Abstract
Lactobacillus reuteri is a heterofermentative lactic acid bacterium that naturally inhabits the gut of humans and other animals. The probiotic effects of L. reuteri have been proposed to be largely associated with the production of the broad-spectrum antimicrobial compound reuterin during anaerobic metabolism of glycerol. We determined the complete genome sequences of the reuterin-producing L. reuteri JCM 1112T and its closely related species Lactobacillus fermentum IFO 3956. Both are in the same phylogenetic group within the genus Lactobacillus. Comparative genome analysis revealed that L. reuteri JCM 1112T has a unique cluster of 58 genes for the biosynthesis of reuterin and cobalamin (vitamin B12). The 58-gene cluster has a lower GC content and is apparently inserted into the conserved region, suggesting that the cluster represents a genomic island acquired from an anomalous source. Two-dimensional nuclear magnetic resonance (2D-NMR) with 13C3-glycerol demonstrated that L. reuteri JCM 1112T could convert glycerol to reuterin in vivo, substantiating the potential of L. reuteri JCM 1112T to produce reuterin in the intestine. Given that glycerol is shown to be naturally present in feces, the acquired ability to produce reuterin and cobalamin is an adaptive evolutionary response that likely contributes to the probiotic properties of L. reuteri.
Key words: Lactobacillus reuteri, Lactobacillus fermentum, reuterin, cobalamin, genome
1. Introduction
Lactobacillus reuteri is a heterofermentative lactic acid bacterium and is frequently found in the gastrointestinal tract of humans and other animals. According to the World Health Organization (WHO), probiotics are defined as live microorganisms which when administered in adequate amounts confer a health benefit on the host. Lactobacillus reuteri has been reported to exhibit “probiotic” properties.1,2 It has been extensively analyzed for probiotic applications, including its safe administration to healthy individuals, its ability to colonize the intestine, as a diarrhea therapeutic agent, as an inhibitor of bacterial pathogens, and the immunological modulation of the gastrointestinal mucosa.3–8 Some strains of L. reuteri have the ability to produce and excrete the broad-spectrum antimicrobial compound reuterin, which is structurally identical to 3-hydroxypropionaldehyde (3-HPA), during anaerobic metabolism of glycerol.9–11 The probiotic effects of L. reuteri have been proposed to be largely associated with the production of reuterin, and this antimicrobial substance is also an effective food preservative agent.12,13 This attribute of reuterin-producing L. reuteri strains is why they are frequently used as probiotic cultures in commercial dairy products worldwide.13
The molecular basis for the probiotic effects of L. reuteri is not yet established. Although reuterin production is proposed to be important for its probiotic effects, the in vivo production of reuterin by L. reuteri has not been demonstrated. Lactobacillus fermentum is a closely related species based on a sequence analysis of the 16S ribosomal RNA gene (95% identity)14 and on phenotypic properties, including being an obligate heterofermentative organism.15 Previously, both these species were classified as a single species, L. fermentum, but were subsequently separated based primarily on DNA hybridization and GC content.15 Despite many similar phenotypic characteristics, probiotic effects have primarily been observed for L. reuteri, with a few studies suggesting probiotic properties for L. fermentum.16–18 To provide the framework for a comprehensive examination of the probiotic mechanisms of L. reuteri, we determined the complete genome sequences of the reuterin-producing L. reuteri JCM 1112T, which was isolated from human feces, and L. fermentum IFO 3956, which was isolated from fermented plant material, and compared them to the genomes of other available Lactobacillus genomes.19–26 Furthermore, we present in vivo evidence for the production of reuterin in the intestine of BALB/c mice.
2. Materials and methods
2.1. Genome sequencing
Lactobacillus reuteri JCM 1112T, which was originally isolated from human feces, is a type of strain and was obtained from the Japan Collection of Microorganisms (JCM), Riken, Japan. Lactobacillus fermentum IFO 3956, which was originally isolated from fermented plant material, was obtained from the Institute for Fermentation, Osaka (IFO), Japan. Each strain was cultured in MRS broth (Oxoid) at 37°C for 18 h, and the genomic DNA was isolated and purified using standard genomic DNA affinity columns. The nucleotide sequences of the L. reuteri JCM 1112T and L. fermentum IFO 3956 genomes were determined by a whole-genome shotgun strategy. We constructed small-insert (2 kb) and large-insert (5 kb) genomic DNA libraries. Template DNA for sequencing was prepared by amplification of the DNA inserts using PCR with Ex-Taq (Takara Bio Inc.). Sequencing was subsequently carried out using the dye terminator strategy before precipitation with ethanol and running on ABI 3730xl (Applied Biosystems) and MegaBACE 4500 (GE Healthcare) capillary sequencers. We generated 27 456 reads (7.4-fold coverage) for JCM 1112T and 35 136 reads (9.5-fold coverage) for IFO 3956, respectively, from both ends of the genomic clones. Data were assembled with the Phred-Phrap-Consed program,27 and gaps were closed by direct sequencing of clones that spanned the gaps or of PCR products amplified with oligonucleotide primers designed to anneal to each end of neighboring contigs. The overall accuracy of the finished sequence was estimated to have an error rate of <1 per 10 000 bases (PHRAP score of ≥40).
