Abstract
Mammals use DNA methylation for the heritable silencing of retrotransposons and imprinted genes and for the inactivation of the X chromosome in females. The establishment of patterns of DNA methylation during gametogenesis depends in part on DNMT3L, an enzymatically inactive regulatory factor that is related in sequence to the DNA methyltransferases DNMT3A and DNMT3B1,2. The main proteins that interact in vivo with the product of an epitope-tagged allele of the endogenous Dnmt3L gene were identified by mass spectrometry as DNMT3A2, DNMT3B and the four core histones. Peptide interaction assays showed that DNMT3L specifically interacts with the extreme amino terminus of histone H3; this interaction was strongly inhibited by methylation at lysine 4 of histone H3 but was insensitive to modifications at other positions. Crystallographic studies of human DNMT3L showed that the protein has a carboxy-terminal methyltransferase-like domain and an N-terminal cysteine-rich domain. Cocrystallization of DNMT3L with the tail of histone H3 revealed that the tail bound to the cysteine-rich domain of DNMT3L, and substitution of key residues in the binding site eliminated the H3 tail-DNMT3L interaction. These data indicate that DNMT3L recognizes histone H3 tails that are unmethylated at lysine 4 and induces de novo DNA methylation by recruitment or activation of DNMT3A2.
Null alleles of Dnmt3L are lethal when transmitted through the maternal germ line as a result of a failure to establish maternal genomic imprints during oogenesis, and mutant males show reactivation of retrotransposons and extreme meiotic defects in spermatocytes. The loss of DNA methylation in DNMT3L-deficient male germ cells also causes hyperacetylation of histones and a loss of methylation from lysine 9 of histone H3 (ref. 3). DNMT3L stimulates de novo methylation by DNMT3A2 (refs 4,5) but does not enhance the binding of DNMT3A2 to DNA, and DNMT3L alone does not bind to DNA5,6. However, the nature of the cues that regulate DNMT3L are unknown.
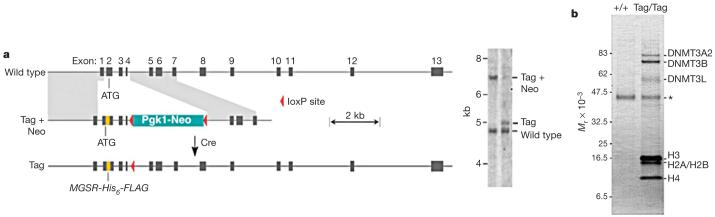
Embryonic stem (ES) cells express both DNMT3L and DNMT3A2, a germ-cell-specific isoform of DNMT3A that is also required for genomic imprinting4,7. ES cells show much higher rates of de novo methylation than do differentiated somatic cells, and methylation imprints are gained and lost at high rates in ES cells8,9. ES cells are therefore an appropriate cell type in which to carry out biochemical identification of proteins that interact with DNMT3L. We introduced a tandem His6-FLAG tag into the N terminus of the endogenous Dnmt3L gene by homologous recombination in ES cells to generate the Dnmt3LTag allele (Fig. 1a and Supplementary Fig. S1). Southern blotting (Fig. 1a) and DNA sequencing confirmed that the tag was targeted correctly, and expression of the tagged DNMT3L protein was confirmed on immunoblots probed with an anti-FLAG antibody (Supplementary Fig. S1). Mice that were homozygous for the tagged allele were of normal phenotype and both sexes were fertile, which indicated that the tag did not interfere with the function of DNMT3L.
Figure 1. Generation of epitope-tagged Dnmt3L locus and DNMT3L protein interaction screen in ES cells.
a, Targeting of the endogenous mouse Dnmt3L locus with a replacement cassette that introduces an N-terminal His6-FLAG epitope tag. The tag was introduced immediately after the first four amino acids of DNMT3L (Supplementary Fig. S1). The neo resistance cassette was deleted by transient exposure to Cre recombinase. Mice homozygous for the Dnmt3LTag allele are fertile and were used to generate homozygous Dnmt3LTag/Tag ES cells. The southern blot on the right shows that homologous recombination and Cre-mediated excision of the marker occurred as predicted. b, Protein interaction screen in wild-type and Dnmt3LTag/Tag ES cells by FLAG immunoprecipitation. Coomassie-stained bands were subjected to MALDI-reTOF and mass spectra were screened against a non-redundant protein database to identify interacting proteins. The protein band marked by the asterisk is actin, a common contaminant in anti-FLAG immunoprecipitation.
