Abstract
Linear mixed effects (LME) models are useful for longitudinal data/repeated measurements. We propose a new class of covariate-adjusted LME models for longitudinal data that nonparametrically adjusts for a normalizing covariate. The proposed approach involves fitting a parametric LME model to the data after adjusting for the nonparametric effects of a baseline confounding covariate. In particular, the effect of the observable covariate on the response and predictors of the LME model is modeled nonparametrically via smooth unknown functions. In addition to covariate-adjusted estimation of fixed/population parameters and random effects, an estimation procedure for the variance components is also developed. Numerical properties of the proposed estimators are investigated with simulation studies. The consistency and convergence rates of the proposed estimators are also established. An application to a longitudinal data set on calcium absorption, accounting for baseline distortion from body mass index, illustrates the proposed methodology.
Keywords: Binning, Covariance structure, Covariate-adjusted regression (CAR), Longitudinal data, Mixed model, Multiplicative effect, Varying coefficient models
1 Introduction
Longitudinal data are common in biomedical and health sciences research, where repeated measurements are obtained over time for each individual. Linear (and nonlinear) mixed effects models are useful for analyzing longitudinal data, providing a simple and effective way to incorporate within-subject and between-subject variation and the correlation structure of longitudinal data. Comprehensive overviews of mixed effects models for analyzing longitudinal data include [1, 2, 3], among others.
When the response and predictors are observed under possibly nonparametric effects of a confounding covariate, a direct application of linear mixed effects (LME) models may lead to biased estimates of the regression relationship of interest. As an example, we consider the relationship between calcium intake and calcium absorption. Insufficient calcium intake has an important influence on bone heath and the risk of osteoporosis [4], especially in women, who comprise ~80% of cases. More recently, low intake has also been associated with the risk of colorectal cancer [5], hypertension [6] and obesity [7]. The resulting health benefits of calcium depend on the body’s ability to absorb the ingested calcium, which is partly affected by body composition, e.g., underweight, overweight, obese. Thus, it is of interest to examine the effect of calcium intake levels on absorption, where the response measurements and the effect of the predictor are both potentially modulated by a common observable confounding factor, body mass index (BMI). BMI is a standard measurement that characterizes body composition taking into account both height and weight (BMI = kg/m2; e.g. overweight: 25.0 to 29.9, obese: 30+). The dual confounding effects of BMI on both the response and the predictor coupled with the uncertainty of the exact form of these effects motivate our proposed approach, which involves a LME model combined with nonparametric modeling of the confounder effects.
More specifically, for the true underlying relationship between repeatedly measured calcium intake (X) and absorption (Y), we assume/postulate an (unobserved) latent linear mixed effects model with random intercept and/or slope parameters to accommodate subject-specific variation. The response and predictor modulating factor, namely U = BMI, is referred to as the “distorting” or confounding covariate. The distorting effects of BMI on both the response and predictor are modeled flexibly through the unknown, unspecified smooth distorting functions ψ(U) and φ(U) as
| (1) |
where Ỹ and X̃ denote the observed response and predictor variables, respectively. The above multiplicative distortion modeling framework was previously proposed for covariate-adjusted regression (CAR) in the context of linear regression models for cross-sectional data [8]. Note that the formulation in (1) allows for the flexibility of nonparametric modeling of the distortion due to U = BMI on absorption and effects of intake, using the general unknown smooth functions ψ(·) and φ(·), because one does not have a priori knowledge of these functional dependencies.
The motivation for the multiplicative distortion form (1) was based on studies that involve adjustment for body configuration measures, such as U = BMI, through division of the main variables of interest by U, i.e. ψ(U) = φ(U) = U so that Y = Ỹ/U and X = X̃/U. For many applications, the multiplicative distortion model (1) can be justified, but alternative distortion models, including additive (Ỹ = ψ(U) + Y, X̃ = φ(U)+X) and no distortion (Ỹ = Y, X̃ = X), can be considered. See [8] for a more extended discussion. Although formulated in terms of general multiplicative distortion (1), the CAR estimation method from [8] is actually adaptive to all three distortion cases (multiplicative, additive and no distortion) in that it yields consistent regression coefficient estimates in all these distortion settings. This flexibility to accommodate a variety of distortion models also generalizes to the method proposed in this paper. This is further discussed in Section 3.2.
