Abstract
Genetic studies in budding and fission yeasts have provided evidence that Rdh54, a Swi2/Snf2-like factor, synergizes with the Dmc1 recombinase to mediate inter-homologue recombination during meiosis. Rdh54 associates with Dmc1 in the yeast two-hybrid assay, but whether the Rdh54-Dmc1 interaction is direct and the manner in which these two recombination factors may functionally co-operate to accomplish their biological task have not yet been defined. Here, using purified Schizosaccharomyces pombe proteins, we demonstrate complex formation between Rdh54 and Dmc1 and enhancement of the recombinase activity of Dmc1 by Rdh54. Consistent with published cytological and chromatin immunoprecipitation data that implicate Rdh54 in preventing the non-specific association of Dmc1 with chromatin, we show here that Rdh54 mediates the efficient removal of Dmc1 from dsDNA. These functional attributes of Rdh54 are reliant on its ATPase function. The results presented herein provide valuable information concerning the Rdh54-Dmc1 protein pair that is germane for understanding their role in meiotic recombination. The biochemical systems established in this study should be useful for the continuing dissection of the action mechanism of Rdh54 and Dmc1.
1. Introduction
Homologous recombination (HR) is an important DNA repair tool, particularly in the elimination of DNA double-strand breaks (DSB)s and DNA inter-strand crosslinks. Furthermore, HR helps restart DNA replication forks that have stalled or are damaged [1-4], and it provides a means of restoring the length of shortened telomeres independently of telomerase [5,6]. Accordingly, HR deficiency is associated with genome destabilization and cancer predisposition. HR plays an indispensable role in meiotic chromosome metabolism as well. Specifically, programmed DSBs that occur at meiotic chromosomal hotspots trigger recombination events that serve to tie homologues together, so as to ensure their proper disjunction in meiosis I [4,6-8].
In HR-mediated DSB repair, single-stranded DNA tails derived from nucleolytic processing of the break are bound by a recombinase enzyme to assemble as a right-handed protein filament, commonly referred to as the presynaptic filament. Assembly of the presynaptic filament requires ATP binding but not its hydrolysis [9-15]. The biochemical steps, including the engagement of the partner chromatid, the search for homology in the partner chromatid, and DNA joint formation upon location of DNA homology, occur within the confines of the presynaptic filament [16-18]. The DNA joint formed as a result of the invasion of a homologous chromatid by the initiating ssDNA substrate is called the displacement loop, or D-loop. The D-loop reaction can be examined using linear ssDNA and negatively supercoiled homologous duplex as substrates [16,19]. The vast majority of eukaryotes, including the budding yeast Saccharomyces cerevisiae and humans, possess two recombinases, Rad51 and Dmc1, that are capable of mediating the D-loop reaction [20,21]. Whereas Rad51 is expressed in both mitotic and meiotic cells and required for HR and DNA repair in both life states, the expression of Dmc1 is strictly restricted to meiosis. [8,15,22].
The Rad51-mediated D-loop reaction is enhanced by the Swi2/Snf2-like factors Rad54 and Rdh54 [23,24]. Rad54 and Rdh54 possess a robust dsDNA-dependent ATPase activity, and at the expense of ATP hydrolysis, these HR factors translocate on dsDNA, inducing dynamic topological changes in the DNA and the transient separation of the strands in the DNA duplex [24-29]. This DNA strand-opening activity of Rad54 and Rdh54 very likely facilitates DNA strand invasion during D-loop formation [24,30,31]. In addition, Rad54 and Rdh54, through their DNA translocase activity, remove Rad51 from dsDNA [26,32], remodel chromatin [33-36], and dissociate branched DNA structures that resemble HR intermediates [27,37]. The ability of Rad54 and Rdh54 to remove Rad51 from dsDNA is thought to prevent the non-specific association of this recombinase with bulk chromatin and to render the primer-template junction in the D-loop available for DNA repair synthesis [26,32].
Rdh54 associates with Dmc1 in the yeast two-hybrid system [38,39], providing the first clue for a functional relationship between these two HR factors. Interestingly, results from chromatin immunoprecipitation (ChIP) and cytological experiments have furnished evidence for a role of the Rdh54 DNA translocase activity in preventing the accumulation of Dmc1 on bulk chromatin [40,41]. The premise of Dmc1 and Rdh54 functionally interacting in the mediation of HR reactions is consistent with the role of Rdh54 in meiotic recombination [39,40,42]. Here, using purified S. pombe proteins, we show a complex of Rdh54 and Dmc1 and functional synergy between these two HR factors in the D-loop reaction. Importantly, we provide evidence for an ability of Rdh54 to remove Dmc1 from dsDNA. The findings and biochemical systems described herein should form the basis for delineating the functional relationship between Rdh54 and Dmc1 in meiotic recombination.
