Abstract
Chronic pancreatitis is a persistent inflammatory disease of the pancreas. The digestive protease trypsin plays a fundamental role in the pathogenesis. Here we analyzed the gene encoding the trypsin-degrading enzyme chymotrypsin C (CTRC) in German subjects with idiopathic or hereditary chronic pancreatitis. Two alterations, p.R254W and p.K247_R254del, were significantly overrepresented in the pancreatitis group and were present in 30/901 (3.3%) affected individuals but only in 21/2,804 (0.7%) controls (OR=4.6; CI=2.6−8.0; P=1.3×10−7). A replication study identified these two variants in 10/348 (2.9%) individuals with alcoholic chronic pancreatitis but only in 3/432 (0.7%) subjects with alcoholic liver disease (OR=4.2; CI=1.2−15.5; P=0.02). CTRC variants were also found in 10/71 (14.1%) Indian subjects with tropical pancreatitis but only in 1/84 (1.2%) control (OR=13.6; CI=1.7−109.2; P=0.0028). Functional analysis of the CTRC variants revealed impaired activity and/or reduced secretion. The results indicate that loss-of-function alterations in CTRC predispose to pancreatitis by diminishing its protective trypsin-degrading activity.
Chronic pancreatitis is a continuing inflammatory disorder characterized by permanent destruction of the pancreatic parenchyma leading to maldigestion and diabetes mellitus due to exocrine and endocrine insufficiency. Penetrating insight into the pathomechanism came from relatively recent studies investigating the genes encoding cationic trypsinogen (PRSS1; OMIM 276000), anionic trypsinogen (PRSS2; OMIM 601564), and the pancreatic secretory trypsin inhibitor (SPINK1; OMIM 167790). Gain-of-function variants in PRSS1 have been linked to autosomal dominant hereditary pancreatitis and subsequently also to idiopathic chronic pancreatitis1-4. Recently, triplication of the PRSS1 locus has been observed in a subset of families with hereditary pancreatitis5. In vitro biochemical studies revealed that the majority of disease predisposing PRSS1 variants increase autocatalytic conversion of trypsinogen to active trypsin and probably promote premature intrapancreatic trypsin activation in vivo6,7. Consistent with the central pathophysiological role of trypsin, p.N34S and other loss-of-function alterations in the trypsin inhibitor SPINK1 predispose to idiopathic, tropical, and alcoholic chronic pancreatitis8-15. In contrast to pathogenic PRSS1 and SPINK1 variations, the p.G191R PRSS2 variant affords protection against chronic pancreatitis due to rapid autodegradation16. Taken together, genetic and biochemical evidence defines a pathological pathway in which a sustained imbalance between intrapancreatic trypsinogen activation and trypsin inactivation results in the development of chronic pancreatitis (Supplementary Fig. 1).
Because trypsin degradation serves as a protective mechanism against pancreatitis, we hypothesized that loss of function in trypsin degrading enzymes increases the risk for pancreatitis. We recently demonstrated that chymotrypsin C (CTRC) degrades all human trypsin and trypsinogen isoforms with high specificity17. From these studies, CTRC emerged as a strong novel candidate for a pancreatitis-associated gene. We performed direct DNA sequencing of all 8 exons of the 8.2 kb long CTRC in 621 individuals with idiopathic or hereditary chronic pancreatitis and in 614 control subjects of German origin. We found several CTRC variants, the large majority of which were in exons 2, 3, and 7. Therefore, we extended our analyses by sequencing an additional 280 affected individuals for these three exons and an additional 2075 controls for exons 2 and 3 and 2190 controls for exon 7. Altogether, we identified 11 missense and 2 deletion variants in CTRC (Table 1). The two most frequent variants, c.760C>T (p.R254W) and c.738_761del24 (p.K247_R254del), both located in exon 7, were found in affected individuals with a frequency of 2.1% and 1.2%, respectively. Taken together, the two alterations were significantly overrepresented in the pancreatitis group (30/901; 3.3%) compared to controls (21/2,804; 0.7%) (OR = 4.6; CI = 2.6−8.0; P = 1.3 × 10−7). Variant c.738_761del24, which causes an in-frame deletion of 8 amino acids from Lys247 through Arg254 (p.K247_R254del), showed the strongest disease association (OR = 11.5; CI = 3.2 − 41.5; P = 0.00003). Subgroup analysis for these two heterozygous variants revealed similar frequency in the hereditary 6/143 (4.2%; 6 × p.R254W) and idiopathic 24/758 (3.2%; 13 × p.R254W; 11 × p.K247_R254del) groups (Table 1). One individual with idiopathic disease was compound heterozygous for p.V235I (inherited from the mother) and p.R254W (inherited from the father).
