element Sp9. (E) EEL on prophage-like element Sp10. (F) EEL on prophage-like element Sp14. (G) EEL on prophage-like element Sp17. (H) EEL on prophage-like element SpLE3. IS and remnant of IS are shown in light green boxes.
EHEC O157 Sakai possesses 24 prophage or prophage-like elements, which make up more than half of the S-loop sequences. The binding loci for Pch and Ler were unevenly distributed in these acquired sequences. They rarely bound to sequences conserved among the phages; rather, they mostly bound to sequences that were unique to each phage element. Nine lambdoid-like phage elements and two phage-like elements possess EEL, which encode the majority of functional effector genes. Binding loci of Pch were detected within the EEL of all phage elements and Ler-binding loci were detected within the EEL of Sp9, Sp14, and SpLE4 (including LEE) (Fig. 4). Among EHEC O157-specific loci, the locus that contains genes for synthesis of polysaccharide moieties of lipopolysaccharide was bound by both Pch and Ler. Other locus that was covered by Pch was ETT2 locus encodes incomplete type III secretion system with truncated substrate genes. Fourteen loci encoding a set of fimbriae biosynthesis genes were identified on EHEC O157 Sakai chromosome. Pch was bound upstream part of 11 fimbriae operons and three of them were also bound by Ler. They include three loci that are conserved in K-12 strain and four loci that are partially conserved in K-12 strain.
Although most regions bound by Pch ranged from 0.3 to 0.8 kb, several loci showed binding that extended to as much as 3.0 kb. Of the 59 Ler-binding loci, 46 overlapped with the Pch-binding loci, and all the Ler-binding loci in S-loops also bound Pch. Because of the low resolution of ChIP-chip analysis, we could not identify a consensus sequence for the Pch-binding sites other than that they are of lower G + C content than average of E. coli chromosome. These results suggested that both Pch and Ler are DNA-binding proteins with a low binding specificity.
3.3. Correlation between gene transcription profiles and binding by Pch and/or Ler
We next considered whether there were differences in binding by Pch and Ler in the L1 and L2 gene classes. Although most of effector genes were transcriptionally activated by Pch, they were classified into two distinctive classes by transcription profiles. Ten effector genes in class L1—seven in the LEE island and three outside it—were regulated coordinately with other LEE genes that function in the biosynthesis of the type III secretion machinery. Both Pch and Ler bound to the upstream or coding sequences of all three L1 prophage-associated effector genes, nleA (ECs1812), espJ (ECs2714), and espF-u/tccP (ECs2715) (Fig. 4D and F), strongly suggesting the direct activation of their transcription through coordinate binding by Pch and Ler.
Twenty-one class L2 effector genes were located on seven prophage-like elements scattered throughout the chromosome. In contrast to class L1 effector genes, although Pch bound in and around the L2 effector gene sequences, Ler binding was not detected in many cases (Fig. 4B–E, G, and H). Positions of binding loci of Pch relative to effector genes vary among genes. For example, Pch bound upstream of nleC, nleA, nleG8-2, whereas binding to coding regions were observed for espX7, espN, espJ, and espL2. In the EEL of Sp10, two Pch-binding loci were found upstream and downstream of a cluster of three nleG genes. Three L2 effector genes, nleG5-2 (ECs2154), nleG6-2 (ECs2155), and nleG2-3 (ECs2156), on Sp11 bound Pch only at upstream of the gene cluster. Since the probes for nleG5-2, nleG6-2, and nleG2-3 used in the DNA microarray for transcriptome analysis were the same as for nleG5-1 (ECs1996), nleG6-1 (ECs1995), and nleG2-2 (ECs1994) on Sp11, respectively, it is likely that the signals we detected originated only from the genes on Sp10 and that the genes on Sp11 were less expressed. These binding data suggested that the effector genes in class L2 were directly activated by Pch without Ler.