The latent LME model for the underlying response and predictor coupled with their nonparametric distortion models described above is referred to as the covariate-adjusted linear mixed effects (CA-LME) model. The main objective of this paper is estimation of the underlying latent relationship between the response and predictor based on the observed (distorted) longitudinal data {Ỹ, X̃, U}. This involves estimation of fixed parameters, random effects and variance components of the model. The previous work of [8] involves estimation of fixed parameters in regression models for cross-sectional data. In the current work we account for correlation between repeated measurements in the estimation of fixed parameters and propose estimation methods for variance components and subject-specific effects, adjusted for the distorting effects.
We note that the above framework has similarities with measurement error modeling [9] if one views ψ(U) and φ(U) as unobserved errors affecting the response and predictor. The proposed modeling can then be viewed as a multiplicative measurement error model where the error is in both the response and predictor. There is a large literature on additive measurement error modeling, although work on multiplicative measurement error models is limited to multiplicative errors only in the predictors [10, 11]. Also, a key difference with the errors in variables setting is that a part of the error, namely U, is observed in the CA-LME model. This additional information from U is utilized in our proposed estimation method.
The paper is organized as follows. We describe the main ideas of the CA-LME models with a single predictor in Section 2. Estimation of the fixed effects, subject-specific effects and variance components and the adjustments to eliminate the distortion effects are described in Section 3. In Section 4, we discuss generalizations of the CA-LME model that allow for (1) multiple predictors under general fixed and random effects structures, as in standard LME modeling of directly observed longitudinal data, and (2) combination of predictors with and/or without distortion in the model. The asymptotic results for the proposed CA-LME estimators are also given in Section 4. An illustration of the proposed CA-LME methodology to longitudinal data on the effects of calcium intake on absorption, as introduced above, is summarized in Section 5. Numerical properties are investigated in Section 6 and technical details are deferred to an appendix.
2 Covariate-Adjusted Linear Mixed Effects Models
To present the main ideas of covariate-adjusted linear mixed models, we first consider the case of a single, possibly time-varying, predictor. Suppose that we have repeated measurements from a longitudinal study. Denote the unobserved true response variable of the ith subject by Yij (i = 1, …, n) at the jth time point (j = 1, …, ni) and the unobserved true predictor variable by Xij. A simple LME model for the outcome Yij is
| (2) |
where eij denotes the mean zero random measurement error term for subject i at occasion j. For convenience we may write model (2) using matrix notation as Yi =
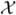 i(γ + γi)+ ei, where the vector of fixed effects or population parameters is γ = (γ0, γ1)T, the vector of subject-specific random effects is γi = (γ0i, γ1i)T, the vector of response values for subject i is Yi = (Yi1, …, Yini)T, and the corresponding matrix of predictor values is
i(γ + γi)+ ei, where the vector of fixed effects or population parameters is γ = (γ0, γ1)T, the vector of subject-specific random effects is γi = (γ0i, γ1i)T, the vector of response values for subject i is Yi = (Yi1, …, Yini)T, and the corresponding matrix of predictor values is
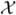 i with the jth row given by
. The error term, ei = (ei1, …, eini)T, is assumed to be ei ~ (0, Ri) and the random effects γi ~ (0, D), where Ri and D = (Dkl) (k, l = 1, 2) denote the covariance matrices of ei and γi, respectively. For simplicity, we may assume that Ri = σ2Ini and define the collection of parameters of interest as θi = (γ, γi, σ2, D).
i with the jth row given by
. The error term, ei = (ei1, …, eini)T, is assumed to be ei ~ (0, Ri) and the random effects γi ~ (0, D), where Ri and D = (Dkl) (k, l = 1, 2) denote the covariance matrices of ei and γi, respectively. For simplicity, we may assume that Ri = σ2Ini and define the collection of parameters of interest as θi = (γ, γi, σ2, D).
The standard LME model (2) provides a reasonable approximation in many longitudinal data settings, given that the underlying observations {Yij, Xij} are observable. In this case, standard estimation of the model parameters, namely the fixed effects (γ), random effects (γi), and variance components (σ2 and D), can be based on maximum likelihood (ML) or restricted maximum likelihood (REML) methods (see e.g. [1]). However, these standard estimation approaches are no longer applicable for distortion-prone response and predictors.
We consider estimation in model (2) under the following longitudinal data distortion framework, where only distorted versions of Yij and Xij are available. More precisely, the estimation must be based on the available distorted response and predictor data,
| (3) |
The unknown distorting functions {ψ(Ui), φ(Ui)} are assumed to be smooth functions of the observable confounder U. The distorted data available for estimation is the collection
for n subjects, where the ni-vector Ỹi and matrix of predictors
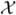 ˜i are defined analogously to Yi and
˜i are defined analogously to Yi and
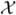 i above. Also, we let T denote the number of distinct observation times, t1, …, tT.
i above. Also, we let T denote the number of distinct observation times, t1, …, tT.