Table 1.
CTRC variants detected in German subjects with idiopathic or hereditary chronic pancreatitis and healthy controls
| Exon | Nucleotide change | Amino acid change | Affected individuals (ICP) | Affected individuals (HP) | Affected individuals (all) | Controls | P value | OR | 95% CI |
|---|---|---|---|---|---|---|---|---|---|
| 2 | c.103G>C | p.D35H | 0/758 (0%) | 0/143 (0%) | 0/901 (0%) | 1/2689 (0.04%) | 1.0 | - | - |
| 2 | c.103G>A | p.D35N | 0/758 (0%) | 0/143 (0%) | 0/901 (0%) | 1/2689 (0.04%) | 1.0 | - | - |
| 2 | c.110G>A | p.R37Q | 5/758 (0.7%) | 1/143 (0.7%) | 6/901 (0.7%) | 10/2689 (0.4%) | 0.25 | - | - |
| 3 | c.143A>G | p.Q48R | 0/758 (0%) | 2/143 (1.4%) | 2/901 (0.2%) | 1/2689 (0.04%) | 0.16 | - | - |
| 4 | c.308delG | p.G103VfsX31 | 0/499 (0%) | 1/122 (0.8%) | 1/621 (0.2%) | 0/614 (0%) | 1.0 | - | - |
| 6 | c.514A>G | p.K172E | 0/499 (0%) | 0/122 (0%) | 0/621 (0%) | 1/614 (0.2%) | 0.5 | - | - |
| 7 | c.649G>A | p.G217S | 2/758 (0.3%) | 0/143 (0%) | 2/901 (0.2%) | 1/2804 (0.04%) | 0.15 | - | - |
| 7 | c.652G>A | p.G218S | 0/758 (0%) | 0/143 (0%) | 0/901 (0%) | 1/2804 (0.04%) | 1.0 | - | - |
| 7 | c.659T>G | p.L220R | 0/758 (0%) | 0/143 (0%) | 0/901 (0%) | 1/2804 (0.04%) | 1.0 | - | - |
| 7 | c.674A>C | p.E225A | 0/758 (0%) | 0/143 (0%) | 0/901 (0%) | 1/2804 (0.04%) | 1.0 | - | - |
| 7 | c.703G>A | p.V235I | 1/758 (0.1%) | 0/143 (0%) | 1/901 (0.1%) | 1/2804 (0.04%) | 0.43 | - | - |
| 7 | c.760C>T | p.R254W | 13/758 (1.7%) | 6/143 (4.2%) | 19/901 (2.1%) | 18/2804 (0.6%) | 0.0004* | 3.3 | 1.7−6.4 |
| 7 | c.738_761del24 | p.K247_R254del | 11/758 (1.5%) | 0/143 (0%) | 11/901 (1.2%) | 3/2804 (0.1%) | 0.00003* | 11.5 | 3.2−41.5 |
P values were determined by Fisher's Exact Test.
all affected individuals against controls
ICP, idiopathic chronic pancreatitis; HP, hereditary chronic pancreatitis
To confirm our findings in an independent cohort with another inflammatory pancreatic disease, we sequenced all 8 exons of CTRC in 96 German individuals affected with alcohol-related chronic pancreatitis. Subsequently, exons 2, 3 and 7 were sequenced in an additional 252 subjects (348 subjects in total) with alcoholic chronic pancreatitis. As controls, we analyzed exons 2, 3 and 7 in 432 German individuals with alcoholic liver disease without chronic pancreatitis. Again, we found a significant enrichment of the two exon 7 variants, p.R254W and p.K247_R254del, in subjects with alcoholic pancreatitis (10/348; 2.9%) versus subjects with alcohol-related liver disease (3/432; 0.7%); (OR = 4.2; CI = 1.2−15.5; P = 0.02) (Table 2). Finally, to investigate the significance of CTRC variants in chronic pancreatitis in subjects of non-European descent, we analyzed 71 individuals affected with tropical pancreatitis and 84 controls of Indian origin. Remarkably, the frequency of CTRC alterations in subjects with pancreatitis was even higher in this cohort (Table 3). Overall, 14.1% of affected individuals but only 1.2% of controls carried a CTRC variant (OR = 13.6; CI = 1.7−109.2; P = 0.0028). Two relatively frequent variants found in Indians were absent in Germans; the c.217G>A (p.A73T) missense alteration and the c.190_193delATTG (p.I64Lfs×69) frame-shift deletion. On the other hand, the p.K247_R254del variant that was relatively frequent in affected individuals from Germany was not found in the Indian population and the enrichment of the p.R254W variant in subjects with tropical pancreatitis did not reach statistical significance. However, due to the significantly smaller size of the Indian cohort relative to the German cohort, caution is warranted in the interpretation of these differences. Nonetheless, the data clearly indicate that CTRC variants increase the risk for tropical pancreatitis as well.