We conducted anti-FLAG immunoprecipitation on lysates derived from Dnmt3LTag/Tag ES cells. Mass spectrometry of polypeptides specific to eluates derived from Dnmt3LTag samples (Fig. 1b) identified DNMT3A2 and DNMT3B, which have been reported to interact with Dnmt3L in vitro4,5,10. DNMT3A2 has been reported to be required for de novo DNA methylation in germ cells, and germ-cell-specific conditional alleles of Dnmt3A2 phenocopy null alleles of Dnmt3L (ref. 7). We also found that DNMT3L was associated with the four core histones (Fig. 1b).
Peptide interaction assays with biotinylated tails of the four core histones showed that recombinant human DNMT3L bound only to the N-terminal tail of histone H3 (Fig. 2a). Peptide interaction assays using different regions of the N-terminal histone H3 tail revealed that DNMT3L binding required the first seven N-terminal amino acids of H3 (Fig. 2a, right). The binding of DNMT3L to histone H3 was abolished by mono-, di- or trimethylation of lysine 4 on histone H3, but was insensitive to modifications at other positions (Fig. 2b). These data indicate that the interaction between DNMT3L and the N-terminal tail of histone H3 depends on the methylation state of H3 lysine 4. Fluorescence polarization data showed that the dissociation constant (Kd) for the interaction between the unmodified H3 N-terminal tail and full-length DNMT3L was 2.1 μM; a single methyl group increased the Kd to 38.5 μM, and no interaction could be detected when the pep tide was di- or trimethylated at lysine 4 (Fig. 2d). Isolation of mononucleosomes by treatment of nuclei with micrococcal nuclease before anti-FLAG immunoprecipitation showed that DNMT3L is associated with nucleosomes that are depleted for H3 methylated at lysine 4 (Fig. 2e). These data confirm that the interactions shown in Fig. 2a, b also occur in vivo.
Figure 2. Interaction of DNMT3L with the N terminus of histone H3 is abolished by methylation of H3 lysine 4.
a, Peptide interaction assays with unmodified histone tails as indicated, incubated with full-length human DNMT3L. DNMT3L interacts only with the N-terminal tail of H3 (left), and this interaction requires only the first seven amino acids of H3 (right). b, Peptide interaction assays with DNMT3L and modified histone tails as indicated. DNMT3L bound only to the H3 tail that is unmodified at lysine 4. c, WDR5 (WD repeat domain 5) and the chromodomain of HP-1α (heterochromatin protein 1α) were used as controls for peptide binding specificity. d, Dissociation constants as determined by fluorescence polarization with C-terminal fluoresceinated peptides. Kd values are shown on the right. The 18-fold increase in Kd caused by monomethylation of H3 lysine 4 prevented detection of DNMT3L binding in b. e, Histones associated with DNMT3L in vivo are depleted in H3 that is trimethylated at lysine 4. Nuclear extract was treated with micrococcal nuclease before immunoprecipitation followed by separation of proteins by SDS-polyacrylamide gel electrophoresis. Immunoblot was performed with the antibodies indicated. A specific depletion in H3 trimethylated at lysine 4 can be seen in the fourth immunoblot from the top.
The results of crystallographic studies have suggested a mechanism for the histone-DNMT3L interaction. The structure of full-length human DNMT3L was determined to a resolution of 3.29 Å. The three endogenous zinc ions within the cysteine-rich region established X-ray phasing (Fig. 3a and Supplementary Table T1). Although the classical methyltransferase fold that is characteristic of S-adenosyl l-methionine-dependent methyltransferases was present11, the DNA recognition domain12 was absent and a cysteine-rich region organized around the three zinc ions was located opposite to the region where the catalytic centre is located in active DNA (cytosine-5) methyltransferases (Fig. 3a).
Figure 3. Structure of DNMT3L and mode of recognition of histone H3 unmethylated at lysine 4.

a, DNMT3L contains a classical methyltransferase fold as well as a cysteine-rich region organized around three zinc ions (indicated in yellow; zinc ions in orange). The structure of DNMT3L alone was determined to a resolution of 3.29 Å. M.HhaI is a bacterial DNA cytosine-5 methyltransferase that contains a classical methyltransferase fold12. b, Structure of DNMT3L after incorporation of the H3 N-terminal tail into the DNMT3L crystal by soaking with peptide. The omit electron density (orange) is contoured at 4.0σ. At the resolution of 3.69 Å, the electron density map alone does not provide an unambiguous position of the bound peptide. The model shown here is based on stereochemical principles and on the strong structural similarity with BHC80, which also binds specifically to histone H3 unmethylated at lysine 4 (Supplementary Fig. S2 and ref. 14). Only the N-terminal seven amino acids of the 24-amino-acid peptide were structured. The side chain of R2 was also disordered in the crystal. c, Detailed interaction between the H3 N terminus (amino acids numbered in red) and DNMT3L (numbered in black). The dashed lines indicate potential interactions between amino-acid side chains. H3 lysine 4 makes contacts with the carboxylates of DNMT3L D90 and D88, and methylation of lysine 4 will occlude these interactions. d, Mutagenesis of residues in the peptide binding site of DNMT3L abolished binding. Arrowheads in c indicate positions of I107W and D90A mutations. Each mutation abolished the interaction of DNMT3L with the unmethylated N-terminal peptide as shown by binding assay (top) and fluorescence polarization (bottom). Kd for binding to the wild type was 2.1 μM and for each of the two mutated proteins was >500 μM.