2.2. Sequence analysis
An initial set of open reading frames (ORFs) that likely encode proteins was identified using Glimmer 2.028 and overlapped ORFs, and those shorter than 120 bp were eliminated. All predicted proteins were searched against a non-redundant protein database (nr, NCBI) using BLASTP. The start codon of each ORF was manually refined from BLASTP alignments. The tRNA genes were predicted by the tRNAscan-SE29 and the rRNA genes were detected using BLASTN in a database of known bacterial RNAs. Functional classification of ORFs was conducted using the NCBI clusters of orthologous groups (COG) and LaCOG databases.25,30 Protein domains were identified using the Pfam database.31 The codon adaptation index for each protein was predicted using the cai program within the EMBOSS package32 based on the codon usage preference of genes encoding ribosomal proteins. Sequences were aligned with ClustalW, and BLASTP was used for protein comparisons. Orthologous genes were identified as the reciprocal best matches using BLASTP. The data were parsed by custom Perl script. Genome sequences of L. reuteri strains DSM 20016T (accession no. CP000705) and 100-23 (accession no. AAPZ00000000), which were determined by Joint Genome Institute (JGI), were obtained from GenBank. The genome sequence data of L. reuteri JCM 1112T and L. fermentum IFO 3956 have been deposited in GenBank/DDBJ/EMBL under accession nos. AP007281 and AP008937, respectively.
2.3. Gnotobiotic mice assays
Lactobacillus reuteri JCM 1112T and LRΔgupCDE were cultured on MRS agar at 37°C for 3 days before pelleting and resuspending in phosphate-buffered saline (pH 7.2). BALB/c female mice (13 weeks old and 20–23 g) raised under germ-free conditions in the Gnotobiote Laboratory, University of Tokyo, were used.33 Three germ-free mice were inoculated orally with 0.5 mL of a suspension of each bacterial strain (108 cfu/mL). The mice bacterial counts of JCM 1112T and LRΔgupCDE were 107 cfu/g feces. To detect in vivo reuterin production from L. reuteri JCM 1112T in mono-associated mice, an intestinal loop assay was conducted under anesthetic.34 The front and back of the cecum was bound with twine, and 100 μL of 2 M 13C3-glycerol solution was injected. After 3 h, each mouse was killed by cervical vertebra dislocation and the cecal contents were collected.
2.4. Analysis of glycerol metabolism in L. reuteri JCM 1112T using 13C3-labeled glycerol and 2D-NMR
Lactobacillus reuteri JCM 1112T and the mutant strain LRΔgupCDE were grown in 30 mL serum vials containing 20 mL of MRS broth with 200 mM 13C3-glycerol. Cultures were incubated in duplicate at 37°C for 10 h before 1 mL of each was sub-inoculated into new tubes for another 2.5 h incubation. An 0.1 mL aliquot of D2O was then added for 2D-NMR analysis. To measure the in vivo glycerol metabolism, metabolites were extracted with 0.5 mL of CD3OD from 1 g (wet weight) of JCM 1112T and LRΔgupCDE mono-associated murine cecal contents, as described previously.35 All NMR spectra of CD3OD-extracted sample solutions were recorded on a Bruker DRU-700 spectrometer equipped with a Z-axis gradient and a Bruker DRX-500 spectrometer equipped with a triple axis gradient. The temperature of the NMR samples was maintained at 298 K. A total of 128 complex f1 (13C) and 1024 complex f2 (1H) points were recorded with 32 scans per f1 increment for 2D-13C-HSQC36 and 2D-13C-edited HSQC37 spectra. The spectral widths were 12 000 and 8400 Hz for f1 and f2, respectively. In order to confirm signal assignments by 3JHH connectivity's, 2D-13C-HSQC-TOCSY spectra were recorded by a total of 64 complex f1 (13C) and 1024 complex f2 (1H) points using 32 scans per f1 increment.38 To quantify the signal intensities, a Lorentzian-to-Gaussian window with a Lorentzian line width of 10 Hz and a Gaussian line width of 15 Hz was applied in both dimensions, prior to Fourier transformation. A fifth-order polynomial baseline correction was subsequently applied in the f1 dimension. The indirect dimension was zero-filled to 2048 points in the final data matrix. 13C3-Glycerol was purchased from Cambridge Isotope Laboratories, Inc., and 1,3-propanediol and 3-hydroxypropionic acid were purchased from Tokyo Chemical Industry Co., Ltd.
3. Results and discussion
3.1. Genome features of L. reuteri JCM 1112T and L. fermentum IFO 3956
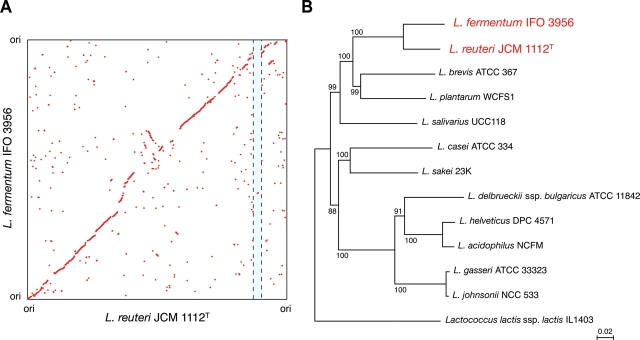
The genomes of L. reuteri JCM 1112T and L. fermentum IFO 3956 each consist of single circular chromosomes of 2 039 414 and 2 098 685 bp, respectively, and both have no plasmids (Supplementary Fig. S1 ). The general features of both genome sequences are summarized in Table 1. The genomes of L. reuteri JCM 1112T and L. fermentum IFO 3956 contain 1820 and 1844 ORFs, respectively, and both shared 1265 ORFs. A high level of synteny is conserved between the two genomes (Fig. 1A). The close phylogenetic relationship is also evident from a comparative analysis of the ribosomal protein sequences from all lactobacilli, whose genomes have been sequenced, and thus the two species constitute a distinct group (Fig. 1B). Despite the close phylogenic relationship of L. reuteri and L. fermentum, the L. fermentum IFO 3956 genome has a significantly higher GC content (52%) than that of the L. reuteri JCM 1112T genome (39%) and the highest among the sequenced Lactobacillus genomes. This can be explained primarily by the GC content at the third codon position between L. reuteri JCM 1112T and L. fermentum IFO 3956 (32 versus 65%; Table 1).
Table 1.