Table 2.
CTRC variants detected in German subjects with alcohol-related chronic pancreatitis and controls with alcoholic liver disease without pancreatitis
| Exon | Nucleotide change | Amino acid change | Affected individuals | Controls | P value | OR | 95% CI |
|---|---|---|---|---|---|---|---|
| 2 | c.110G>A | p.R37Q | 0/348 (0%) | 3/432 (0.7%) | 0.26 | - | - |
| 7 | c.649G>C | p.G217R | 1/348 (0.3%) | 0/432 (0%) | 0.45 | - | - |
| 7 | c.703G>A | p.V235I | 1/348 (0.3%) | 1/432 (0.2%) | 1.0 | - | - |
| 7 | c.746C>T | p.P249L | 1/348 (0.3%) | 0/432 (0%) | 0.45 | - | - |
| 7 | c.760C>T | p.R254W | 8/348 (2.3%) | 2/432 (0.5%) | 0.03 | 5.1 | 1.1−24.0 |
| 7 | c.738_761del24 | p.K247_R254del | 2/348 (0.6%) | 1/432 (0.2%) | 0.58 | - | - |
P values were determined by Fisher's Exact Test.
Table 3.
CTRC variants detected in subjects with tropical pancreatitis and healthy controls of Indian origin
| Exon | Nucleotide change | Amino acid change | Affected individuals | Controls | P value | OR | 95% CI |
|---|---|---|---|---|---|---|---|
| 3 | c.190_193delATTG | p.I64LfsX69 | 2/71 (2.8%) | 0/84 (0%) | 0.21 | - | - |
| 3 | c.217G>A | p.A73T | 4/71 (5.6%) | 0/84 (0%) | 0.04 | 11.3 | 0.6−213 |
| 7 | c.703G>A | p.V235I | 1/71 (1.4%) | 0/84 (0%) | 0.46 | - | - |
| 7 | c.760C>T | p.R254W | 2/71 (2.8%) | 1/84 (1.2%) | 0.60 | - | - |
| 7 |
c.778G>A |
p.D260N |
1/71 (1.4%) |
0/84 (0%) |
0.46 |
- |
- |
| All | 10/71 (14.1%) | 1/84 (1.2%) | 0.0028 | 13.6 | 1.7−109.2 |
P values were determined by Fisher's Exact Test.
Chronic pancreatitis is a complex multigenic disease and affected individuals often carry mutations in several disease-associated genes. To elucidate the relationship between CTRC alterations and PRSS1 and SPINK1 variants, we investigated all German subjects with idiopathic or hereditary pancreatitis for p.A16V, p.N29I and p.R122H in PRSS1 and for p.N34S in SPINK1. In total, 52/901 (5.8%) individuals carried a heterozygous PRSS1 variant, whereas 138/901 (15.3%) were positive for p.N34S (121 heterozygotes, 17 homozygotes). In the hereditary pancreatitis subgroup 32/143 (22.4%) subjects carried a heterozygous PRSS1 mutation; 30/143 (21%) carried a heterozygous p.N34S SPINK1 mutation, and 2/143 (1.4%) were homozygous for p.N34S. In the idiopathic pancreatitis subgroup, 20/758 (2.6%) individuals tested positive for a heterozygous PRSS1 mutation, 91/758 (12%) were heterozygous for the p.N34S SPINK1 mutation and 15/758 (2%) were homozygous for p.N34S. One subject with idiopathic disease was trans-heterozygous for the PRSS1 p.R122H variant (inherited from the mother) and the CTRC p.G217S variant (inherited from the father). None of the 17 SPINK1 p.N34S homozygotes carried a CTRC variant. On the other hand, 9/121 (7.4%) p.N34S heterozygotes were also heterozygous for one of two pancreatitis-associated CTRC variants (7 × p.R254W, 2 × p.K247_R254del). In contrast, only 21/763 (2.8%) of affected individuals without p.N34S were heterozygous for p.R254W or p.K247_R254del (P = 0.014; p.N34S heterozygous vs. p.N34S wild-type). Sixteen out of 902 (1.8%) control subjects were p.N34S heterozygous, but none of the 16 carried a CTRC variant. The alcoholic chronic pancreatitis