2. Materials and methods
2.1. DNA substrates
For the D-loop assay, the 90-mer oligonucleotide D1 [43], being complementary to positions 1932 to 2022 of pBluescript SK DNA, was 5’ end-labeled with T4 polynucleotide kinase (Promega) and [γ-32P] ATP (Amersham Bioscience). The biotinylated 600 bp dsDNA used in the Rdh54-mediated Dmc1 removal experiment was prepared by PCR amplification of pBluescript SK DNA using a biotinylated primer and a non-biotinylated primer, as described [26]. pBluescript SK DNA was made in E. coli DH5α and purified using the HiSpeed Plasmid Purification Kit (Qiagen). The biotinylated DNA was immobilized on streptavidin magnetic beads (Roche Molecular Biochemicals) to give a preparation that contained 50 ng of DNA per μl of suspended volume of the beads, as described [26]. The pBluescript SK dsDNA included as the Dmc1 trap in the assay that measures the removal of Dmc1 from dsDNA was linearized by treatment with EcoRV.
2.2. Affinity pulldown using Affi-SpDmc1 beads
SpDmc1 or BSA was covalently coupled to Affi-Gel 15 beads (Bio-Rad) according to the manufacturer’s instructions, to generate matrices containing 3 mg/ml SpDmc1 or 12 mg/ml BSA. To examine binding of Rdh54 to the Affi beads, 4.5 μg of it was mixed with 8 μl of Affi-beads, Affi-SpDmc1 or Affi-BSA beads in 30 μl of buffer A (20 mM KH2PO4 pH 7.4, 10% glycerol, 0.5 mM EDTA, 0.01 % Igepal, 2 mM mercaptoethanol) containing 75 mM KCl. The reaction mixtures were incubated for 40 min on ice with gentle mixing every 30 seconds. The beads were collected by centrifugation, and the supernatant was removed. After being washed three times with 30 μl of buffer A containing 120 mM KCl, the beads were treated with 20 μl of 2% SDS to elute bound proteins. The supernatant, third wash, and SDS eluate, 8 μl each, were subject to SDS-PAGE analysis with Coomassie Blue staining to determine their protein content.
2.3. D-loop assay
The 32P-labeled 90-mer oligonucleotide substrate (2.4 μM nucleotides) was incubated with SpDmc1 (0.8 μM) in 10.5 μl buffer C (35 mM Tris-HCl pH 7.5, 1 mM DTT, 50 mM KCl) containing 2 mM ATP, 5 mM MgCl2, 250 μM CaCl2, 100 μg/ml BSA and an ATP-regenerating system for 5 min at 37°C. The indicated amount of SpRdh54 was then added in 1 μl, followed by a 1-min incubation at 23°C. The D-loop reaction was initiated by adding pBluescript SK DNA (35 μM base pairs) in 1 μl. The reaction mixtures were incubated for 10 min at 30°C, deproteinized, and subject to electrophoresis in 0.9% agarose gels in TAE buffer. The gels were dried onto a sheet of DEAE paper and the radiolabeled D-loop was visualized and quantified in the phosphorimager.
2.4. Assay to monitor SpDmc1 removal from dsDNA
To assemble SpDmc1-dsDNA nucleoprotein filaments, SpDmc1 (3.8 μM) was incubated with magnetic beads containing biotinylated dsDNA (15 μM base pairs) in 18 μl buffer C containing 2 mM ATP, 5 mM MgCl2 and an ATP-regenerating system for 5 min at 37°C. After the incorporation of the indicated amount of Rdh54 in 1 μl and a 3-min incubation at 23°C, the reactions were completed by adding linear pBluescript SK dsDNA (75 μM base pairs) as SpDmc1 trap. Following a 25 min incubation at 30°C, the beads were captured with a Magnetic Particle Separator (Boehringer Mannheim), and the supernatants were set aside for further analysis. Bound proteins were eluted from the beads with 20 μl of 2% SDS. The various supernatants and SDS eluates (8 μl each) were analyzed by SDS-PAGE and Coomassie Blue staining to determine their content of proteins.