General features of the L. reuteri and L. fermentum genomes
| L. reuteri JCM 1112T | L. fermentum IFO 3956 | |
|---|---|---|
| Chromosome size (bp) | 2 039 414 | 2 098 685 |
| GC content (%) | 38.9 | 51.5 |
| Total ORFs | 1820 | 1844 |
| Functionally assigned | 1211 | 1212 |
| Conserved hypothetical | 413 | 360 |
| Non-conserved hypothetical | 196 | 272 |
| GC content at each codon position (%) | ||
| First position | 51.4 | 56.2 |
| Second position | 35.3 | 38.0 |
| Third position | 32.0 | 64.7 |
| Coding density (%) | 83.6 | 80.4 |
| tRNAs | 58 | 54 |
| rRNA operons | 6 | 5 |
| Phage-related ORFs | 53 | 24 |
| Transposases | 55 | 106 |
| Group II introns | 12 | 0 |
Figure 1.
Genome-based phylogenetic analysis of L. reuteri JCM 1112T and L. fermentum IFO 3956. (A) Synteny between the L. reuteri JCM 1112T and L. fermentum IFO 3956 chromosomes. Each dot represents an orthologous gene that was defined by bidirectional best hits based on BLASTP comparisons. Genome numbering was initiated at dnaA in both chromosomes. The shaded region indicates the location of the pdu-cbi-cob-hem cluster in the L. reuteri JCM 1112T genome. (B) Phylogenetic relationships between the genomes of sequenced Lactobacillus species and other lactic acid bacteria, including Lactococcus lactis, inferred from 34 concatenated ribosomal protein amino acid sequences. The scale bar represents an evolutionary distance. Sequences were aligned with ClustalW with a bootstrap trial of 1000 and bootstrap values (%) are indicated at the nodes. An unrooted tree was generated using NJplot.
During the preparation of this manuscript, the genome sequence (1 999 618 bp) of another L. reuteri strain DSM 20016T was deposited in public databases, and this strain also produces reuterin. It was expected that there would be minimum sequence differences between the two strains, because both are derived from the same original isolate, L. reuteri F275.39 However, an alignment of both genome sequences shows two large unique regions in L. reuteri JCM 1112T that result in a 40-kb increase in its genome (Supplementary Table S1). This highlights the tendency of bacteria to undergo significant genome changes during regular laboratory culturing and transfers. One unique region (JCM 1112T genome coordinates 442 995 to 451 429; unique region I) consists of 8435 bp and is flanked by a copy of IS4 on either end (LAR_0380 and LAR_0385), and the remnant (155 bp) of IS4 is present between orthologous genes (Lreu_0389 and Lreu_0390) at this position in the DSM 20016T genome. It is difficult to imagine that a pure culture stock picks up additional DNA, and thus DSM 20016T might have lost these genes. The unique region I contains the genes for enzymes involved in glycolysis (Supplementary Table S1). The other unique region (JCM 1112T genome coordinates 1 064 161 to 1 094 397; unique region II) consists of 30 237 bp and is bordered by an IS4 element (LAR_935) at one end and an IS200 element (LAR_959) on the other, both of which are also present at this position (Lreu_1000 and Lreu_1004) in the DSM 20016T genome. The unique region II contains the gene cluster encoding nitrate reductase subunits and the genes involved in molybdopterin biosynthesis (Supplementary Table S1). This gene cluster (LP_1473-1500) is also conserved in the genome of Lactobacillus plantarum WCFS1,19 and it is one of the variable regions in L. plantarum strains.40
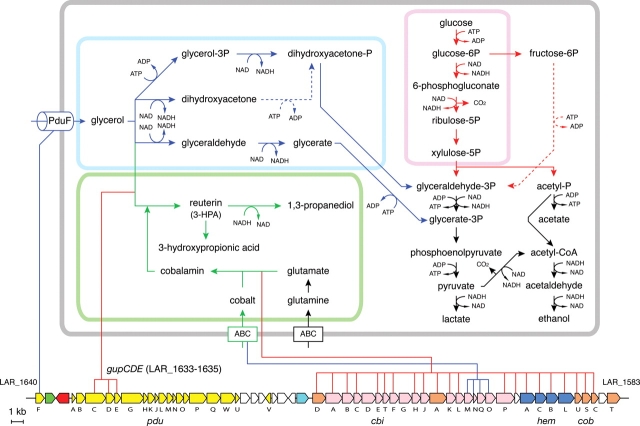
Biological roles were assigned to 1211 (67%) of the predicted ORFs for L. reuteri JCM 1112T, and 1212 (66%) for L. fermentum IFO 3956. Obligately heterofermentative lactobacilli, such as L. reuteri, L. fermentum, and Lactobacillus brevis, produce CO2, ethanol, acetate, and lactate from metabolism of glucose, whereas facultative heterofermentative and obligately homofermentative lactobacilli produce only lactate. The primary catabolic pathway for glucose metabolism in Lactobacillus is believed to be the glycolytic pathway. Surprisingly, the pfk gene encoding 6-phosphofructokinase is absent from the L. reuteri JCM 1112T and L. fermentum IFO 3956 genomes, and the fba gene for fructose-bisphosphate aldolase is also missing in IFO 3956. The pfk and fba genes are essential for the glycolytic pathway. The L. brevis genome also lacks the pfk and fba genes,25 suggesting that pfk and fba are key genes distinguishing homo- and hetero-fermentation. On the other hand, both have the complete gene set for the pentose phosphate pathway. These findings indicate that L. reuteri JCM 1112T and L. fermentum IFO 3956 use the pentose phosphate pathway to metabolize glucose (Fig. 2). Of all the ORFs in both genomes, 181 (10%) in JCM 1112T and 187 (10%) in IFO 3956 have no homolog in the other sequenced Lactobacillus genomes. Most of these genes encode conserved hypothetical proteins, but some with predicted functions are present. Genes encoding cobalamin biosynthesis are found only in JCM 1112T (described later) and genes encoding allantoate amidohydrolase (LAF_0160), isocitrate dehydrogenase (LAF_0939), isopropylmalate isomerase (LAF_0963-0965), and nitric oxide reductase (LAF_1644) are found only in IFO 3956. A COG analysis showed that L. reuteri JCM 1112T and L. fermentum IFO 3956 contain fewer glycosidases and no sugar ATP-binding cassette transporter when compared with other lactobacilli (data not shown). JCM 1112T contains the phosphoenolpyruvate:carbohydrate phosphotransferase systems (PTS) subunit IIC for cellobiose (LAR_0225) and galactitol (LAR_1559), but completely lacks other PTS enzyme II subunits. IFO 3956 has only two complete PTS enzyme II complexes for mannose (LAF_0391-0393) and sucrose (LAF_1579), but its gene-encoding mannose-6-phosphate isomerase is disrupted. Lactobacillus reuteri JCM 1112T and L. fermentum IFO 3956 apparently have a reduced capacity to utilize carbohydrates compared with other lactobacilli.