cohort was analyzed for the SPINK1 p.N34S variant. Twenty-two of 348 (6.3%) subjects were heterozygous for p.N34S, and 1 subject (1/22, 4.6%) was trans-heterozygous for SPINK1 p.N34S and CTRC p.K247_R254del. The Indian subjects were screened for PRSS1 (p.A16V, p.N29I and p.R122H) and SPINK1 (p.N34S) variants as reported previously12. None of the affected individuals or control subjects carried a PRSS1 variant. Three controls (3.6%) were heterozygous for p.N34S but carried no CTRC alterations. Among the subjects with tropical pancreatitis, 29/71 (40.9%) were positive for p.N34S (22 heterozygotes, 7 homozygotes). Remarkably, none of the homozygotes, but 6/22 (27.3%) of the heterozygotes carried a CTRC variant (2 × p.I64Lfs×69, 2 × p.A73T, 2 × p.R254W, and 1 × p.D260N; 1 individual was compound heterozygous for p.A73T and p.D260N). In contrast, only 3/42 (7.1%) individuals with wild-type SPINK1 were heterozygous for a CTRC variant (2 × p.A73T, 1 × p.V235I) (P = 0.051; p.N34S heterozygous vs. p.N34S wild-type).
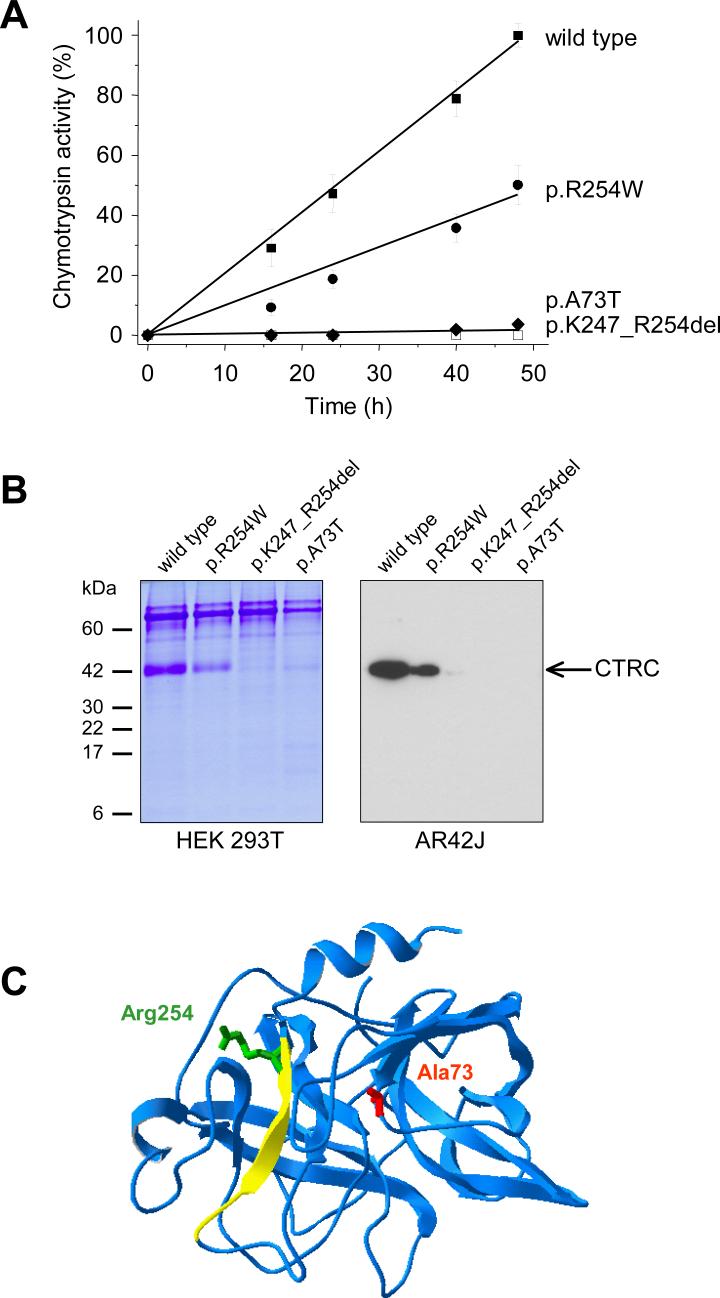
As observed previously for other pancreatitis-associated gene alterations, the majority of the identified CTRC variants do not alter the reading frame of the translated CTRC protein. The two exceptions are the c.190_193delATTG (p.I64Lfs×69) and the c.308delG (p.G103Vfs×31) variants, which cause a shift in the reading frame and may result in a truncated polypeptide chain. Alternatively, the mRNA of these mutants may undergo nonsense mediated decay and thus result in no CTRC protein whatsoever. In any event, these frame-shift variants are expected to cause a complete loss of CTRC function. To investigate the functional consequences of the CTRC missense alterations and the p.K247_R254del in-frame deletion, we expressed wild-type and mutant CTRC in human embryonic kidney (HEK) 293T cells via transient transfection. We found that secretion of p.K247_R254del and p.A73T was severely diminished relative to the wild-type, as evidenced by the loss of secreted CTRC activity and the faint protein bands on gels (Figs. 1A and 1B; Supplementary Fig 2C, Supplementary Table 1). When the specific activity of these two mutants was determined, p.K247_R254del proved completely inactive, whereas p.A73T exhibited significant protease activity (Supplementary Table 1). In contrast to the nearly complete loss of function observed with p.K247_R254del and p.A73T, CTRC activity secreted by cells expressing the p.R254W mutant was reduced to about 50% of wild-type (Fig. 1A). SDS-PAGE of conditioned media revealed that secreted levels of p.R254W were about 40% of wild-type CTRC, suggesting that the functional defect in this mutant is related to decreased production rather than impaired catalytic activity (Fig. 1B; Supplementary Fig 2C, Supplementary Table 1). This conclusion was strengthened by enzyme kinetic analysis of purified wild-type and p.R254W mutant CTRC. Kinetic parameters compared on two different chromogenic peptide substrates were essentially identical (Supplementary Table 2), indicating that reduced function of p.R254W is solely due to decreased production. In addition to the frequent CTRC alterations mentioned above, we