A peptide corresponding to the first 24 amino acids of histone H3 was soaked into the DNMT3L crystal shown in Fig. 3a. Crystallographic analysis of the complex showed electron density for approximately seven amino acids bound to the zinc-binding domain; the remainder of the peptide was unstructured (Fig. 3b). On the basis of the binding data (Fig. 2) and the structural comparison with the PHD-like domain of BHC80, a component of the LSD1 histone H3 lysine 4 demethylation complex13, we deduced that the structured portion of the peptide is the first seven amino acids of H3. BHC80 selectively binds unmethylated H3 lysine 4 peptides, and this binding is mediated by a PHD-like domain that has strong structural similarities to the cysteine-rich domain of DNMT3L (Supplementary Fig. S2 and ref. 14). Mutations that introduce a bulky tryptophan side chain into the H3 binding site of DNMT3L (I107W) or that disrupt the interaction of an aspartic acid side chain with the amino group of H3 lysine 4 (D90A) eliminate the interaction of the H3 tail with DNMT3L (Fig. 3c). The basis for methylation-sensitive binding of H3 to DNMT3L is steric occlusion of the interaction between aspartic acid 90 in DNMT3L and lysine 4 of histone H3 (Fig. 3b, c and data not shown).
These data indicate that Dnmt3L responds to states of histone modification to regulate de novo DNA methylation. Under this model, DNMT3L triggers de novo DNA methylation by activation or recruitment of DNMT3A2 upon contact with nucleosomes that contain unmethylated H3 lysine 4, while other sequences are masked from DNMT3L by the inhibitory effects of nucleosomes that contain histone H3 methylated at lysine 4.
Methylation of lysine 9 on histone H3 is required for DNA methylation in vegetative cells of the ascomycete Neurospora crassa, and for methylation of some non-CpG sequences in Arabidopsis thaliana15,16. Mouse ES cells that lack the H3 lysine 9 methyltransferases Suv39h1 and Suv39h2 show slight demethylation of satellite DNA17. In each of these cases, the signal for DNA methylation is the presence of methylation at H3 lysine 9 rather than the absence of methylation at H3 lysine 4. Methylation of lysine 4 on histone H3 has recently been suggested to protect gene promoters from de novo DNA methylation in mammalian somatic cells18,19, and this suggestion is fully compatible with the findings presented here.
DNMT3L is required for the de novo methylation of imprinting control regions in female germ cells and for the de novo methylation of dispersed repeated sequences in male germ cells. The data presented here point towards a novel mechanism whereby DNMT3L could convert patterns of histone H3 lysine 4 methylation, which are not known to be transmitted by mitotic inheritance, into patterns of DNA methylation that mediate the heritable transcriptional silencing of the affected sequences.
METHODS SUMMARY
Generation of Dnmt3Ltag allele
A synthetic oligonucleotide coding for His6 followed by the FLAG epitope was introduced into the Dnmt3L gene by homologous recombination in ES cells. Once we had confirmed correct targeting, we excised the neomycin resistance cassette by transient exposure to Cre, before injecting the cells into blastocysts to generate chimaeric mice. Dnmt3LTag/Tag ES cells were derived from blastocysts from homozygous breedings.
Immunoprecipitation
We immunoprecipitated clarified lysates from wild-type and Dnmt3LTag/Tag ES cells using EZview Red Anti-FLAG M2 Affinity Gel (Sigma) and eluted them with FLAG peptide (Sigma) according to the manufacturer’s protocol.
Peptide interaction assays
The interaction of DNMT3L with biotinylated histone tails was assayed essentially as described20.