There are some important consequences of distortion-prone data on the standard ML or REML estimation methods for the LME model (2). As will be illustrated subsequently, the estimates for γ can be severely biased and variance components estimates will also be off-target. Additionally, efficiency considerations, such as incorporating an estimate of the covariance structure between repeated measurements into the standard estimation procedure, do not reduce the apparent bias resulting from the distortion. Because of the data contamination, common methods for estimating the covariance structure in longitudinal data, including parametric and nonparametric approaches (e.g., [12]), can-not capture the true covariance structure of the response, , due to biases in variance components estimation. We will propose an estimation method to resolve these problems due to the data distortion in Section 3.
Some constraints on the unknown distortion functions are needed for the identifiability of the estimation problem. Similar to the identifiability condition used for covariate-adjusted regression for cross-sectional data, we use the condition that the distortion is mean preserving, i.e., the means of the observed variables E(Ỹij) and E(X̃rij) are the same as that of the underlying variables, E(Yij) and E(Xrij), respectively. This identifiability condition (IC) is equivalent to the following constraint on the distorting functions [8]:
| (4) |
Note that since the mean preserving distortion is an identifiability condition, it is not testable. This is analogous to the zero mean errors for additive distortion in the measurement error literature. That is, the IC for multiplicative distortion on X is E(X̃) = E(Xφ(U)) = E(X) since E(φ(U)) = 1 and for additive distortion, it is E(X̃*) = E(X + φ*(U)) = E(X) since E(φ*(U)) = 0.
3 Estimation Procedure
3.1 Observable Mixed Effects Varying Coefficient Model
To motivate the proposed estimation method, we first note that the conditional mean of the observable response variable Ỹij, given the subject-specific effects γi and data
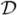 ≡ {
≡ {
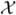 ˜i, Ui; i = 1, …, n} is,
˜i, Ui; i = 1, …, n} is,
| (5) |
where the fixed and subject-specific coefficient functions in (5) are
| (6) |
| (7) |
respectively. Thus, the above relationship (5) suggests that the regression of the available distorted data {Ỹij, X̃ij, Ui} leads to the following observable varying coefficient model with both fixed and random coefficient functions,
| (8) |
where εij ≡ eijψ(Ui). Model (8), written more succinctly in matrix notation, is
| (9) |
with the vectors of functions β(Ui) = {β0(Ui), β1(Ui)}T and bi(Ui) = {b0i(Ui), b1i(Ui)}T, where εi = (εi1, …, εini)T. Also note that given Ui = u, bi(u) = {b0i(u), b1i(u)}T ~ (0, D̃(u)) and εi ~ (0, R̃i(u)), where R̃i(u) ≡ Var(εi) and D̃ (u) ≡ Var{bi(u)}.
We note that the above observable mixed varying coefficient model shares some common features with the class of “random varying coefficient” models [13], where the index of the fixed and subject-specific coefficient functions is the observation time, tij, instead of Ui. The observation that model (8) is a varying coefficient model is useful for developing the estimation technique proposed to target γ and γi. More specifically, we will first use a computationally efficient method based on binning to target the varying coefficient functions in (8). Our proposed estimators for γ and γi will then be derived using the relations between the varying coefficient functions and the underlying parameters given in (6) and (7).
Note that even though the connection between the CA-LME model in (2) and the varying coefficient model (VCM) in (8) facilitates estimation in the CA-LME model, the proposed CA-LME model is distinct from the VCM in that it is free from or adjusted for the effects of U. The VCM can be viewed as a stratified analysis (with respect to U) and such a model aims to address the question, “What is the relationship between Ỹ and X̃ at varying levels of U?” This is an important question in itself, however this is not the object of inference of interest with respect to the proposed CA-LME model. The object of inference in the CA-LME model is the relationship between X and Y (not directly observed). This relationship corresponds to the relation between Ỹ and X̃ with the effects of U removed, i.e. free of U, which is quite different than the target relationship of interest in the VCM. We discuss in further detail the distinction between CA-LME and VCM as well as their limitations, advantages and disadvantages in the Discussion Section.
Thus, the main contribution of this paper (given in Sections 3.2, 3.3. and 3.4) is the proposal of and the estimation and inference procedures for a latent variable model that explains the regression relation between the variables of primary interest adjusted for U in the context of repeated (correlated) measurements. Previous work in covariate adjusted modelling considers a linear regression model for cross-sectional/uncorrelated data, whereas the CA-LME model is designed for correlated data. The proposed estimation procedure is also new where the specific contributions include developing consistent point estimates for fixed effects, estimation of random effects and (within and between subject) variance components from distorted data.