Figure 2.
Proposed glycerol and glucose metabolic pathways in L. reuteri JCM 1112T. The pathways are color coded as follows: blue, glycerol metabolism; green, biosynthesis of reuterin; red, glucose metabolism. Dashed lines indicate an unidentified enzyme in L. reuteri JCM 1112T. The bottom part of the figure shows the structure of the pdu-cbi-cob-hem gene cluster in L. reuteri JCM 1112T. Genes are depicted with arrows indicating the transcription direction with the following colors: yellow, pdu including gupCDE genes; pink, cbi genes; orange, cob genes; blue, hem genes; red, pocR; green, eut; sky blue, transposase gene; and white, other genes. Several lines connect corresponding genes between the pathway and the cluster (enzymes, red; transporters, blue).
3.2. Gene cluster for reuterin and cobalamin production in L. reuteri JCM 1112T
Synthesis of reuterin is mediated by glycerol dehydratase (EC 4.2.1.30), which catalyzes the conversion of glycerol to 3-HPA, and this enzyme exists as a dimer of the three subunits α2, β2, and γ2 that exhibit significant sequence and structural similarities to the corresponding subunits of propanediol dehydratase.41 The glycerol dehydratase of some L. reuteri strains has been purified and characterized,42 but not at the sequence level. JCM 1112T contains three genes (LAR_1633-1635) with dehydratase subunit motifs (Pfam PF02286-02288) in the propanediol utilizing operon (LAR_1616-1640) (Fig. 2). The encoded proteins of these three genes showed sequence similarity to PduCDE (propanediol dehydratase) of Salmonella typhimurium43 and thus the three genes were designated gupCDE (glycerol utilization gene candidates in the pdu operon) in JCM 1112T. The pdu operon from Lactobacillus collinoides has been sequenced,44 and it is also present in the L. brevis genome,25 but the enzymatic activity has not been examined. Homologues for PduCDE have also been reported for Lactobacillus hilgardii and Lactobacillus diolivorans.45 A comparison of the pdu operon in L. reuteri JCM 1112T with L. brevis and L. collinoides reveals a conserved gene order (Supplementary Fig. S2). The encoded amino acid sequences of the gupCDE genes exhibit the highest identity to their homologues from L. brevis ATCC 367, showing 81, 66, and 57% identities, respectively (Supplementary Fig. S2). There is no synteny between the flanking region of the pdu operon of L. reuteri JCM 1112T and L. brevis ATCC 367.
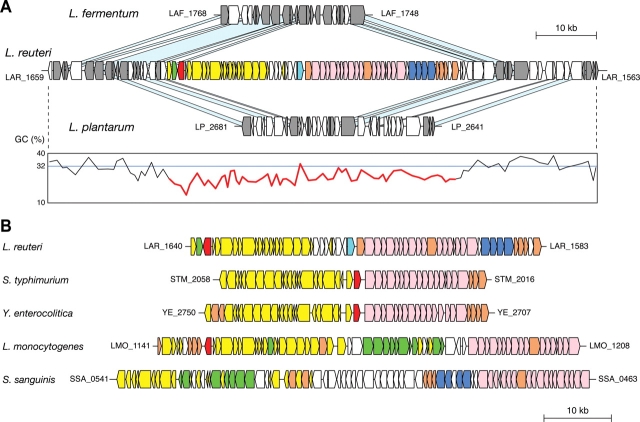
Both propanediol and glycerol dehydratase enzymes require cobalamin (vitamin B12) as a cofactor.41 JCM 1112T possesses the gene sets (cbi, cob, and hem) for cobalamin biosynthesis, and these are located adjacent to the pdu operon (Fig. 2). The structure of this gene cluster represents a close association of two operons (pdu and cbi-cob), and this is probably a reflection of the cobalamin requirement for glycerol dehydratase activity.41,46 These genes for cobalamin biosynthesis have not been identified in other lactobacilli including L. brevis. A comparison of this genetic locus with the corresponding locus in L. fermentum IFO 3956 and L. plantarum WCFS1 revealed that this pdu-cbi-cob-hem gene cluster comprising 58 ORFs (LAR_1583-1640), including 20 pdu genes (including gupCDE), 17 cbi genes, 6 cob genes, 4 hem genes, 1 pocR gene, 1 putative transposase gene, and 9 non-conserved ORFs, was apparently inserted into a locus that is common to all three bacteria (Fig. 3A). The pocR gene encodes a positive regulator of both the pdu and cbi-cob operons.47 The average GC content (25%) at the third codon of the ORFs in this gene cluster is significantly lower than that (32%) of the remaining ORFs in the JCM 1112T genome (Fig. 3A). In addition, there is a putative transposase gene (LAR_1612) at the boundary between the pdu and cbi-cob-hem operons (Fig. 2) and four IS elements within 20 kb of both the flanking regions of the pdu-cbi-cob-hem cluster in JCM 1112T. These data strongly suggest that the pdu-cbi-cob-hem cluster in L. reuteri JCM 1112T is a genomic island that has been acquired through lateral gene transfer. There is no direct repeat or tRNA gene in the flanking regions of this cluster, and the mechanism of the acquisition of this island is unclear. This finding is further supported from a comparison with the draft genome sequence data (total, 2.1 Mb) of L. reuteri 100-23, a rodent-specific strain, in which the pdu-cbi-cob-hem gene cluster is absent.