3. Results
3.1. Purification and characterization of SpRdh54
We expressed the SpRdh54 protein in E. coli and purified it to near homogeneity (Fig. 1A; Supplementary Materials and Methods). Like ScRdh54 [24,26], SpRdh54 binds dsDNA (Supplementary Fig. S1A), hydrolyzes ATP (Supplementary Fig. S1B), supercoils DNA (Supplementary Fig. S2A), and promotes DNA strand opening (Supplementary Fig. S2B).
Figure 1. SpDmc1 binding and promotion of the D-loop reaction by purified SpRdh54.

(A) Purified SpRdh54 and SpDmc1 (1 μg) were analyzed by SDS-PAGE with Coomassie Blue staining.
(B) SpRdh54 was mixed with Affi-Gel 15 beads or beads containing covalently conjugated BSA or SpDmc1. After washing, the bound SpRdh54 was eluted with SDS (E) and analyzed along with the supernatant (S) and wash (W) by SDS-PAGE and Coomassie Blue staining.
(C) The reaction schematic of the D-loop reaction is shown in panel I. In panel II, the ability of SpDmc1 to form D-loop was tested in the absence or presence of SpRdh54 (150 and 250 nM in lanes 4 and 5, and 380 nM in lanes 2, 6, 7, and 9) or ATP, as indicated. The results were plotted in panel III.
3.2. Purification and characterization of SpDmc1
SpDmc1 was expressed in E. coli and purified to near homogeneity (Fig. 1A; Supplementary Materials and Methods). Consistent with the published studies [12, 44], the purified SpDmc1 protein hydrolyzes ATP (Supplementary Fig. S3A), binds DNA in an ATP-dependent manner (Supplementary Fig. S3, A and B), forms extended filaments on DNA (Supplementary Fig. S3C) and induces topological changes in DNA (Supplementary Fig. S3D).
3.3. SpDmc1 and SpRdh54 physically interact
Genetic studies in S. cerevisiae and S. pombe have provided evidence for a role of Rdh54 in meiotic inter-homologue HR, a characteristic that has been ascribed to Dmc1 as well [4,39,45,46]. Furthermore, yeast two-hybrid analyses have found that Rdh54 associates with Dmc1 [38,39]. For these reasons, we asked whether purified SpDmc1 and SpRdh54 proteins physically interact. For this, we covalently conjugated the purified SpDmc1 protein to Affi-Gel 15 beads (Affi-Dmc1) to use as affinity matrix in pulldown experiments with purified SpRdh54. We employed Affi-gel beads harboring covalently conjugated bovine serum albumin (Affi-BSA) as control in the pulldown experiment. As shown in Figure 1B, a significant fraction of the input SpRdh54 was retained on the Affi-Dmc1 beads, while, as expected, there was no binding to the Affi-BSA beads. Thus, in congruence with the yeast two-hybrid data [38,39], our results help establish a direct interaction between Dmc1 and Rdh54.
3.4. SpDmc1 and SpRdh54 functionally co-operate in the D-loop reaction
The homologous DNA pairing reaction mediated by various recombinases invariably requires ATP. A D-loop assay (see Fig. 1C, panel I for reaction schematic) was employed to assess the functional role of Rdh54 in the reaction. The results showed that SpDmc1 on its own has only a modest capability of making D-loops (Fig. 1C, panel II and panel III). Since ScRdh54 greatly stimulates the ScRad51-mediated D-loop reaction [24,26], we asked whether SpRdh54 can similarly enhance the D-loop forming ability of SpDmc1. Importantly, while SpRdh54 alone is devoid of homologous pairing activity, its addition to the Dmc1-mediated reaction led to a strong enhancement of the reaction efficiency (Fig. 1C). We note that the enhancement of the homologous pairing reaction occurs at amounts of SpRdh54 (150, 250, 380 nM) substoichiometric to that of SpDmc1 (800 nM) (Fig. 1C). Taken together, the results indicate that, as in the case with Rad51 [24,26], SpRdh54 functionally synergizes with SpDmc1 in the D-loop reaction.
3.5. SpRdh54 removes SpDmc1 from duplex DNA
Biochemical studies with S. cerevisiae proteins have found that Rad54 and Rdh54 catalyze the removal of Rad51 from dsDNA [26,32,47,48]. That Rdh54 may also be capable of removing Dmc1 from dsDNA has been suggested from cytological and chromatin immunoprecipitation experiments done in meiotic S. cerevisiae cells [41]. We employed a previously devised assay system [26] to determine whether SpRdh54 can remove SpDmc1 protein from dsDNA. Briefly, SpDmc1 is preloaded onto a biotinylated duplex DNA molecule immobilized on streptavidin magnetic beads, before being incubated with SpRdh54 and a non-biotinylated DNA molecule that serves as the trap for SpDmc1 protein molecules dislodged from the biotinylated DNA (Fig. 2A). The supernatant containing SpDmc1 bound to the DNA trap and the SDS eluate of the magnetic beads harboring SpDmc1 that remains associated with the biotinylated DNA are analyzed by SDS-PAGE.