3.2 CA-LME Parameter Estimation
Estimation of the underlying parameters of interest, namely θi = (γ, γi, σ2, D), requires (a) estimation of the fixed and random varying coefficient functions in the mixed effects varying coefficient model (8), (b) estimation and incorporation of the covariance structure among repeated measurements (distinguishing within- and between-subject variation) and (c) adjustment for the distortion effects. The estimation method has three main steps: (1) binning (or “stratification”) of the data with respect to the confounding variable U, (2) fitting a LME model within each bin to obtain bin-specific estimates and (3) aggregating or averaging bin-specific estimates to obtain the covariate-adjusted LME estimators of θi. The basic approach to eliminating the distorting effects of U is to localize the model fitting by using data in each bin only. Details of each step are provided next.
The observable data available for estimation is the collection . Assuming that the confounding covariate U is bounded, a ≤ U ≤ b where a < b are real numbers, the initial step of the estimation procedure divides the interval [a, b] into H equidistant intervals, denoted B1, …, BH and referred to as bins. Let Lv be the number of subjects falling into bin v, for v = 1, …, H. To track the data corresponding to subjects falling into a given bin, observations in any given bin are marked by a prime. More specifically, the data for which Ui ∈ Bv, for i = 1, …, n, is given by the collection { , k = 1, …, Lv,}. Let denote the number of repeated measurements for subject k in bin v, then the data corresponding to the kth subject falling in bin v is , where is the × 2 matrix of predictor values, and is the confounder. For example, if subjects 2 and 3 fall into the bin v = 7, then k = 1, 2, L7 = 2 and with , and . Similarly, and .
After binning the data, we approximate the mixed effects varying coefficient model (9), specifically Ỹi =
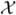 ˜i {β(Ui) + bi(Ui)} + εi, for i = 1, …, n, local to bin v (v = 1, …, H). This is achieved by fitting a LME model using the data within each bin. More precisely, for each bin v = 1, …, H, we fit the following LME model,
˜i {β(Ui) + bi(Ui)} + εi, for i = 1, …, n, local to bin v (v = 1, …, H). This is achieved by fitting a LME model using the data within each bin. More precisely, for each bin v = 1, …, H, we fit the following LME model,
| (10) |
where βv = (β0v, β1v)T, bvk = (b0vk, b1vk)T and denote bin-specific vectors of fixed effects, subject-specific effects and errors for subject k in bin v. Also, denote the bin-specific covariance matrices by D̃v ≡ Var(bvk) and .
Model (10) can be interpreted as an approximation to model (9) “local” to bin v. Note that because model (10) is based on data local/specific to bin v, the resulting fixed effects and variance components estimates in model (10), denoted β̂v, and target β(·), D̃(·)and R̃k(·) evaluated in the specific neighborhood associated with bin Bv. It follows from standard LME model theory that the estimator of βv and bvk are
| (11) |
| (12) |
where and . When D̃v and R̃vk are unknown, they can be estimated using the REML (or ML) method [3]. The REML estimators of D̃v and R̃vk, denoted and , are substituted into (11) and (12).
The covariate-adjusted linear mixed effects estimators of the fixed effects, γ0 and γ1, involve averaging of the bin-specific estimated fixed varying coefficient functions, namely , to eliminate the distortion across U. The CA-LME estimators are given by
| (13) |
where is the overall mean of the predictor variable (with X0ij ≡ 1, X1ij = Xij). Here mj denotes the number of subjects observed at time tj. Also, Ij denotes the set of indices of subjects that are observed at time tj. Similarly, Ivj is the set of indices of subjects in bin Bv observed at time tj. Note that the distortion effects of U cancel in the CA-LME estimators given in (13) and γ̂r targets the fixed effect parameter γr, since E{βr(U)X̃rj} = γrE{ψ(U)Xrj} = γrE(Xrj) = γrE(X̃rj), for all j = 1, …, T and r = 0, 1. The CA-LME estimators, γ̂r, can be interpreted as method of moments estimators, because targets E(X̃r) and targets E{βr(U)X̃r}.
We note here that the proposed estimators in (13) are consistent under multiplicative distortion (Theorem 1). Note also that an alternative way to handle multiplicative distortion in practice would be to transform it into additive distortion by taking logarithms where the underlying latent model would then be holding between the logarithm of the underlying variables. However, if the distortion is in fact additive taking logarithms would not deal with the problem. Even though the proposed estimators are formulated for multiplicative distortion, they are also consistent under additive distortion (Ỹ = ψ(U) + Y and X̃r = φr(U) + Xr) as well as the trivial case of no distortion as is shown next. The fact that the proposed estimators are consistent under different types of distortion and no distortion is an advantage of the proposed method over taking logarithms, since it is difficult (and more typically not possible) to justify one type of distortion over another a priori.