Figure 3.
(A) A comparison of the genomic location that contains the pdu-cbi-cob-hem gene cluster of L. reuteri JCM 1112T (center) with the corresponding location of L. fermentum IFO 3956 (upper) and L. plantarum WCFS1 (lower). Genes in the pdu-cbi-cob-hem gene cluster are depicted by arrows indicating the transcription direction with the same color codes as described in the Fig. 2. Genes conserved between the three genomes are colored gray and light blue bars indicate orthologous regions. The GC content at the third codon position of the ORFs in L. reuteri JCM 1112T is indicated under each ORF. Red lines represent the GC content at the third codon position of the ORFs in the pdu-cbi-cob-hem cluster (LAR_1583-1640). The blue horizontal line indicates the average GC content (32%) at the third codon position of the remaining ORFs in the L. reuteri JCM 1112T genome excluding the pdu-cbi-cob-hem gene cluster. (B) The pdu-cbi-cob gene cluster arrangement in L. reuteri JCM 1112T, S. typhimurium LT2, L. monocytogenes EGD-e, Y. enterocolitica subsp. enterocolitica 8081, and S. sanguinis SK36 are shown using the same color coding as described in (A).
The pdu-cbi-cob gene cluster has been identified in the genomes of pathogenic bacteria such as Salmonella spp.,48 Listeria spp.,49 Shigella sonnei,50 Yersinia enterocolitica,51 and Streptococcus sanguinis52 (Fig. 3B and Supplementary Fig. S2). However, the hem genes are clustered at different positions in the genomes of Yersinia, Shigella, and Listeria, and are scattered in the Salmonella genome. Only one gene (eutJ) of 12 ethanolamine utilizing genes (eut) in Listeria is encoded between pocR and pduF in JCM 1112T. Given the different gene organization of the pdu and hem operons, as well as a different codon adaptation index (CAI)53 between the pdu (0.55) and cbi-cob/hem (0.47) operons of JCM 1112T, it suggests that the pdu and cbi-cob/hem operons may have been independently inserted in these genomes and these clusters in L. reuteri JCM 1112T may have been generated by at least two independent insertion events rather than by a single event. However, the possibility that the pdu-cbi-cob gene cluster might have been formed prior to the integration into the JCM 1112T genome cannot be excluded.
3.3. Production of reuterin and cobalamin by L. reuteri JCM 1112T
The gupCDE genes from L. reuteri JCM 1112T were cloned in Escherichia coli to experimentally examine their encoded glycerol dehydratase activity. Some glycerol dehydratase activity was detected in E. coli carrying gupCDE, but not in E. coli derivatives containing the individual genes (Supplementary Table S2), indicating that the gene products of gupCDE are the subunits of glycerol dehydratase. Wild-type L. reuteri JCM 1112T and a gupCDE knockout mutant (LRΔgupCDE) (Supplementary Fig. S3), grown in MRS broth supplemented with glucose and 13C3-glycerol, were analyzed using 2D-NMR to determine whether the proteins encoded by gupCDE can convert glycerol to 3-HPA. After 7.5 h incubation, 3-HPA was detected in JCM 1112T, but not in LRΔgupCDE under the same conditions (Supplementary Fig. S4). These results demonstrate that the glycerol dehydratase encoded by gupCDE in JCM 1112T can convert glycerol to reuterin, consistent with previous studies using L. reuteri 1063.9
Lactobacillus reuteri JCM 1112T also possesses the gene (LAR_0029) encoding 1,3-propanediol dehydrogenase (Fig. 2). While 1,3-propanediol was detected in JCM 1112T after 2.5 h incubation (Supplementary Fig. S4), 3-HPA was first detected after 7.5 h incubation during culture in the MRS broth. These results reveal that 1,3-propanediol accumulated faster and more abundantly through the fermentation of glycerol than 3-HPA in L. reuteri cells. Negative effects of accumulated 1,3-propanediol together with a lower pH and an increase in NAD+ may be involved in the subsequent accumulation and extracellular release of 3-HPA by L. reuteri, as was previously observed in the fermentation of glycerol involving 1,3-propanediol dehydrogenase in Enterobacter agglomerans.54 JCM 1112T produced neither 13C-labeled lactic acid nor 13C-labeled ethanol, which are fermentation end products (Supplementary Fig. S4). JCM 1112T cannot be grown in MRS broth in which glycerol is the sole carbon source (data not shown). Taken together, these observations indicate that fermentation of glycerol for energy and growth did not occur in L. reuteri, which is consistent with the lack of the gene for dihydroxyacetone kinase in JCM 1112T for conversion of glycerol to dihydroxyacetone phosphate prior to metabolism via the Embden–Meyerhof–Parnas pathway (Fig. 2).
As discussed above, JCM 1112T contains the cbi-cob-hem gene cluster for cobalamin biosynthesis. Cobalamin production by L. reuteri was analyzed as described previously55 and was found to be present in the cell extract of JCM 1112T (Supplementary Table S3). This confirmed that JCM 1112T has the ability to produce both reuterin and cobalamin, consistent with the predicted gene products of the pdu-cbi-cob-hem gene cluster. It was previously demonstrated that adenosylcobalamin has a higher affinity for the glycerol dehydratase of L. reuteri than that of other Lactobacillus species.56 A high affinity of glycerol dehydratase for cobalamin may accelerate reuterin production by L. reuteri.