Of the 57 genes in class L1, only three S-loop genes and two K-12 common genes showed no binding by PchA or Ler (Supplementary Table S1). Rather, 68.6% of the L1 S-loop genes bound both Pch and Ler, as did 50% of the L1 backbone or K-12 common genes (Fig. 5). Therefore, most class L1 genes, including those on the backbone, are probably directly regulated by Pch and Ler. However, among the L2 genes, more than half bound neither Pch nor Ler, suggesting that their transcription was regulated indirectly by these proteins. As shown for the effector genes, the non-effector class L2 genes that bound Pch, but not Ler, were much more abundant than those that bound both (Fig. 5 and Table 2). 66.6% of Pch-bound genes were not bound by Ler. This suggested that the transcriptional regulation of the class L2 genes was, overall, directly regulated by Pch without the involvement of Ler.
Figure 5.
Correlation between Pch and/or Ler binding with transcription profiles. The binding of Pch and Ler to loci in and around genes was determined as in Fig. 2 and Fig. 3. (A) The number of effector genes assigned to class L1 and class L2. Effector genes not in either class are labeled NA (not applicable). (B) The number of genes in classes L1, L2, and N. Genes in S-loops (EHEC O157 strain-specific regions) and in the E. coli backbone (regions conserved with E. coli K-12) are identified as O157 and K12, respectively. Binding of Pch and/or Ler is denoted as follows: grey, bound by Ler and Pch; black, bound by Pch, but not Ler; dark grey, bound by Ler, but not Pch; white, no binding by Pch or Ler.
Table 2.
ORFs in clusters L2 with binding sites for Pch/Ler
| ECs# | Gene/ORF | Product | Locus | Binding** |
|---|---|---|---|---|
| ECs0240 | Hypothetical membrane protein | S-loop13 | P + L | |
| ECs0241 | H repeat-associated protein | S-loop13 | P + L*** | |
| ECs0242 | Rhs core protein | S-loop13 | P + L *** | |
| ECs0243 | Hypothetical protein | S-loop13 | P + L*** | |
| ECs0244 | Hypothetical protein | S-loop13 | P + L*** | |
| ECs0245 | Putative H repeat-associated protein | S-loop13 | L | |
| ECs0293 | Hypothetical protein | S-loop19 | P + L | |
| ECs0294 | Hypothetical protein | S-loop19 | P + L | |
| ECs0295 | Hypothetical protein | S-loop19 | P + L | |
| ECs0815 | Antitermination protein | Sp3 | P + L | |
| ECs0816 | Hypothetical membrane protein | Sp3 | P + L*** | |
| ECs0846 | nleB2-1′ | NleB-like protein, pseudogene | Sp3 | P |
| ECs0848* | nleH1-1 | Effector protein NleH1 | Sp3 | P |
| ECs0849 | Hypothetical protein | Sp3 | P | |
| ECs0949 | Hypothetical protein | S-loop74 | P | |
| ECs1560* | espX7 | Effector protein EspX7 | Sp6 | P |
| ECs1561* | espN | Effector protein EspN | Sp6 | P |