The consistency of the estimators given in (13) for the additive distortion and no distortion cases follows from the fact that both of these cases also lead to a mixed varying coefficient model between the observed variables, as described for the multiplicative distortion in Section 3.1 above. More precisely, in the mixed varying coefficient models derived from these two cases, β1(U) = γ1 (in both models), β0(U) = γ0 for the no distortion case and β0(U) = γ0 − γ1φ(U) + ψ(U) for the additive distortion case. Hence, it is follows that E{βr(U)X̃rj} = γrE(X̃rj) holds for all cases, because E{φ(U)} = E{ψ(U)} = 0 under additive effects guaranteeing no distortion on average, i.e. E(X̃) = E(X) and E(Ỹ) = E(Y). Using this information Sentürk and Müller (2005) proposed a diagnostic tool to check whether the underlying distortion is additive by checking whether the slope function of the fitted varying coefficient model is equal to a constant. Similarly, for the case of no-distortion, one can check whether the slope and the y-intercept functions are both constant functions. This diagnostic tool provides information on the type of distortion based on the data and is implemented in Section 5 via the proposed bootstrap hypothesis test of Sentürk and Müller. This is helpful in identifying cases where a simpler estimation approach can be adopted when the model reduces to additive or no distortion.
Next, we consider the estimators (predictors) of the subject-specific effects for the CA-LME model, namely γri for the ith individual. Recall from (7) that we have
where the last equality above follows directly from equation (6). Assume without loss of generality that the ith subject is the kth individual in bin v. Then the estimator of the random effect coefficient function, bri(·), is targeted by the subject-specific estimator b̂rvk from bin v, given by (12). Targeting βr(Ui) and γr by β̂rv and γ̂r, we arrive at the following plug-in estimator of γri for individual k in bin v
| (14) |
3.3 Covariate-Adjusted Estimators of Variance Components
Similar to the estimation of the fixed and random effects, adjustments are also needed for the estimation of the variance components. The effects of the data distortion on the variance components estimation can be seen from the mixed effects varying coefficient model (8). Thus, given Ui = u, the within-subject covariance matrix is R̃i(u) ≡ Var(εi) = ψ2(u)σ2Ini and the between-subject covariance matrix is
The above calculation shows the direct relationship between the variance components of the true unobserved LME model (i.e. σ2 and D) and the corresponding ones in the mixed effects varying coefficient model (8), at Ui = u. Thus, the bin-specific (REML) estimators of the variance components, namely and , target D̃(·) and R̃i(·) evaluated in the specific neighborhood of U, respectively.
Consider the average of the unadjusted bin-specific between-subject variance components estimators, i.e. for l, l′ = 1, 2, which target
where λ11 = E{ψ2(U)}, λ12 = E{ψ2(U)/φ(U)} and λ22 = E{ψ2(U)/φ2(U)}. To obtain the underlying between-subject variance components, Dll′, we estimate the coefficients λll′ and make the required adjustments. The estimated adjustment coefficients are
where and . Hence, the covariate-adjusted between-subject variance and covariance estimators are obtained as , for l, l′ = 1, 2. Similarly, for the within-subject variance, averaging the bin-specific within-subject variances, namely , will target λ11σ2, where is the bin-specific within-subject variance estimate obtained from bin v. Therefore, the adjusted within-subject variance estimator that targets σ2 is .
Finally, we note that it is not difficult to see that the estimators of the required adjustment factors, namely {λ̂11, λ̂22, λ̂12} given above, are sample averages/moments corresponding to the expectations {λ11, λ22, λ12}, which follows from (6). That is, we have from (6) that and . We note that showing the consistency of λ̂ll′ for λll′ involves similar arguments as in the proof of Theorem 2 in [14] and, therefore, is omitted in this work.
4 Model Generalization and Consistency
For simplicity of exposition, we have considered a LME model with a random intercept and slope and one predictor variable as the underlying model (2). Generalization to a model containing (a) multiple predictors, (b) a mixture of distorted and undistorted predictors, and/or (c) more complex random effects and covariance structures would broaden the applicability of the proposed method. We now consider a more general CA-LME model that incorporates extensions (a), (b) and (c). Denote p distorted predictors by X̃rij, for r = 1, …, p, where X̃rij = φr(Ui)Xrij. Note that the distortion on each predictor is modeled flexibly by allowing a different distorting function, φr(·), corresponding to the rth predictor. Furthermore, to accommodate undistorted predictors in the regression model, such as age or time, let Wrij, r = 1, …, q, be a set of q undistorted predictors. For instance, descriptive and graphical analysis may suggest a model with baseline age and a quadratic trend in time, . In such a case, one may choose to include the terms and W2ij = agei in the model as undistorted predictors.
