Abstract
The genome of Sorangium cellulosum has recently been completely sequenced, and it is the largest bacterial genome sequenced so far. In their report, Schneiker et al. (in Complete genome sequence of the myxobacterium Sorangium cellulosum, Nat. Biotechnol., 2007, 25, 1281–1289) concluded that ‘In the absence of the GC-skew inversion typically seen at the replication origin of bacterial chromosomes, it was not possible to discern the location of oriC’. In addition, the complete genome of Microcystis aeruginosa NIES-843 has also been recently sequenced, and in this report, Kaneko et al. (in Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843, DNA Res., 2007, 14, 247–256) concluded that ‘there was no characteristic pattern, according to GC skew analysis’. Therefore, oriC locations of the above genomes remain unsolved. Using Ori-Finder, a recently developed computer program, in both genomes, we have identified candidate oriC regions that have almost all sequence hallmarks of bacterial oriCs, such as asymmetrical nucleotide distributions, being adjacent to the dnaN gene, and containing DnaA boxes and repeat elements.
Key words: Sorangium cellulosum, Microcystis aeruginosa, origin of replication, Z-curve
Replication of chromosomes is one of the central events in the cell cycle. Identification of replication origin in a genome is important not only in understanding the mechanisms of DNA replication but also in gaining insights into the structure and function of the genome. In bacteria, chromosome replication initiates at a single chromosome locus, called the replication origin (oriC), from which replication proceeds bidirectionally to the terminus. At the beginning of replication, ATP binds DnaA, resulting in a large oligomeric complex consisting of DnaA monomers bound to a series of 9-mer consensus elements termed DnaA boxes.1 Typical bacterial oriCs have many conserved sequence features, including (i) having single oriC in an intergenic region, (ii) having asymmetrical nucleotide distributions around oriCs, (iii) sequence between oriC and terminus being about half in length of the entire chromosome, (iv) containing multiple copies of DnaA boxes, (v) close to replication related genes, such as dnaA or dnaN, and (vi) containing repeat sequences.
The genome of Sorangium cellulosum has recently been completely sequenced.2 In their report, Schneiker et al.2 concluded that ‘In the absence of the GC-skew inversion typically seen at the replication origin of bacterial chromosomes, it was not possible to discern the location of oriC’. Additionally, we also note that the complete genome of Microcystis aeruginosa NIES-843 has been determined recently.3 Kaneko et al. concluded that ‘there was no characteristic pattern, according to GC skew analysis’.3 Therefore, oriC locations of the above genomes remain unsolved.
To identify oriC regions of unannotated bacterial genomes, we recently developed an online tool, Ori-Finder, based on an integrated method comprising de novo gene identification, the Z-curve method,4 distribution of DnaA boxes, occurrence of gene frequently close to oriCs and phylogenetic relationships.5
Using this software, in the genome of S. cellulosum, we have identified an oriC, which is within an intergenic region between a kinase gene (sce8163) and the dnaN gene, rather than the dnaA gene, from 11 354 923 to 11 355 551 nt of the genome. Around this oriC, there are clear asymmetrical base distributions of A/T, G/C, M/K, and R/Y (Fig. 1A). The DnaA box motif is TTATCCCCC, probably due to the high genomic GC content (71.4%), rather than TTATCCACA, the DnaA box motif of E. coli. The dif-like sequence (GGATCGCATAAGAAACATTATGTCAACT) has been found between 5 024 594 and 5 024 621 nt, which matches 20 sites compared with the 28-nt E. coli dif sequence (GGTGCGCATAATGTATATTATGTTAAAT), which is usually present in replication termini. Consequently, the sequence lengths between the predicted oriC and dif-like sequence are about 6 331 kb (48.6%) and 6 703 kb (51.4%), each of which is equal roughly to half of the genome size. The oriC regions usually contain multiple copies of repeat sequences, which are generally believed to facilitate the binding of the complex of enzymes to these DNA sequences.6 In the oriC of S. cellulosum, we found four copies of perfect reverse repeats using the software REPuter7 (Fig. 1B). Therefore, it is very likely that the intergenic region between sce8163 and dnaN genes, which has almost all the hallmarks of a bacterial oriC, is the replication origin of S. cellulosum. Note that the asymmetrical nucleotide distribution around oriC region of S. cellulosum can also be discerned by performing the cumulative GC skew analysis.
Figure 1.