also analyzed the variants p.R37Q, p.Q48R, p.G217S, and p.V235I by transient transfections in HEK 293T cells (Supplementary Fig. 2; Supplementary Table 1). Variant p.R37Q exhibited essentially normal activity and secretion (∼82−88 % of wild-type). In contrast, no CTRC activity was measurable from conditioned media of cells transfected with mutant p.Q48R and SDS-PAGE revealed a significant secretion defect (∼30 % of wild type). We also found that mutant p.Q48R underwent degradation during trypsin-mediated activation, presumably because the mutation introduced a new trypsin sensitive site (not shown). Mutant p.G217S showed significant catalytic impairment (specific activity only ∼7 % of wild type) and a modest decrease in secretion (∼70 % of wild type). Finally, CTRC activity secreted by cells transfected with variant p.V235I was moderately reduced (∼65 % of wild type); which seemed to be due to a combination of slightly lower specific activity and slightly reduced secretion of CTRC protein. Michaelis-Menten kinetic parameters determined for mutant p.V235I were not significantly different from those of wild type CTRC (Supplementary Table 2). Thus, from the 4 relatively rare CTRC variants analyzed, p.Q48R and p.G217S are loss-of-function CTRC alterations, whereas p.R37Q and p.V235I exhibit normal or slightly diminished function, respectively.
Figure 1.
Effects of pancreatitis associated CTRC variants on the secretion of chymotrypsinogen C. A. HEK 293T cells were transfected with the indicated constructs, and aliquots of conditioned media were withdrawn at the given times. CTRC activity was determined after activation with trypsin, as described in Methods, and expressed as percentage of the maximal activity, which corresponded to 14.7 μM p-nitroaniline released per minute. B. Aliquots (0.15 mL) of conditioned media of transfected HEK 293T cells were precipitated with 10% trichloroacetic acid (final concentration) and analyzed by SDS-PAGE and Coomassie Blue staining (left panel). Alternatively, AR42J cells were transfected with Glu-Glu tagged versions of wild-type and mutant CTRC constructs and cells were stimulated with 1 nM cerulein. Aliquots (0.1 mL) of the conditioned media were then precipitated with trichloroacetic acid, and analyzed by Western blotting using an HRP-conjugated goat polyclonal antibody against the Glu-Glu tag (Abcam). As a control, the cerulein-induced endogenous chymotrypsin secretion was also measured from 0.1 mL medium with 0.1 mM Suc-Ala-Ala-Pro-Phe-p-nitroaniline substrate (final concentration). The activity values determined were 24, 23, 21 and 21 μM p-nitroaniline / min for cells expressing wild type; p.R254W; p.K247_R254del or p.A73T, respectively. C. Ribbon diagram of chymotrypsinogen C. The bovine chymotrypsinogen C molecule is shown, which was crystallized as part of a ternary complex with proproteinase E and procarboxypeptidase A (Protein Data Bank file 1PYT). The position of Ala73 (mutated in p.A73T) is shown in red and Arg254 (mutated in p.R254W) is indicated in green. The yellow peptide segment is deleted in mutant p.K247_R254del. The image was rendered using DeepView/Swiss-PdbViewer v. 3.7 (www.expasy.org/spdbv/).