Mass spectrometry
Protein bands were excised and digested with trypsin and then batch fractionated on a RP micro-tip, and the peptide mixtures were analysed by matrix assisted laser resorption/ionization reflectron time-of-flight (MALDI-reTOF) mass spectrometry (UltraFlex TOF/TOF; Bruker Daltonics) as described21, 22. We took selected experimental masses (m/z) to search a non-redundant protein database (‘NR’; ∼3.7 × 106 entries; National Center for Biotechnology Information; Bethesda, Maryland), using the PeptideSearch algorithm (M. Mann, Southern Denmark University, Odense, Denmark). Mass spectrometric sequencing of selected peptides was done by MALDI-TOF/TOF (MS/MS) analysis on the same prepared samples, using the UltraFlex instrument in ‘LIFT’ mode. We used fragment ion spectra to search the NR database using the MASCOT MS/MS Ion Search program (Matrix Science Ltd.).
X-ray crystallography
We solved the structure of full-length human DNMT3L by three-wavelength Zn anomalous diffraction data (Supplementary Table T1).
Supplementary Material
Acknowledgements
We thank C.S. Lin for advice and assistance, D. Bourc’his, M. Damelin, C. Schaefer, X. Zhang and R. E. Collins for helpful discussions, J. R. Horton for collection of X-ray diffraction data, A. Ruthenberg for recombinant WDR5 protein, and K. Anderson, D. Bourc’his and C. Schaefer for criticism of the manuscript. This work was supported by grants from the National Institutes of Health to C. D. A., X. C., P. T. and T. H. B. and by a fellowship from the European Molecular Biology Organisation to S.K.T.O.
Appendix
METHODS
Generation of Dnmt3LTag allele
We synthesized the right and left homologous arms of the Dnmt3L targeting construct by cloning the indicated PCR products into pBluescript SK (Stratagene). We designed forward and reverse primers using the available mouse genome sequence data. The synthetic oligonucleotide (5′-GG CAC CAT CAC CAC CAT CAC GAC TAC AAA GAC GAT GAC GAT AAA TCC C-3′) encoding a His6 tag followed by FLAG epitope was introduced into a SmaI site within exon 2, which contains the ATG start codon. An additional loxP site in inverse orientation was introduced into a ClaI site between exons 1 and 2 using the synthetic oligonucleotide 5′-ATA ACT TCG TAT AAT GTA TGC TAT ACG AAG TTA TAT CG-3′. The floxed neomycin resistance cassette was introduced from plasmid pEasyFlox (a gift from T. Ludwig) into NotI and XbaI sites within the pBluescript SK multiple cloning site between exons 3 and 4. Following gene targeting and G418 selection of 129Sv/Ev ES cell, we identified clones that had been correctly targeted by homologous recombination at the endogenous Dnmt3L locus by southern blotting with a 516-bp PCR-amplified probe, generated with 5′-AGA ATT CGC GGG CCC TAT GGA GAT ATA CAA GT-3′ forward and 5′- ATG GAT CCG AAG TTC AGG AAG GTG TGT GTG T-3′ reverse primers. The neomycin cassette was removed by Cre-mediated excision after transfection of the Cre-expressing plasmid pCAGGS-Cre (a gift from V. E. Papaioannou). Cre-mediated site-specific recombination was verified by southern blotting and sequencing of PCR products that spanned LoxP sites. Targeted clones that had deleted the neomycin cassette were injected into blastocysts and chimaeric mice were identified by southern blotting. Both male and female animals that are homozygous for the Dnmt3LTag allele are viable and fertile. Dnmt3LTag/Tag ES cells were derived from blastocysts from homozygous breedings.
Footnotes
The authors declare no competing financial interests
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 2.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 3.Webster KE, et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc. Natl Acad. Sci. USA. 2005;102:4068–4073. doi: 10.1073/pnas.0500702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J.Biol. Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 5.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 6.Kareta MS, Botello ZM, Ennis JJ, Chou C, Chedin F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 2006;281:25893–25902. doi: 10.1074/jbc.M603140200. [DOI] [PubMed] [Google Scholar]
- 7.Kaneda M, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 8.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 9.Humpherys D, et al. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 10.Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chedin F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell. Biochem. 2005;95:902–917. doi: 10.1002/jcb.20447. [DOI] [PubMed] [Google Scholar]
- 11.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 12.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Lan FF, et al. Recognition of un-methylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. doi: 10.1038/nature06034. doi:10.1038/nature06034 (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamaru H, et al. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nature Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- 16.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 17.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 18.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 19.Appanah R, Dickerson DR, Goyal P, Groudine M, Lorincz MC. An unmethylated 3′ promoter-proximal region is required for efficient transcription initiation. PLoS Genet. 2007;3:e27. doi: 10.1371/journal.pgen.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 21.Sebastiaan Winkler G, et al. Isolation and mass spectrometry of transcription factor complexes. Methods. 2002;26:260–269. doi: 10.1016/S1046-2023(02)00030-0. [DOI] [PubMed] [Google Scholar]
- 22.Erdjument-Bromage H, et al. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.