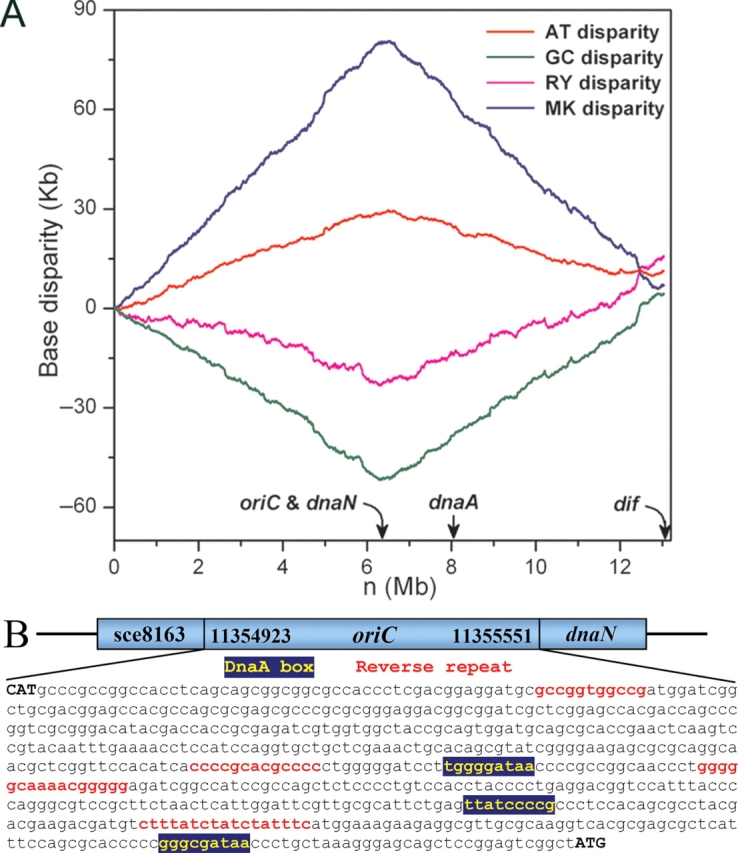
(A) AT, GC, RY, and MK disparity curves for the rotated S. cellulosum genome sequence beginning and ending in the dif site. The locations of dnaA, dnaN, oriC, and the dif site are indicated by arrows. (B) Schematic diagram of the replication origin of S. cellulosum. The oriC is located in the intergenic region between a kinase gene (ID: sce8163) and the dnaN gene, from 11 354 923 to 11 355 551 nt of the genome. Within this region, there are four copies of perfect reverse repeats (red) and three DnaA boxes (yellow).
Based on Ori-Finder,5 we also identified a candidate oriC region in M. aeruginosa. The oriC is within an intergenic region between the dnaN and the hemL genes, from 3 542 737 to 3 543 291 nt of the genome. The DnaA box motif is TTTTCCACA rather than TTATCCACA. The features that the ‘species-specific’ DnaA box is TTTTCCACA, and oriC is adjacent to the dnaN gene are universal for the bacteria of the phylum Cyanobacteria, such as Prochlorococcus marinus, Synechococcus, etc. For more details, visit DoriC, a database of oriC regions in bacterial genomes,8 which is available at http://tubic.tju.edu.cn/doric/. In the oriC of M. aeruginosa, we also found two copies of perfect palindromic repeats using the software REPuter (Fig. 2). Therefore, it is very likely that the intergenic region between the dnaN and the hemL genes, which has common oriC features among the bacteria of the phylum Cyanobacteria, is the replication origin of M. aeruginosa.
Figure 2.
Schematic diagram of the replication origin of M. aeruginosa. The oriC is located in the intergenic region between the dnaN and the hemL gene, from 3 542 737 to 3 543 291 nt of the genome. Within this region, there are two copies of perfect palindromic repeats (blue) and three DnaA boxes (yellow).
Funding
The present work was supported in part by NNSF of China (Grant No. 90408028 to CT Zhang and 10747150 to F Gao). Funding for open access charge is supported by the National Natural Science Foundation of China (NNSF).
Acknowledgments
We would like to thank Dr. Ren Zhang for invaluable assistance. We are also indebted to both referees for their constructive comments, which are critical for improving the quality of the paper.
References
- 1.Robinson N. P., Bell S. D. Origins of DNA replication in the three domains of life. FEBS J. 2005;272:3757–3766. doi: 10.1111/j.1742-4658.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- 2.Schneiker S., Perlova O., Kaiser O., et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 2007;25:1281–1289. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- 3.Kaneko T., Nakajima N., Okamoto S., et al. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. 2007;14:247–256. doi: 10.1093/dnares/dsm026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang R., Zhang C. T. Identification of replication origins in archaeal genomes based on the Z-curve method. Archaea. 2005;1:335–346. doi: 10.1155/2005/509646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao F., Zhang C. T. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinform. 2008;9:79. doi: 10.1186/1471-2105-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chew D. S., Choi K. P., Leung M. Y. Scoring schemes of palindrome clusters for more sensitive prediction of replication origins in herpesviruses. Nucleic Acids Res. 2005;33:e134. doi: 10.1093/nar/gni135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurtz S., Choudhuri J. V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao F., Zhang C. T. DoriC: a database of oriC regions in bacterial genomes. Bioinformatics. 2007;23:1866–1867. doi: 10.1093/bioinformatics/btm255. [DOI] [PubMed] [Google Scholar]