To demonstrate that the clinically significant CTRC mutants are also poorly secreted in cells that more closely resemble pancreatic acinar cells, we performed transfection experiments with the AR42J rat acinar cell line18. Wild-type CTRC and mutants p.A73T, p.K247_R254del and p.R254W were tagged with a Glu-Glu affinity tag to allow specific detection in the background of the native chymotrypsinogens secreted by AR42J cells. Stimulation of transfected cells with the cholecystokinine analog cerulein resulted in secretion of immunoreactive wild-type and somewhat less p.R254W mutant, whereas p.A73T and p.K247_R254del were not secreted to detectable levels (Fig. 1B).
Molecular modeling indicates that p.K247_R254del eliminates the last of the 6 β-strands of the C-terminal antiparallel β-barrel domain (Fig. 1C). A change of this magnitude in a structurally conserved part is expected to cause a folding defect, which might lead to the observed loss of catalytic function and the diminished secretion. Interestingly, p.R254W is also located within this deleted peptide segment. The mechanism of the decreased CTRC production caused by the p.A73T mutation is not readily apparent but may also involve misfolding. It is intriguing to note that mutations at different positions in the protein structure can lead to similar secretion problems. A possible explanation for this phenomenon is that local misfolding exposes hydrophobic parts of the protein or the mutation itself renders the hydrophilic surface more hydrophobic. The hydrophobic surfaces may in turn interact with various chaperons resident in the endoplasmic reticulum resulting in retention and eventual degradation.
In summary, the genetic and functional data presented in this study identify CTRC as a novel pancreatitis associated gene. Our observations provide further support for the trypsin-dependent pathogenic model of chronic pancreatitis in humans (Supplementary Fig. 1) by demonstrating that trypsin/trypsinogen degradation by CTRC is an important mechanism in the maintenance of the physiological protease-antiprotease balance in the pancreas.
METHODS
Study population
This study was approved by the medical ethical review committee of the University of Leipzig and the medical ethical review committee of the Charité University Hospital. All affected individuals gave informed consent. We enrolled 1,320 unrelated individuals with the diagnosis of hereditary (n = 143) or idiopathic chronic pancreatitis (n = 758) and alcoholic chronic pancreatitis (n = 348) originating from Germany. In addition, we also investigated subjects affected with tropical calcific pancreatitis originating from India (n = 71). The diagnosis of chronic pancreatitis was based on two or more of the following findings: presence of a typical history of recurrent pancreatitis, pancreatic calcifications and/or pancreatic ductal irregularities revealed by endoscopic retrograde pancreaticography or by magnetic resonance imaging of the pancreas and/or pathological sonographic findings. Hereditary chronic pancreatitis was diagnosed when one first-degree relative or two or more second-degree relatives suffered from recurrent acute or chronic pancreatitis without apparent precipitating factors. Affected individuals were classified as having idiopathic chronic pancreatitis when precipitating factors, such as alcohol abuse, trauma, medication, infection, metabolic disorders or a positive family history consistent with hereditary pancreatitis were absent. Alcohol-induced chronic pancreatitis was diagnosed in patients who consumed more than 60 g (females) or 80 g (males) of ethanol per day for more than two years. Control subjects free of pancreatic diseases were recruited from Germany (n = 2,804) and India (n = 81). The German controls included parents of children recruited to the German Multicenter Allergy Study; healthy controls recruited for a genetic study on type-2 diabetes at the University Hospital at Leipzig, blood donors, medical students and staff and subjects recruited to the Berlin Aging Study. In addition, 432 German subjects with alcoholic liver disease but without pancreatic disease were recruited as controls for the alcoholic chronic pancreatitis group.