3.4. In vivo production of reuterin by L. reuteri JCM 1112T
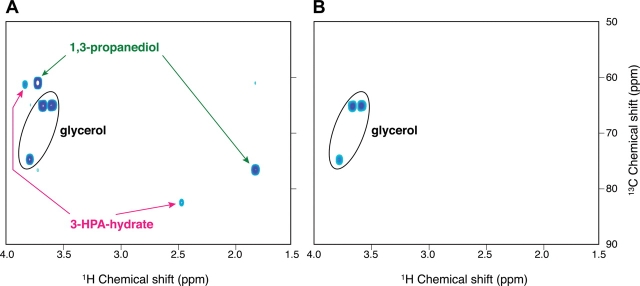
In vivo reuterin production by L. reuteri JCM 1112T was investigated using gnotobiotic mice mono-associated with JCM 1112T or its mutant derivative LRΔgupCDE. The intestinal loop assay of mono-associated mice was conducted 7 days after injection of a 13C3-glycerol solution into the murine cecum. The results revealed that reuterin was detected in samples taken from the cecum of gnotobiotic mice mono-associated with JCM 1112T, but not in mice mono-associated with LRΔgupCDE (Fig. 4). More rapid and abundant accumulation of 1,3-propanediol than that of 3-HPA was also observed in vivo, similar to the in vitro analysis (Supplementary Fig. S4). These results clearly demonstrated that glycerol dehydratase in L. reuteri converts glycerol into reuterin in vivo, supporting the hypothesis that the proposed probiotic effects of reuterin may occur in the intestine when glycerol is present. An analysis of the feces of untreated mice using NMR spectrometry and gas chromatography revealed 7–10 mM glycerol was present, suggesting the presence of glycerol in the intestine (data not shown). This is the first demonstration that the antimicrobial substance (reuterin) produced by L. reuteri can be produced in vivo in the gut under physiological conditions. This supports the genomic analysis as the genomic island encoding this phenotype, in this intestinal microbe, would likely not have been acquired unless it conferred a competitive advantage.
Figure 4.
In vivo detection of 3-HPA-hydrate derived from 13C3-glycerol in the cecal contents of mice colonized by wild-type L. reuteri JCM 1112T (A) and its mutant derivative, LRΔgupCDE (B), as detected by 2D 1H, 13C-HSQC NMR. δ1H (ppm) and δ13C (ppm) of 3-HPA-hydrate 2 (Hh-2) and 3-HPA-hydrate 3 (Hh-3) were observed at 2.43 and 42.6, and 3.79 and 61.6 at pH 7.0, respectively. Although the spot derived from 3-HPA-hydrate 1 (Hh-1) in 1H chemical shift was detected, the spot was not shown in (A), because the range of 1H chemical shift showed between 4.0 to 1.5 ppm. See Supplementary Figure S5 for details.
3.5. Conclusions
The complete genome sequences of L. reuteri JCM 1112T and L. fermentum IFO 3956 were determined. A comparative genome analysis suggested that L. reuteri JCM 1112T contains a specific gene cluster for reuterin and cobalamin production, and that the gene cluster is encoded on a genomic island that was acquired through lateral gene transfer. The pdu-cbi-cob gene cluster has also been identified in the genomes of some pathogenic bacteria, and previous studies have suggested that S. typhimurium acquired the pdu-cbi-cob gene cluster by horizontal transfer to enable it to degrade propanediol.43 As the pdu, cbi, and cob operons are not universally present in all salmonellae,57 it supports the hypothesis that the genes for propanediol utilization (pdu) and cobalamin biosynthesis (cbi-cob) may have been acquired via horizontal gene in response to environmental evolutionary adaptation. Genes for anaerobic metabolism of glycerol are located in the dihydroxyacetone (dha) regulon, whereas genes for the anaerobic degradation of propanediol are part of the pdu operon.41 The dehydratases of L. collinoides and L. brevis are general diol dehydratases (EC 4.2.1.28) rather than glycerol-specific dehydratases,56 but the dehydratase of L. reuteri has a higher affinity for glycerol than for propanediol,42 thus making it more efficient for conversion of glycerol to reuterin. The gene products of the homologues of pduCDE might determine properties of the pdu-cbi-cob gene cluster, and this cluster including gupCDE might provide a selective advantage to L. reuteri. Given the positive attributes of this phenotype, the pdu-cbi-cob-hem gene cluster might contribute to the probiotic properties of lactic acid bacteria. Genomic islands involved in pathogenesis or symbiosis are believed to be significant in the evolution of different bacterial species. In this study, the L. reuteri genome has a genomic island that may be important in the evolution of L. reuteri strains as health promoting bacteria in the human gut.
Funding
This research was supported by the ‘Academic Frontier’ Project for Private Universities: Matching Fund Subsidy, 2002-2006 (Azabu University), the Grants-in-Aid of Scientific Research (C) (18580275), Grant-in-Aid for Scientific Research on Priority Areas “Comprehensive Genomics” (17020007), and Grant-in-Aid for Encouragement of Young Scientists (B) (17710191) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charge: the ‘Academic Frontier’ Project for Private Universities: Matching Fund Subsidy (Azabu University).
Supplementary Data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
Supplementary Material
Acknowledgments
We are grateful to the consortium members of Azabu University Academic Frontier Project. We thank A. Yamashita, K. Furuya, C. Yoshino, H. Inaba, Y. Yamashita, A. Tamura, and N. Ito (Kitasato University), E. Okada and Y. Arita (Hitachi High-Tech Science Systems Co., Ltd.), and J.H. Lee (University of Minnesota) for technical assistance. We also thank DOE JGI for use of unpublished data of L. reuteri 100-23 and L. reuteri DSM 20016T.