| ECs1562 | Hypothetical protein | Sp6 | P*** | |
| ECs1567* | espO1-1 | Effector protein EspO1-1 | Sp6 | P |
| ECs1568* | espK | Effector protein EspK | Sp6 | P |
| ECs1814* | nleH1-2 | Effector protein NleH1-2 | Sp9 | P*** |
| ECs1815* | nleF | Effector protein NleF | Sp9 | P |
| ECs1821 | espO1-2 | Effector protein EspO1-2 | Sp9 | P + L |
| ECs1822 | Hypothetical protein | Sp9 | P + L | |
| ECs1823 | Hypothetical protein | Sp9 | P + L | |
| ECs1824* | nleG | Effector protein NleG | Sp9 | P + L*** |
| ECs1994/ECs2156* | nleG2-2/nleG2-3 | Effector protein NleG2-2/NleG2-3 | Sp10/Sp11 | P*** |
| ECs1995/ECs2155* | nleG6-1/nleG6-2 | Effector protein NleG6-1/NleG6-2 | Sp10/Sp11 | P*** |
| ECs1996/ECs2154* | nleG5-1/nleG5-2 | Effector protein NleG5-1/NleG5-2 | Sp10/Sp11 | P |
| ECs2701 | yedR | Predicted inner membrane protein | K12 | P + L |
| ECs2917 | yehD | Putative fimbrial-like protein | K12 | P + L |
| ECs3140 | ais | Aluminum-inducible protein | K12 | P |
| ECs3367 | b2505 | Putative outer membrane lipoprotein | K12 | L |
| ECs3486* | nleG8-2 | Effector protein NleG8-2 | Sp17 | P |
| ECs3487* | espW | Effector protein EspW | Sp17 | P |
| ECs3508 | Hypothetical protein | Sp17 | P | |
| ECs3509 | Hypothetical protein | Sp17 | P | |
| ECs3510 | Hypothetical protein | Sp17 | P | |
| ECs3511 | Hypothetical protein | Sp17 | P | |
| ECs3512 | Putative site-specific recombinase | Sp17 | P | |
| ECs3517 | ypjC | Hypothetical protein | K12 | P + L |
| ECs3616 | ybdY | Hypothetical protein | K12 | P + L |
| ECs3703 | yqeH | Hypothetical protein | K12 | P |
| ECs3713 | b2857 | Hypothetical protein | K12 | P |
| ECs3718 | eprI | Type III secretion protein EprI | ETT2 | P |
| ECs3720 | Putative transcriptional regulator | ETT2 | P | |
| ECs3721 | epaS | Type III secretion protein EprS | ETT2 | P |
| ECs3855* | espL2 | Effector protein EspL2 | SpLE3 | P |
| ECs3857* | nleB1 | Effector protein NleB1 | SpLE3 | P |
| ECs3858* | nleE | Effector protein NleE | SpLE3 | P |
| ECs4000 | yhaB | Hypothetical protein | K12 | P |
| ECs4001 | yhaC | Hypothetical protein | K12 | P |
| ECs4292 | Hypothetical membrane protein | S-loop87 | P | |
| ECs4569 | escV | T3SS EscV protein | LEE | P |
| ECs4589 | Hypothetical protein | LEE | P + L | |
| ECs4653* | espY4 | Effector protein EspY4 | S-loop39 | P |
| ECs4656 | Hypothetical protein | S-loop39 | P | |
| ECs4657 | espY5′ | Pseudogene | S-loop39 | P |
| ECs4658 | Hypothetical protein | S-loop39 | P | |
| ECs5314 | Hypothetical protein | S-loop | P |
*Gene encoding type-III effector protein (Tobe et al., 2006).
**P, Pch; L, Ler; P + L; Pch and Ler.
***Bound at upstream gene in operon.
Table 1.