Mutation screening
Oligonucleotide sequences, PCR and cycle sequencing conditions are available online as Supplementary Methods. Briefly, we digested the PCR products with shrimp alkaline phosphatase and exonuclease I and performed cycle sequencing using BigDye terminator mix (Applied Biosystems). The reaction products were purified with ethanol precipitation or Sephadex G-50 and loaded onto an ABI 3100-Avant or an ABI 3730 sequencer (Applied Biosystems).
Plasmid construction and mutagenesis
We constructed the pcDNA3.1(−)_CTRC expression plasmid using the IMAGE clone #5221216 (GenBank: BI832476), as described in17. CTRC mutants and the Glu-Glu-tagged CTRC constructs were generated by overlap extension PCR mutagenesis and ligated into pcDNA3.1(−). The Glu-Glu tag sequence (EYMPME) is derived from the polyoma virus medium T antigen.
Transfection of HEK 293T and AR42J cells
Human embryonic kidney (HEK) 293T cells were cultured and transfected as described in14. Briefly, approximately 106 cells per well were plated in 6-well tissue culture plates in D-MEM culture medium (Invitrogen) supplemented with 10% fetal bovine serum and 4 mM glutamine. Transfections were performed using 4 μg pcDNA3.1(−)_CTRC plasmid and 10 μL Lipofectamine 2000 (Invitrogen) in 2 mL OptiMEM medium (Invitrogen) supplemented with 2 mM glutamine. After 5 h incubation at 37 °C, 2 mL D-MEM with 20% fetal bovine serum and 4 mM glutamine was added to each well and cells were incubated for an additional 24 h. Cells were then washed with 1 mL OptiMEM, 2 mM glutamine and the transfection medium was replaced with 2 mL OptiMEM, 2 mM glutamine. Time-courses of expression were measured starting from this medium change and were followed for 48 h. AR42J cells were maintained as subconfluent cultures in D-MEM containing 20% fetal bovine serum, 4 mM glutamine and 1% penicillin/streptomycin solution. Cells (106) were plated into 35-mm wells and were grown in the presence of 100 nM dexamethasone for 48 hours. Transfections were performed using the Glu-Glu-tagged CTRC constructs, as described above. After 48 h, the medium was replaced with fresh OptiMEM, 2 mM glutamine; and 1 nM cerulein (final concentration) was added to stimulate secretion. After 15 min incubation the medium was collected and analyzed by Western blotting.
Chymotrypsin C activity assay
The conditioned medium was supplemented with 0.1 M Tris-HCl (pH 8.0) and 10 mM CaCl2 (final concentrations) and CTRC was activated with 100 nM human cationic trypsin (final concentration) for 20 min at 37 °C. All CTRC variants were fully activated by trypsin within this time period (reviewed but not shown). We measured CTRC activity with Suc-Ala-Ala-Pro-Phe-p-nitroanilide or Glt-Ala-Ala-Pro-Leu-p-nitroanilide (0.15 mM final concentration) in 0.1 M Tris-HCl (pH 8.0), 1 mM CaCl2, at room temperature.
Western blotting
Samples were run on 15% Tris-glycine gels under reducing conditions and transferred onto an Immobilon-P membrane (Millipore) at 300 mA for 1 h. The membrane was blocked with 5% milk powder solution overnight and incubated with an HRP-conjugated goat polyclonal antibody against the Glu-Glu tag (Abcam, Cambridge, Massachusetts) at a concentration of 0.1 μg/mL for 1 h at room temperature. HRP was detected using the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Statistics
The significance of the differences between mutation frequencies in affected individuals and controls was tested by two-tailed Fisher's Exact Test. Additional odds ratios were calculated using SAS/STAT software (v 9.1) and GraphPad Prism (v 4.03).
Accession codes
Entrez nucleotide: chymotrypsin C (CTRC): NT_004873 (chromosome 1 genomic contig); NM_007272 (human CTRC mRNA sequence); BI832476 (IMAGE clone used for plasmid construction). Protein Data Bank: chymotrypsinogen C crystal structure, 1PYT.
URLs
DeepView/Swiss-PdbViewer v. 3.7: http://www.expasy.org/spdbv/.
Supplementary Material
ACKNOWLEDGMENTS
We thank the individuals who have participated in this study. We also thank Antje Schulzki, Markus Braun and Vera Sahin-Tóth for excellent technical assistance. This work was supported by NIH grant DK058088 (to M.S.-T.), a scholarship from the Rosztoczy Foundation (to B.O.), by the Medical Faculty of the University of Leipzig formel.1 (to J.R.), and the Deutsche Forschungsgemeinschaft (DFG) grant Te 352/2-1 (to N.T.) and grants Wi 2036/2-1 & Wi 2036/2-2 (to H.W.).