References
- 1.Casas I. A., Dobrogosz W. J. Lactobacillus reuteri: an overview of a new probiotic for humans and animals. Microecol. Therap. 1997;26:221–231. [Google Scholar]
- 2.Casas I. A., Dobrogosz W. J. Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microb. Ecol. Health Dis. 2000;12:247–285. [Google Scholar]
- 3.Shornikova A. V., Casas I. A., Isolauri E., Mykkänen H., Vesikari T. Lactobacillus reuteri as a therapeutic agent in acute diarrhea in young children. J. Pediatr. Gastroenterol. Nutr. 1997;24:399–404. doi: 10.1097/00005176-199704000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Valeur N., Engel P., Carbajal N., Connolly E., Ladefoged K. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl. Environ. Microbiol. 2004;70:1176–1181. doi: 10.1128/AEM.70.2.1176-1181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang Y. H., Kim J. K., Kim H. J., Kim W. Y., Kim Y. B., Park Y. H. Selection of a potential probiotic Lactobacillus strain and subsequent in vivo studies. Antonie van Leeuwenhoek. 2001;80:193–199. doi: 10.1023/a:1012213728917. [DOI] [PubMed] [Google Scholar]
- 6.Rosenfeldt V., Michaelsen K. F., Jakobsen M., et al. Effect of probiotic Lactobacillus strains on acute diarrhea in a cohort of nonhospitalized children attending day-care centers. Pediatr. Infect. Dis. J. 2002;21:417–419. doi: 10.1097/00006454-200205000-00013. [DOI] [PubMed] [Google Scholar]
- 7.Weizman Z., Asli G., Alsheikh A. Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents. Pediatrics. 2005;115:5–9. doi: 10.1542/peds.2004-1815. [DOI] [PubMed] [Google Scholar]
- 8.Niv E., Naftali T., Hallak R., Vaisman N. The efficacy of Lactobacillus reuteri ATCC 55730 in the treatment of patients with irritable bowel syndrome: a double blind, placebo-controlled, randomized study. Clin. Nutr. 2005;24:925–931. doi: 10.1016/j.clnu.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Talarico T. L., Casas I. A., Chung T. C., Dobrogosz W. J. Production and isolation of reuterin: a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 1988;32:1854–1858. doi: 10.1128/aac.32.12.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung T. C., Axelsson L., Lindgren S. E., Dobrogosz W. J. In vitro studies on reuterin synthesis by Lactobacillus reuteri. Microb. Ecol. Health Dis. 1989;2:137–144. [Google Scholar]
- 11.Talarico T. L., Dobrogosz W. J. Chemical characterization of an antimicrobial substance produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 1989;33:674–679. doi: 10.1128/aac.33.5.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lüthi-Peng Q., Schärer S., Puhan Z. Production and stability of 3-hydroxypropionaldehyde in Lactobacillus reuteri. Appl. Microbiol. Biotechnol. 2002;60:73–80. doi: 10.1007/s00253-002-1099-0. [DOI] [PubMed] [Google Scholar]
- 13.Vollenweider S., Lacroix C. 3-Hydroxypropionaldehyde: applications and perspectives of biotechnological production. Appl. Microbiol. Biotechnol. 2004;64:16–27. doi: 10.1007/s00253-003-1497-y. [DOI] [PubMed] [Google Scholar]
- 14.Ennahar S., Cai Y., Fujita Y. Phylogenetic diversity of lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl. Environ. Microbiol. 2003;69:444–451. doi: 10.1128/AEM.69.1.444-451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein G., Pack A., Bonaparte C., Reuter G. Taxonomy and physiology of probiotic lactic acid bacteria. Int. J. Food Microbiol. 1998;41:103–125. doi: 10.1016/s0168-1605(98)00049-x. [DOI] [PubMed] [Google Scholar]
- 16.Strompfová V., Marcináková M., Simonová M., Bogovic-Matijasić B., Lauková A. Application of potential probiotic Lactobacillus fermentum AD1 strain in healthy dogs. Anaerobe. 2006;12:75–79. doi: 10.1016/j.anaerobe.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Peran L., Camuesco D., Comalada M., et al. Lactobacillus fermentum, a probiotic capable to release glutathione, prevents colonic inflammation in the TNBS model of rat colitis. Int. J. Colorectal Dis. 2006;21:737–746. doi: 10.1007/s00384-005-0773-y. [DOI] [PubMed] [Google Scholar]
- 18.Geier M. S., Butler R. N., Giffard P. M., Howarth G. S. Lactobacillus fermentum BR11, a potential new probiotic, alleviates symptoms of colitis induced bydextran sulfate sodium (DSS) in rats. Int. J. Food Microbiol. 2007;114:267–274. doi: 10.1016/j.ijfoodmicro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Kleerebezem M., Boekhorst J., van Kranenburg R., et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pridmore R. D., Berger B., Desiere F., et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA. 2004;101:2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altermann E., Russell W. M., Azcarate-Peril M. A., et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA. 2005;102:3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaillou S., Champomier-Vergès M. C., Cornet M., et al. Complete genome sequence of the meat-borne lactic acid bacterium Lactobacillus sakei 23K. Nat. Biotechnol. 2005;23:1527–1533. doi: 10.1038/nbt1160. [DOI] [PubMed] [Google Scholar]