Without loss of generality, we may assume that random effects are associated with the first p1 distorted predictors (p1 ≤ p) and the first q1 (q1 ≤ q) undistorted predictors. Then the underlying linear mixed effects model for individual i at time tj is
or equivalently
| (15) |
where
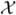 i is a ni ×(p+q+1) predictor matrix for the fixed effects with the jth row given by (1, X1ij, …, Xpij, W1ij, …, Wqij),
i is a ni ×(p+q+1) predictor matrix for the fixed effects with the jth row given by (1, X1ij, …, Xpij, W1ij, …, Wqij),
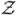 i is and ni×(p1+q1+1) predictor matrix corresponding to the random effects with the jth row given by (1, X1ij, …, Xp1ij, W1ij, …, Wq1ij), γ = (γ0, γ1, …, γp, δ1, …, δq)T and γi = (γ0i, γ1i, …, γp1i, δ1i, …, δq1i)T. The corresponding observable mixed effects varying coefficient model, generalizing (8), is
i is and ni×(p1+q1+1) predictor matrix corresponding to the random effects with the jth row given by (1, X1ij, …, Xp1ij, W1ij, …, Wq1ij), γ = (γ0, γ1, …, γp, δ1, …, δq)T and γi = (γ0i, γ1i, …, γp1i, δ1i, …, δq1i)T. The corresponding observable mixed effects varying coefficient model, generalizing (8), is
| (16) |
where X̃0ij ≡ 1 and the fixed and random varying coefficient functions are
with φ0(Ui) ≡ 1. The LME model local to bin v, approximating model (16) is
| (17) |
where βv = (β0v, β1v, …, βpv, η1v, …, ηqv)T and bvk = (b0vk, b1vk, …, bp1vk, g1vk, …, gq1vk)T are coefficient vectors,
is the vector of bin-specific errors, and
and
are the fixed and random predictor matrix for individual k in bin v analogous to the predictor matrices
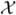 i and
i and
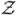 i defined above. The coefficients β’s and η’s correspond to the distorted and undistorted fixed effects and the b’s and g’s correspond to distorted and undistorted random effects.
i defined above. The coefficients β’s and η’s correspond to the distorted and undistorted fixed effects and the b’s and g’s correspond to distorted and undistorted random effects.
The solution to (17) is similar to the single-predictor case given by (11)–(12) in Section 3.2. More specifically, the estimator of βv is as given by (11), except that the matrix is replaced by the one defined above for the more general fixed and random effects structure (after equation 15). The estimator for the subject-specific effects given by (12) is modified accordingly as , where and . As before, we replace the unknown D̃v and with their R̃vk REML estimators, and , based on data within bin v. Note that the implementation requires no new ideas from the simple case because the fitting can be achieved by common software packages (e.g. R, SPLUS, SAS) as in the simple case. One only needs to specify the additional columns of data corresponding to the predictors in and .
The CA-LME estimators of the fixed and subject-specific effects are computed similar to (13) and (14), respectively, as
The covariate-adjusted estimation of the variance components proceeds similarly as described in Section 3.3 and details for the more general case are in Appendix A.
We state the consistency result of the proposed CA-LME model estimators, γ̂0, …, γ̂p, δ̂1, …, δ̂q, when the total number of subjects n and the number of subjects observed at time j, mj (j = 1, …, T) tend to infinity for a fixed number of total time points T. The proof is deferred to Appendix B. Consistency is established for data missing completely at random (unbalanced data). For m0 = infj mj, assume that m0 → ∞. As is typical for smoothing, the total number of bins H satisfies H → ∞ and m0/{H log(m0)}→ ∞ as m0 → ∞. The CA-LME model estimators are averages of the bin-specific varying coefficient function estimators: , v = 1, …, H. These estimators for the varying coefficient functions and therefore must exist for each bin in order for the proposed estimators to be well-defined Therefore, it is required that Mv and Ṽvk be nonsingular.