ORFs in cluster L1 with binding sites for Pch/Ler
| ECs# | Gene/ORF | Product | Locus | Bound by*** |
|---|---|---|---|---|
| ECs0207 | dniR | Transcriptional regulator for nitrite reductase (cytochrome c552) | K12 | P |
| ECs0335 | Putative oxidoreductase | S-loop57 | P | |
| ECs0354 | Putative invertase | S-loop54 | P + L | |
| ECs0355 | Putative invertase | S-loop54 | P + L | |
| ECs0356 | Hypothetical protein | S-loop54 | P + L | |
| ECs0512 | ylaD | Maltose O-acetyltransferase | K12 | P + L**** |
| ECs0513 | hha | Hemolysin expression modulating protein | K12 | P + L**** |
| ECs0514 | ybaJ | Hypothetical protein | K12 | P + L |
| ECs0814 | Putative outer membrane protein | Sp3 | P + L | |
| ECs1126/2715* | espF-M/U | Type-III secretion effector EspF-U/TccP | Sp4/Sp14 | P + L**** |
| ECs1812* | nleA/espI | Type-III secretion effector NleA/EspI | Sp9 | P + L |
| ECs2714* | espJ | Type-III secretion effector EspJ | Sp14 | P + L |
| ECs3500 | Hypothetical membrane protein | Sp17 | P | |
| ECs4550* | espF | Effector protein EspF | LEE | L**** |
| ECs4551 | L0017 | T3SS protein | LEE | L**** |
| ECs4552 | escF | T3SS EscF protein | LEE | L**** |
| ECs4553 | cesD2 | T3SS chaperone CesD2 | LEE | L**** |
| ECs4554* | espB | Translocator protein EspB | LEE | L**** |
| ECs4555* | espD | Translocator protein EspD | LEE | L**** |
| ECs4556* | espA | Translocator protein EspA | LEE | L**** |
| ECs4557 | sepL | T3SS SepL protein | LEE | L |
| ECs4558 | escD | T3SS EscD protein | LEE | P + L |
| ECs4559 | eae | Gamma intimin | LEE | P + L**** |
| ECs4560 | cesT | T3SS chaperon CesT | LEE | P + L |
| ECs4562* | map | Effector protein Map | LEE | L |
| ECs4563 | cesF | T3SS chaperon CesF | LEE | P + L**** |
| ECs4564* | espH | Effector protein EspH | LEE | P + L**** |
| ECs4565 | sepQ | T3SS SepQ protein | LEE | P + L**** |
| ECs4566 | L0032 | Hypothetical protein | LEE | P + L**** |
| ECs4567 | L0033 | Hypothetical protein | LEE | P + L**** |
| ECs4568 | escN | T3SS protein EscN | LEE | P + L |
| ECs4570 | L0036 | Hypothetical protein | LEE | P + L |
| ECs4571* | sepZ | Effector protein SepZ/EspZ | LEE | P + L |
| ECs4572 | L0038 | Hypothetical protein | LEE | P + L**** |
| ECs4573 | escJ | T3SS EscJ protein | LEE | P + L**** |
| ECs4574 | sepD | T3SS protein SepD | LEE | P + L**** |
| ECs4575 | escC | T3SS EscC protein | LEE | P + L**** |
| ECs4576 | cesD | T3SS chaperone CesD | LEE | P + L**** |
| ECs4577 | grlA | Transcription regulator protein GrlA | LEE | P + L |
| ECs4578 | grlR | Transcription regulator protein GrlR | LEE | P + L |
| ECs4579 | L0045 | Hypothetical protein | LEE | P + L |
| ECs4580 | escU | T3SS EscU protein | LEE | P + L |
| ECs4581 | escT | T3SS EscT protein | LEE | P + L |
| ECs4582 | escS | T3SS EscS protein | LEE | P + L |
| ECs4583 | escR | T3SS EscR protein | LEE | P + L |
| ECs4584 | L0050 | Hypothetical protein | LEE | P + L |
| ECs4585 | L0051 | Hypothetical protein | LEE | P + L |
| ECs4586 | L0052 | Hypothetical protein | LEE | P + L |
| ECs4587 | L0053 | Hypothetical protein | LEE | P + L |
| ECs4590* | espG | Effector protein EspG | LEE | P + L |
*Gene encoding type-III effector or translocator protein.
**ECs4588 (ler) was not included because of artificially controlled expression.
***P, Pch; L, Ler; P + L, Pch and Ler.
****Bound at upstream gene in operon.
Although 146 genes were classified as class N, that is, their expression was negatively correlated with Pch and Ler activity, only 35% showed Pch and/or Ler binding in or near their DNA sequences (Fig. 5). These 40 backbone or K-12 common genes and 11 S-loop genes seemed to be directly regulated by Pch and/or Ler (Table 3). They include genes encoding regulators, yhiF, gadE, gadX, and gadW, and their regulating genes, gadB, hdeA, hdeB, and hdeD. Two genes, yhiF and gadE, inhibit the expression of LEE genes,27 suggesting there is a reciprocal negative interaction between the Pch–Ler virulence regulatory system and the YhiF–GadE regulatory system.