Figure 2. Dissociation of the SpDmc1-dsDNA complex by SpRdh54.

(A) The reaction schematic is shown.
(B) Nucleoprotein complexes of SpDmc1 and magnetic bead-bound DNA were assembled with either ATP or AMP-PNP as nucleotide cofactor and then incubated without or with SpRdh54 (200 nM in lane 3, 400 nM in lanes 4 and 8, 600 nM in lane 5, and 750 nM in lanes 6 and 9). The supernatant and bead fractions were analyzed by SDS-PAGE and Coomassie Blue staining for their protein content. CK, creatine kinase used in the ATP regenerating system. The results were plotted.
As shown in Figure 2B, SpRdh54 possesses the ability to remove SpDmc1 from DNA. To ascertain the role of the SpRdh54 ATPase activity, we assembled the SpDmc1 protein filament using AMP-PNP as nucleotide cofactor and then incorporated SpRdh54. In this case, little or no removal of SpDmc1 occurred. The results thus provided evidence that the dissociation of SpDmc1 from dsDNA requires the ATP hydrolysis by SpRdh54. This premise was further supported by our observation with an ATPase defective variant of ScRdh54, to be presented below. Consistent with our published observation that ssDNA does not activate the Rdh54 ATPase function [24], SpRdh4 is incapable of dissociating the SpDmc1-ssDNA nucleoprotein filament (data not shown).
3.6. Relevance of the Rdh54 ATPase activity in Dmc1 removal from dsDNA
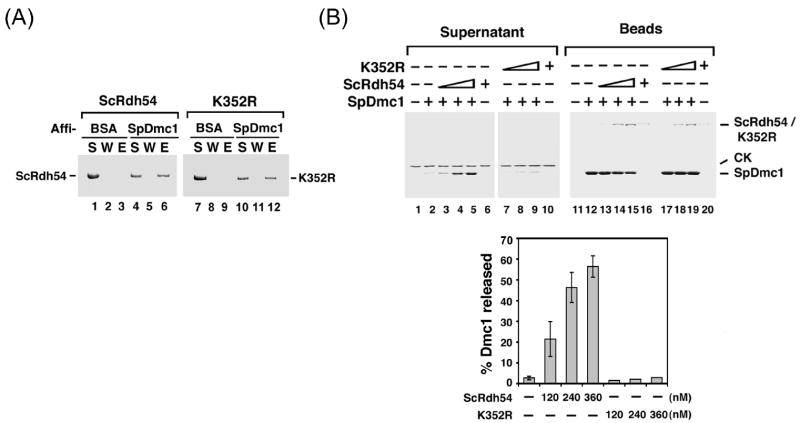
A high degree of sequence conservation exists between the S. pombe and S. cerevisiae Rdh54 orthologues [39]. Consistent with this, in an affinity pulldown experiment, ScRdh54 was efficiently retained on Affi-SpDmc1 beads but not on the control Affi-BSA beads, indicating that SpDmc1 is well capable of complex formation with ScRdh54 (Fig. 3A). Furthermore, we found that ScRdh54 can enhance the SpDmc1-mediated D-loop reaction (data not shown) and can also efficiently remove SpDmc1 from dsDNA (Fig. 3B).
Figure 3. Dependence of functional interactions on the Rdh54 ATPase activity.
(A) ScRdh54 or its K352R variant was mixed with Affi-Gel 15 beads containing covalently conjugated BSA or SpDmc1. After washing, the bound Rdh54 or rdh54 K352 protein was eluted with SDS (E) and analyzed along with the supernatant (S) and wash (W) by SDS-PAGE and Coomassie Blue staining.
(B) The ability of ScRdh54 (120, 240, and 360 nM in lanes 3-5, respectively) or its K352R variant (120, 240, and 360 nM in lanes 7 to 9, respectively) to dissociate the SpDmc1-dsDNA nucleoprotein complex, as in Figure 2. The results were plotted.