Theorem 1
Under the technical conditions given in Appendix B,
5 CA-LME Model of Calcium Absorption and Intake
The relationship between the effects of calcium intake and absorption is important for a variety of human health issues and disease prevention. For example, adequate calcium intake is important in preventing osteoporosis, a condition characterized by porous and fragile bones associated with increased risks of bone fractures especially in women. More recent studies, although preliminary, have linked increased calcium intake to lowered blood pressure, decreased risk of colon adenomas, and reduced weight gain over time. Although the outcome of complex diseases are typically multifactorial in nature, one relationship of interest is between calcium intake and absorption, which we explore in this section.
In particular, we are interested in examining the relationship between the effect of calcium intake levels on calcium absorption, where the response measurements and the effect of the predictor are both potentially modulated by body mass index (BMI). Because different body compositions (e.g. underweight, overweight, obese) may partly influence individuals’ ability to absorb the ingested calcium, BMI is a suitable marker for body composition that accounts for both height and weight (BMI = kg/m2). Also, because absorption continually declines with age our underlying LME model for calcium absorption for individual i at measurement occasion j is
where the parameters of interest include γ = (γ0, γ1)T, γi = (γ0i, γ1i)T, δ1, σ2 = Var(eij) and D = Cov(γi) as previously described in Section 2. Estimation is based on the observed data, denoted { , ageij} and body mass index for each subject (bmii). Note that age is incorporated as an undistorted fixed effect. The data is from a longitudinal study on n = 188 female subjects [15], between age 35 and 45 at the beginning of the study and repeated measurements were taken every 5-year intervals for a total of four occasions (1 ≤ ni ≤ 4, j = 1, …, ni). The number of subjects with 1, 2, 3 and 4 repeated measurements were 31, 41, 50 and 66, respectively.
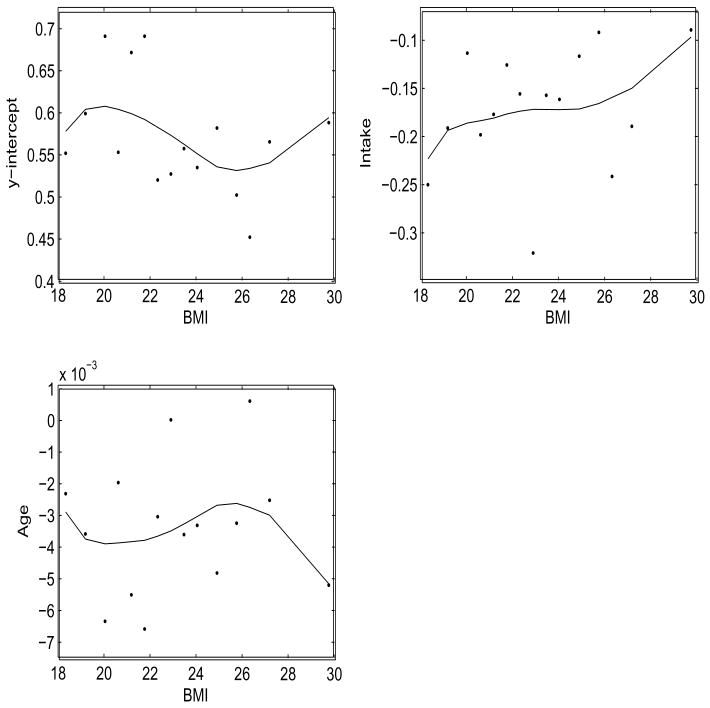
Figure 1 displays the varying coefficient estimates utilized in arriving at the parameter estimates of the CA-LME model. The observed calcium intake and age both have a negative relationship with the observed calcium absorption, while the effect of the observed intake seem to decline with increased body mass index. The y-intercept and slope varying coefficient functions for observed calcium intake are both found to be different than a constant function using the bootstrap test of ªentürk and Müller (2005) (p-values: 0.0239, 0.0153, respectively). This implies that the distortion does not reduce to the additive or the no distortion case from the goodness of fit discussion outlined in Section 3.2.
Figure 1.
Plot of β̂0(·), β̂1(·) and η̂(·) for the calciuim absorption data.