Table 3.
ORFs in cluster N with binding sites for Pch/Ler
| ECs# | Gene/ORF | Product | Locus | Binding* |
|---|---|---|---|---|
| ECs0079 | leuL | Leu operon leader peptide | K12 | P + L |
| ECs0324 | ykgK | Putative regulator | K12 | L |
| ECs0538 | ybaS | Putative glutaminase | K12 | P |
| ECs0539 | ybaT | Putative amino acid/amine transport protein | K12 | P |
| ECs1041 | ompA | Outer membrane protein 3a | K12 | P |
| ECs1272 | Rtn-like protein | S-loop | P | |
| ECs2603 | yecG | Putative regulator | K12 | P |
| ECs2712 | yodB | Putative cytochrome | K12 | P |
| ECs2846 | galF | Putative Galf transferase | K12 | P |
| ECs2974 | stx1A | Shiga toxin I subunit A precursor | Sp15 | P |
| ECs3104 | ompC | Outer membrane protein C OmpC | K12 | P + L |
| ECs3271 | b2392 | High affinity manganese transport protein | K12 | P + L |
| ECs3272 | nupC | Permease of transport system for 3 nucleosides | K12 | P + L |
| ECs3284 | zipA | Cell division protein involved in FtsZ ring | K12 | P |
| ECs3289 | crr | Glucose-specific PTS system IIA component | K12 | P |
| ECs4045 | pnp | Polynucleotide phosphorylase | K12 | P |
| ECs4112 | yhcO | Hypothetical protein | K12 | L |
| ECs4377 | slp | Huter membrane protein Slp | K12 | P + L |
| ECs4379 | Hypothetical protein | S-loop | P + L | |
| ECs4380 | Heme utilization/transport protein | S-loop | P + L | |
| ECs4389 | hdeB | Hypothetical protein | K12 | P + L |
| ECs4390 | hdeA | Hypothetical protein | K12 | P + L |
| ECs4391 | hdeD | Hypothetical protein | K12 | P + L |
| ECs4392 | yhiE | Hypothetical protein | K12 | P + L |
| ECs4393 | yhiU | Putative membrane protein | K12 | P + L |
| ECs4395 | gadW | Putative ARAC-type regulatory protein | K12 | P + L |
| ECs4396 | gadX | Putative ARAC-type regulatory protein | K12 | P + L |
| ECs4427 | Putative fimbrial protein precursor | S-loop | L |
*P, Pch; L, Ler; P + L, Pch and Ler.
3.4. Global regulation to promote the efficient expression of virulence traits
Pch is a 12-kDa protein without significant homology to any transcriptional regulatory proteins except PerC (also called BfpW),30,31 which is encoded on a virulence plasmid of enteropathogenic E. coli. The pch genes encode a transcriptional activator for the LEE1 promoter, and the deletion of pchA and pchB genes greatly reduces the production and secretion of EspB and EspA.8 The introduction of multiple copies of the pchA gene into EHEC O157 enhances the production and secretion of effector proteins, including many non-LEE encoded effectors,7 indicating that Pch is involved in the regulation of non-LEE effector genes as well as of LEE genes. Our study clearly demonstrated that Pch acts as a global regulator of many genes scattered throughout the EHEC O157 chromosome and plasmids. As expected, most effector genes in the EEL on prophage-like elements were positively regulated by Pch. The molecular mechanism of transcriptional activation by Pch may not be simple, since the position of its binding site varied from gene to gene, as did the number of binding sites. Therefore, Pch probably uses a transcriptional activating mechanism that is different from that of a typical transcriptional activator, like an AraC-like protein, which activates transcription by binding upstream of the promoter and interacting with RNA polymerase. The binding of Pch in the coding region or downstream of its target genes suggested that Pch affects promoter activity without directly interacting with RNA polymerase, most likely by inducing changes in the chromosomal structure around the promoter. We could not identify a consensus sequence for Pch binding, besides low G + C content, from the ChIP on chip data, because of low resolution. To characterize more precisely, further analysis such as footprinting must be required.