Footnotes
Competing interests statement:
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Whitcomb DC, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat. Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 2.Gorry MC, et al. Mutations in the cationic trypsinogen gene are associated with recurrent acute and chronic pancreatitis. Gastroenterology. 1997;113:1063–1068. doi: 10.1053/gast.1997.v113.pm9322498. [DOI] [PubMed] [Google Scholar]
- 3.Teich N, Mössner J, Keim V. Mutations of the cationic trypsinogen in hereditary pancreatitis. Hum. Mutat. 1998;12:39–43. doi: 10.1002/(SICI)1098-1004(1998)12:1<39::AID-HUMU6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 4.Witt H, Luck W, Becker M. A signal peptide cleavage site mutation in the cationic trypsinogen gene is strongly associated with chronic pancreatitis. Gastroenterology. 1999;117:7–10. doi: 10.1016/s0016-5085(99)70543-3. [DOI] [PubMed] [Google Scholar]
- 5.Le Maréchal C, et al. Hereditary pancreatitis caused by triplication of the trypsinogen locus. Nat. Genet. 2006;38:1372–1374. doi: 10.1038/ng1904. [DOI] [PubMed] [Google Scholar]
- 6.Teich N, et al. Mutations of human cationic trypsinogen (PRSS1) and chronic pancreatitis. Hum. Mutat. 2006;27:721–730. doi: 10.1002/humu.20343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahin-Tóth M. Biochemical models of hereditary pancreatitis. Endocrinol. Metab. Clin. North Am. 2006;35:303–312. doi: 10.1016/j.ecl.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witt H, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat. Genet. 2000;25:213–216. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 9.Pfützer RH, et al. PSTI/SPINK1 polymorphisms act as disease modifiers in familial and idiopathic chronic pancreatitis. Gastroenterology. 2000;119:615–623. doi: 10.1053/gast.2000.18017. [DOI] [PubMed] [Google Scholar]
- 10.Witt H, et al. Mutation in the SPINK1 trypsin inhibitor gene, alcohol use, and chronic pancreatitis. JAMA. 2001;285:2716–2717. doi: 10.1001/jama.285.21.2716-a. [DOI] [PubMed] [Google Scholar]
- 11.Chandak GR, Idris MM, Reddy DN, Bhaskar S, Sriram PV, Singh L. Mutations in the pancreatic secretory trypsin inhibitor gene (PSTI/SPINK1) rather than the cationic trypsinogen gene (PRSS1) are significantly associated with tropical calcific pancreatitis. J. Med. Genet. 2002;39:347–351. doi: 10.1136/jmg.39.5.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhatia E, et al. Tropical calcific pancreatitis: strong association with SPINK1 trypsin inhibitor mutations. Gastroenterology. 2002;123:1020–1025. doi: 10.1053/gast.2002.36028. [DOI] [PubMed] [Google Scholar]
- 13.Schneider A, et al. SPINK1/PSTI mutations are associated with tropical pancreatitis and type II diabetes mellitus in Bangladesh. Gastroenterology. 2002;123:1026–1030. doi: 10.1053/gast.2002.36059. [DOI] [PubMed] [Google Scholar]
- 14.Király O, Wartmann T, Sahin-Tóth M. Missense mutations in pancreatic secretory trypsin inhibitor (SPINK1) cause intracellular retention and degradation. Gut. 2007 May 24; doi: 10.1136/gut.2006.115725. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boulling A, et al. Functional analysis of pancreatitis-associated missense mutations in the pancreatic secretory trypsin inhibitor (SPINK1) gene. Eur. J. Hum. Genet. 2007 Jun 13; doi: 10.1038/sj.ejhg.5201873. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16.Witt H, et al. A degradation-sensitive anionic trypsinogen (PRSS2) variant protects against chronic pancreatitis. Nat. Genet. 2006;38:668–673. doi: 10.1038/ng1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szmola R, Sahin-Tóth M. Chymotrypsin C (caldecrin) promotes degradation of human cationic trypsin: Identity with Rinderknecht's enzyme Y. Proc. Natl. Acad. Sci. USA. 2007;104:11227–11232. doi: 10.1073/pnas.0703714104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jessop NW, Hay RJ. Characteristics of two rat pancreatic exocrine cell lines derived from transplantable tumors. In Vitro. 1980;16:212. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.