The physical and functional interactions between SpDmc1 and ScRdh54 allowed us to utilize the ATP hydrolysis-defective K352R mutant variant of the latter protein [26] to ascertain the role of the Rdh54 ATPase activity in the the mediation of Dmc1 removal from dsDNA. We first verified, by affinity pulldown using Affi-SpDmc1 beads, that the S. cerevisiae rdh54 K352 mutant protein retains the ability to associate with SpDmc1 (Fig. 3A). Importantly, the results from the SpDmc1 removal assay provided unequivocal evidence that both the Dmc1-dsDNA nucleoprotein complex dissociative function of Rdh54 is completely reliant on its ATPase activity (Fig. 3B). As expected, the S. cerevisiae rdh54 K352 mutant is inactive in the D-loop reaction (data not shown).
4. Discussion
In meiosis, inter-homologue HR events lead to the formation of chromosomal arm crossovers that link the homologue pairs, so as to orchestrate chromosome segregation during the first meiotic division. Genetic, cytological, and yeast two-hybrid studies have provided evidence that Dmc1 and Rdh54 function together in the promotion of inter-homologue recombination. However, to date, there has been no biochemical study directed at delineating the manner in which Dmc1 and Rdh54 physically and functionally interact to promote HR. In our effort to help fill this knowledge gap, we have devised protein expression vectors and procedures for the purification of SpDmc1 and SpRdh54 to near homogeneity and have carried out a variety of biochemical studies with these proteins. Our results confirm that SpRdh54 has similar biochemical attributes as its S. cerevisiae counterpart. Importantly, we have presented results to reveal that SpRdh54 and SpDmc1 interact directly, and that the two HR factors functionally co-operate in the catalysis of D-loop formation.
By ChIP and cytological analyses, Dmc1 has been found to localize to Spo11-made DSBs. Interestingly, in the spo11Δ rdh54Δ double mutant, foci of Dmc1 assemble spontaneously [40,41], and results from ChIP experiments have strongly suggested that these aberrant Dmc1 foci reflect non-specific association of Dmc1 with bulk chromatin [41]. Analysis of the rdh54 K352A mutant has yielded evidence that ATP hydrolysis by Rdh54 is indispensable for the avoidance of Dmc1’s non-specific association with chromatin [41]. Consistent with the in vivo observations, the use of highly purified SpRdh54, ScRdh54, and also the K352R mutant variant of ScRdh54 has allowed us to demonstrate that Rdh54 removes Dmc1 from dsDNA in a manner that requires its ATPase activity. This function of Rdh54 likely ensures the timely delivery of Dmc1 to meiotic DSBs to initiate repair by HR.
Supplementary Material
Acknowledgments
We are grateful to Hiroshi Nojima (Osaka University, Japan) for providing SpDMC1 cDNA and Susan Forsburg (University of Southern California, California) for providing SpRDH54 cDNA. This work was supported by NIH grants RO1GM57814, RO1ES07061, RO1GM053738, and R01GM074739, NSF grant SCEPSCoR 2004 RII-EPS-0447660, NIH postdoctoral fellowship F32GM084587, and Marie Curie Cancer Care.
Footnotes
Conflict of interest The author declares that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Flores-Rozas H, Kolodner RD. Links between replication, recombination and genome instability in eukaryotes. Trends Biochem Sci. 2000;25:196–200. doi: 10.1016/s0968-0004(00)01568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein HL, Kreuzer KN. Replication, recombination, and repair: going for the gold. Mol Cell. 2002;9:471–480. doi: 10.1016/s1097-2765(02)00493-8. [DOI] [PubMed] [Google Scholar]
- 3.Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc Natl Acad Sci USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 6.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- 8.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Hong EL, Shinohara A, Bishop DK. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J Biol Chem. 2001;276:41906–41912. doi: 10.1074/jbc.M105563200. [DOI] [PubMed] [Google Scholar]
- 11.Lee MH, Chang YC, Hong EL, Grubb J, Chang CS, Bishop DK, Wang TF. Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J Biol Chem. 2005;280:40980–40984. doi: 10.1074/jbc.M505896200. [DOI] [PubMed] [Google Scholar]
- 12.Sauvageau S, Stasiak AZ, Banville I, Ploquin M, Stasiak A, Masson JY. Fission yeast rad51 and dmc1, two efficient DNA recombinases forming helical nucleoprotein filaments. Mol Cell Biol. 2005;25:4377–4387. doi: 10.1128/MCB.25.11.4377-4387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung P, Stratton SA. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J Biol Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- 14.Bugreev DV, Golub EI, Stasiak AZ, Stasiak A, Mazin AV. Activation of human meiosis-specific recombinase Dmc1 by Ca2+ J Biol Chem. 2005;280:26886–26895. doi: 10.1074/jbc.M502248200. [DOI] [PubMed] [Google Scholar]