The analysis of transcription profiles revealed that the transcriptional responses of genes in class L1 to changes in the levels of Pch and Ler were slightly different from those of the class L2 genes. Ler binding may be required to activate most class L1 promoters, and these promoters were activated in the absence of Pch as efficiently as in its presence. Although the transcript levels of several L2 genes could be increased in a Pch-negative background by overexpressing Ler, the increase was much smaller than that obtained by the overexpression of Pch. It is likely that Ler was primarily a regulator for a restricted group of genes that are functionally related to the A/E phenotype as compared to Pch. It is supported by one of the non-LEE-encoded effectors, TccP/EspFu, which is required for actin polymerization at the cytoplasmic domain of Tir.22,23 In addition, the nleA (espI) gene of C. rodentium, a mouse A/E pathogen, is involved in the colonization of mouse colon and the induction of hyperplasia.21,32 Finally, since the ler gene is LEE gene, the Ler target genes might primarily be LEE genes.
In the process of establishing the pathogenicity of EHEC, genes encoding proteins with A/E phenotype-related functions might be selected and placed under the direct control of Ler. Pch seems to have a broader role in regulating gene expression, since its binding sites were scattered over the chromosome. EHEC O157:H7 Sakai possesses five pch genes, including at least two encoding active proteins and two truncated pseudogenes, on different prophage-like elements.8,13 Furthermore, multiple pch genes are found among EHEC O157 strains and other serotypes of EHEC strains, such as O26, O111, and O103 (Ogura et al., unpublished). These findings suggest that Pch is required for the expression of virulence genes and that the pch gene itself is rather unstable.
Horizontally transferred genes are thought to be initially repressed by the binding of H-NS to the DNA around them.18,33 Pch might counteract the effect of H-NS, both by activating the horizontally acquired genes and influencing the expression of genes in the backbone or K-12 common chromosome. The global effect of Pch on both EHEC-specific and K-12 common genes must be to promote the integration of the acquired genes with a virulence regulon and to govern the optimization of virulence expression. Because changing the expression levels of some K-12 common genes could have an adverse effect on bacterial physiology, the virulence-regulatory system must be strictly controlled and activated only when it is required. In fact, the evidence shows that the activation of the virulence-regulatory system is strictly regulated in response to environmental signals, through the expression of both pch and ler.12,13 Thus, the virulence-regulatory system involving Pch and Ler is a good model for understanding the process of the evolution of pathogenicity and the emergence of novel pathogens.
Supplementary data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas (C) from The Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Supplementary Material
References
- 1.Gal-Mor O., Finlay B. B. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 2.Kaper J. B., Nataro J. P., Mobley H. L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 3.Hayashi T., Makino K., Ohnishi M., et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 4.Frankel G., Phillips A. D., Rosenshine I., et al. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 5.Hayward R. D., Leong J. M., Koronakis V., et al. Exploiting pathogenic Escherichia coli to model transmembrane receptor signalling. Nat. Rev. Microbiol. 2006;4:358–370. doi: 10.1038/nrmicro1391. [DOI] [PubMed] [Google Scholar]
- 6.Deng W., Puente J. L., Gruenheid S., et al. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tobe T., Beatson S. A., Taniguchi H., et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. USA. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyoda S., Watanabe H. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology. 2004;150:2357–2571. doi: 10.1099/mic.0.27100-0. [DOI] [PubMed] [Google Scholar]
- 9.Elliott S. J., Sperandio V., Giron J. A., et al. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bustamante V. H., Santana F. J., Calva E., et al. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 2001;39:664–678. doi: 10.1046/j.1365-2958.2001.02209.x. [DOI] [PubMed] [Google Scholar]