- 23.Claesson M. J., Li Y., Leahy S., et al. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA. 2006;103:6718–6723. doi: 10.1073/pnas.0511060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van de Guchte M., Penaud S., Grimaldi C., et al. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. USA. 2006;103:9274–9279. doi: 10.1073/pnas.0603024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarova K., Slesarev A., Wolf Y., et al. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Callanan M., Kaleta P., O'Callaghan J., et al. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J. Bacteriol. 2008;190:727–735. doi: 10.1128/JB.01295-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon D., Desmarais C., Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delcher A. L., Harmon D., Kasif S., White O., Salzberg S. L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe T., Eddy S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatusov R. L., Natale D. A., Garkavtsev I. V., et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bateman A., Coin L., Durbin R., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice P., Longden I., Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 33.Tamura M., Hirayama K., Itoh K. Comparison of colonic bacterial enzymes in gnotobiotic mice monoassociated with different intestinal bacteria. Microb. Ecol. Health Dis. 1996;9:287–294. [Google Scholar]
- 34.Sakaguchi T., Sakaguchi S., Nakamura I., Kudo Y., Ichiman Y., Yoshida K. Positive reaction in mouse ligated intestinal loop assay with nonenterotoxigenic and nonhemolytic strains of Staphylococcus aureus. J. Clin. Microbiol. 1988;26:600–601. doi: 10.1128/jcm.26.3.600-601.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kikuchi J., Hirayama T. Practical aspects of stable isotope labeling of higher plants for a hetero-nuclear multi-dimensional NMR-based metabolomics. Methods Mol. Biol. 2006;358:273–286. doi: 10.1007/978-1-59745-244-1_15. [DOI] [PubMed] [Google Scholar]
- 36.Vuister G. W., Bax A. Resolution enhancement and spectral editing of uniformly C-13-enriched proteins by homonuclear broad-band C-13 decoupling. J. Magn. Reson. 1992;98:428–435. [Google Scholar]
- 37.Parrella F., Sanchez-Ferrando A., Virgili A. Improved sensitivity in gradient-based 1D and 2D multiplicity-edited HSQC experiments. J. Magn. Reson. 1997;126:274–277. [Google Scholar]
- 38.Schmieder P., Kurz M., Kessler H. Determination of heteronuclear long-range couplings to heteronuclei in natural abundance by two- and three-dimensional NMR spectroscopy. J. Biomol. NMR. 1991;1:403–420. doi: 10.1007/BF02192863. [DOI] [PubMed] [Google Scholar]
- 39.Kandler O., Stetter K. O., Kohl R. Lactobacillus reuteri sp. nov., a new species of heterofermentative lactobacilli. Zentbl. Bakteriol. Mikrobiol. Hyg. I Abt. Orig., 1980;C1:264–269. [Google Scholar]
- 40.Molenaar D., Bringel F., Schuren F. H., et al. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 2005;187:6119–6127. doi: 10.1128/JB.187.17.6119-6127.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daniel R., Bobik T. A., Gottschalk G. Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes. FEMS Microbiol. Rev. 1998;22:553–566. doi: 10.1111/j.1574-6976.1998.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 42.Talarico T. L., Dobrogosz W. J. Purification and characterization of glycerol dehydratase from Lactobacillus reuteri. Appl. Environ. Microbiol. 1990;56:1195–1197. doi: 10.1128/aem.56.4.1195-1197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bobik T. A., Xu Y., Jeter R. M., et al. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J. Bacteriol. 1997;179:6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauvageot N., Muller C., Hartke A., Auffray Y., Laplace J. M. Characterisation of the diol dehydratase pdu operon of Lactobacillus collinoides. FEMS Microbiol. Lett. 2002;209:69–74. doi: 10.1111/j.1574-6968.2002.tb11111.x. [DOI] [PubMed] [Google Scholar]
- 45.Gorga A., Claisse O., Lonvaud-Funel A. Organisation of the genes encoding glycerol dehydratase of Lactobacillus collinoides, Lactobacillus hilgardii and Lactobacillus diolivorans. Sci. Aliments. 2002;22:151–160. [Google Scholar]
- 46.Lawrence J. G. Gene organization: selection, selfishness, and serendipity. Annu. Rev. Microbiol. 2003;57:419–440. doi: 10.1146/annurev.micro.57.030502.090816. [DOI] [PubMed] [Google Scholar]
- 47.Bobik T. A., Ailion M., Roth J. R. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J. Bacteriol. 1992;174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McClelland M., Sanderson K. E., Spieth J., et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 49.Glaser P., Frangeul L., Buchrieser C., et al. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 50.Yang F., Yang J., Zhang X., et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 2005;33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomson N. R., Howard S., Wren B. W., et al. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu P., Alves J. M., Kitten T., et al. Genome of the opportunistic pathogen Streptococcus sanguinis. J. Bacteriol. 2007;189:3166–3175. doi: 10.1128/JB.01808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharp P. M., Li W. H. The codon adaptation index: a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barbirato F., Larguier A., Conte T., Astruc S., Bories A. Sensitivity to pH, product inhibition and inhibition by NAD+ of 1,3-propanediol dehydrogenase purified from Enterobacter agglomerans CNCM 1210. Arch. Microbiol. 1997;168:160–163. doi: 10.1007/s002030050482. [DOI] [PubMed] [Google Scholar]
- 55.Taranto M. P., Vera J. L., Hugenholtz J., de Valdez G. F., Sesma F. Lactobacillus reuteri CRL1098 produces cobalamin. J. Bacteriol. 2003;185:5643–5647. doi: 10.1128/JB.185.18.5643-5647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sauvageot N., Pichereau V., Louarme L., Hartke A., Auffray Y., Laplace J. M. Purification, characterization and subunits identification of the diol dehydratase of Lactobacillus collinoides. Eur. J. Biochem. 2002;269:5731–5737. doi: 10.1046/j.1432-1033.2002.03288.x. [DOI] [PubMed] [Google Scholar]
- 57.Porwollik S., Wong R.M., McClelland M. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA. 2002;99:8956–8961. doi: 10.1073/pnas.122153699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.