- 15.Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- 16.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 17.Cox MM. Motoring along with the bacterial RecA protein. Nat Rev Mol Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 18.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 19.Van Komen S, Macris M, Sehorn MG, Sung P. Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol. 2006;408:445–463. doi: 10.1016/S0076-6879(06)08028-1. [DOI] [PubMed] [Google Scholar]
- 20.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 21.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 22.Sheridan S, Bishop DK. Red-Hed regulation: recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev. 2006;20:1685–1691. doi: 10.1101/gad.1447606. [DOI] [PubMed] [Google Scholar]
- 23.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 24.Petukhova G, Sung P, Klein H. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Chi P, Kwon Y, Seong C, Epshtein A, Lam I, Sung P, Klein HL. Yeast recombination factor Rdh54 functionally interacts with the Rad51 recombinase and catalyzes Rad51 removal from DNA. J Biol Chem. 2006;281:26268–26279. doi: 10.1074/jbc.M602983200. [DOI] [PubMed] [Google Scholar]
- 27.Nimonkar AV, Amitani I, Baskin RJ, Kowalczykowski SC. Single molecule imaging of Tid1/Rdh54, a Rad54 homolog that translocates on duplex DNA and can disrupt joint molecules. J Biol Chem. 2007;282:30776–30784. doi: 10.1074/jbc.M704767200. [DOI] [PubMed] [Google Scholar]
- 28.Prasad TK, Robertson RB, Visnapuu ML, Chi P, Sung P, Greene EC. A DNA-translocating Snf2 molecular motor: Saccharomyces cerevisiae Rdh54 displays processive translocation and extrudes DNA loops. J Mol Biol. 2007;369:940–953. doi: 10.1016/j.jmb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ristic D, Wyman C, Paulusma C, Kanaar R. The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc Natl Acad Sci USA. 2001;98:8454–8460. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazin AV, Bornarth CJ, Solinger JA, Heyer WD, Kowalczykowski SC. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 31.Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 32.Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- 33.Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat Struct Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 34.Alexiadis V, Kadonaga JT. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 2002;16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaskelioff M, Van Komen S, Krebs JE, Sung P, Peterson CL. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J Biol Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 36.Kwon Y, Seong C, Chi P, Greene EC, Klein H, Sung P. ATP-dependent Chromatin Remodeling by the Saccharomyces cerevisiae Homologous Recombination Factor Rdh54. J Biol Chem. 2008;283:10445–10452. doi: 10.1074/jbc.M800082200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nat Struct Mol Biol. 2007;14:746–753. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- 38.Dresser ME, Ewing DJ, Conrad MN, Dominguez AM, Barstead R, Jiang H, Kodadek T. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Catlett MG, Forsburg SL. Schizosaccharomyces pombe Rdh54 (TID1) acts with Rhp54 (RAD54) to repair meiotic double-strand breaks. Mol Biol Cell. 2003;14:4707–4720. doi: 10.1091/mbc.E03-05-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinohara M, Gasior SL, Bishop DK, Shinohara A. Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc Natl Acad Sci USA. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holzen TM, Shah PP, Olivares HA, Bishop DK. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 2006;20:2593–2604. doi: 10.1101/gad.1447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, Ogawa H, Shinohara A. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smirnova M, Van Komen S, Sung P, Klein HL. Effects of tumor-associated mutations on Rad54 functions. J Biol Chem. 2004;279:24081–24088. doi: 10.1074/jbc.M402719200. [DOI] [PubMed] [Google Scholar]
- 44.Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat Struct Mol Biol. 2006;13:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- 45.Arbel A, Zenvirth D, Simchen G. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson DA, Stahl FW. Genetic control of recombination partner preference in yeast meiosis. Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics. 1999;153:621–641. doi: 10.1093/genetics/153.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiianitsa K, Solinger JA, Heyer WD. Terminal association of Rad54 protein with the Rad51-dsDNA filament. Proc Natl Acad Sci USA. 2006;103:9767–9772. doi: 10.1073/pnas.0604240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Zhang XP, Solinger JA, Kiianitsa K, Yu X, Egelman EH, Heyer WD. Rad51 and Rad54 ATPase activities are both required to modulate Rad51-dsDNA filament dynamics. Nucleic Acids Res. 2007;35:4124–4140. doi: 10.1093/nar/gkm412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.