- 11.Abe H., Tatsuno I., Tobe T., et al. Bicarbonate ion stimulates the expression of locus of enterocyte effacement-encoded genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 2002;70:3500–3509. doi: 10.1128/IAI.70.7.3500-3509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tobe T., Ando H., Ishikawa H., et al. Dual regulatory pathways integrating the RcsC–RcsD–RcsB signalling system control enterohaemorrhagic Escherichia coli pathogenicity. Mol. Microbiol. 2005;58:320–333. doi: 10.1111/j.1365-2958.2005.04828.x. [DOI] [PubMed] [Google Scholar]
- 13.Nakanishi N., Abe H., Ogura Y., et al. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol. Microbiol. 2006;61:194–205. doi: 10.1111/j.1365-2958.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- 14.Umanski T., Rosenshine I., Friedberg D. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology. 2002;148:2735–2744. doi: 10.1099/00221287-148-9-2735. [DOI] [PubMed] [Google Scholar]
- 15.Datsenko K. A., Wanner B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uzzau S., Figueroa-Bossi N., Rubino S., et al. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogura Y., Kurokawa K., Ooka T., et al. Complexity of the genomic diversity in enterohemorrhagic Escherichia coli O157 revealed by the combinational use of the O157 Sakai Oligo-DNA microarray and the whole genome PCR scanning. DNA Res. 2006;13:3–14. doi: 10.1093/dnares/dsi026. [DOI] [PubMed] [Google Scholar]
- 18.Oshima T., Ishikawa S., Kurokawa K., et al. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 19.Friedberg D., Umanski T., Fang Y., et al. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 1999;34:941–952. doi: 10.1046/j.1365-2958.1999.01655.x. [DOI] [PubMed] [Google Scholar]
- 20.Sharp F. C., Sperandio V. QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect. Immun. 2007;75:2432–2440. doi: 10.1128/IAI.02003-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mundy R., Petrovska L., Smollett K., et al. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect. Immun. 2004;72:2288–2302. doi: 10.1128/IAI.72.4.2288-2302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campellone K. G., Robbins D., Leong J. M. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell. 2004;7:217–228. doi: 10.1016/j.devcel.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Garmendia J., Phillips A. D., Carlier M. F., et al. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell Microbiol. 2004;6:1167–1183. doi: 10.1111/j.1462-5822.2004.00459.x. [DOI] [PubMed] [Google Scholar]
- 24.Garmendia J., Ren Z., Tennant S., et al. Distribution of tccP in clinical enterohemorrhagic and enteropathogenic Escherichia coli isolates. J. Clin. Microbiol. 2005;43:5715–5720. doi: 10.1128/JCM.43.11.5715-5720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garmendia J., Frankel G. Operon structure and gene expression of the espJ–tccP locus of enterohaemorrhagic Escherichia coli O157:H7. FEMS Microbiol. Lett. 2005;247:137–145. doi: 10.1016/j.femsle.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 26.Sharma V. K., Zuerner R. L. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 2004;186:7290–7301. doi: 10.1128/JB.186.21.7290-7301.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatsuno I., Nagano K., Taguchi K., et al. Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 2003;71:2598–2606. doi: 10.1128/IAI.71.5.2598-2606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyoda S., Watanabe H. ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J. Bacteriol. 2005;187:4086–4094. doi: 10.1128/JB.187.12.4086-4094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadler C., Shifrin Y., Nov S., et al. Characterization of enteropathogenic Escherichia coli mutants that fail to disrupt host cell spreading and attachment to substratum. Infect. Immun. 2006;74:839–849. doi: 10.1128/IAI.74.2.839-849.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Duarte O. G., Kaper J. B. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobe T., Schoolnik G. K., Sohel I., et al. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 1996;21:963–975. doi: 10.1046/j.1365-2958.1996.531415.x. [DOI] [PubMed] [Google Scholar]
- 32.Gruenheid S., Sekirov I., Thomas N. A., et al. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 2004;51:1233–1249. doi: 10.1046/j.1365-2958.2003.03911.x. [DOI] [PubMed] [Google Scholar]
- 33.Navarre W. W., Porwollik S., Wang Y., et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.