Table 1(A) summarizes the parameter estimates for the CA-LME model, γ̂0, γ̂1 and δ̂1. As a baseline comparison, Table 1(A) also displays the estimates of the underlying regression parameters using a standard LME (restricted maximum likelihood; REML estimation) with the same fixed and random effects structure as the the CA-LME model. The only difference is that the effects of BMI are ignored and estimates are obtained from an unadjusted LME model. We first note that given calcium intake, there is a significant natural decline in absorption with age that is consistent between the CA-LME analysis (δ̂1 = −0.0035) and the unadjusted-LME analysis (age slope estimate also −0.0035), where the ratios of estimates to standard errors (S.E.) are approximately −4.807 and −5.151, respectively. The significant, but small, absolute magnitude in the estimated coefficient for age reflects the age range of the study subjects (between 35 and 64 across all measurement occasions), unlike in young adulthood where there is typically a rapid decline in absorption after reaching peak bone mass and density for skeletal development (e.g., see [16]). Our main parameter of interest is γ1 for calcium intake and the unadjusted-LME model yields a significant estimate of −0.1489 (S.E., 0.0114). Similarly, the CA-LME model which accounts for the possible distortion effects of body mass index, yields the estimate of γ̂1 = −0.1705. Although both estimates of the effect of calcium intake on calcium absorption is clearly significant and in the same direction, the estimate from the CA-LME model is approximately 1.89 standard errors lower, relative to the unadjusted estimate. Thus, the CA-LME model analysis suggests a stronger inverse (negative) relationship between calcium intake and absorption after adjusting for body configuration, BMI. We note that this inverse relationship between calcium intake and absorption is consistent with previous studies (e.g., see [17]). Since bin specific estimators given in (11) are heteroskedastic, we use the wild bootstrap method with 400 boostrap replications to obtain the standard errors for the CA-LME estimates (Table 1). Details of the boostrap implementation are provided in Appendix C.) For this data we note that the precision did not improve with increased replication beyond ~100.
Table 1.
Estimation of fixed effects and variance components for the unobserved latent model of calcium absorption as a function of calcium intake and age: E(ca. absij) = γ0 + γ0i + (γ1 + γ1i)intakeij + δ1ageij, based on observed data for n = 188 women and 1 ≤ ni ≤ 4. Standard errors (S.E.) corresponding to CA-LME model estimates are obtained using 400 bootstrap samples.
| CA-LME | Unadjusted-LME | |||
|---|---|---|---|---|
| (A) Fixed effects estimates and standard error (S.E.) | ||||
| Parameter | Estimate | S.E. | Estimate | S.E. |
| Intercept (γ0) | 0.5698 | 0.0059 | 0.5575 | 0.0262 |
| Intake (γ1) | −0.1705 | 0.0041 | −0.1489 | 0.0114 |
| Age (δ1) | −0.0035 | 0.0001 | −0.0035 | 0.0005 |
| (B) Variance components estimates | ||||
| d11 = Var(γ0i) | 0.0067 | 0.0042 | ||
| d22 = Var(γ1i) | 0.0084 | 0.0012 | ||
| d12 = Cov(γ0i,γ1i) | −0.0062 | −0.0020 | ||
| σ2 | 0.0042 | 0.0046 | ||
The within- and between-subject variance parameter estimates are given in Table 1(B). The estimate of within-subject variance is smaller than the variation in the calcium intake slope parameter among individuals (as well as the intercept) for the CA-LME model. Both CA-LME and the unadjusted-LME models suggest a strong correlation between subject-specific intake slopes and intercepts and −0.92, respectively). The within-subject variance estimates are similar for both models (σ̂2 ≈ 0.004), although variation in the calcium intake slope is appreciably lower when not adjusting for BMI. Also, the number of bins used in the analysis of the calcium data was 15 for a sample size of n = 188. The coefficients estimates are similar for different number of bins used and this is further explored in the simulation study given in the next section.
6 Numerical Properties
To evaluate the performance of the proposed method, we consider a model with a mixture of fixed and random effects and distorted and undistorted predictors. The underlying LME model, analogous to the model used for calcium absorption is
| (18) |
where (γ0, γ1, δ1) = (1.5, 2.0, 0.75) and the subject-specific effects are obtained as bivariate normal random variates: γi ~ Normal(0, D) with D11 = Var(γ0i) = 0.5625, D22 = Var(γ1i) = 1.0, and D12 = Cov(γ0i, γ1i) = 0.375; thus, Corr(γ0i, γ1i) = 0.5. The undistorted predictor, Wij, is taken to be the time points tij. The within-subject covariance matrix is Ri = 0.52Ini. The predictor values are obtained as Xij ~ Normal[1.5(tij +1)2, 1], where the sequence of time points are tij = j/(T + 1), for j = 1, …, T = 6.
To mimic unbalanced data, a common feature in longitudinal data, we randomly removed a proportion, π, of observations from the complete data. We implemented the simulation study for different configurations of sample size and missing rate, (n, π), with n ranging from 100 to 800 and π from 0.1 to 0.4. Each simulation configuration was replicated 1000 times. The distorted data are Ỹij = ψ(Ui)Yij and X̃ij = φ(Ui)Xij with ψ(u) = u(u/4 + 3)/v1 and φ(u) = (3u − 1)2/v2. The constants v1 and v2 are normalizing constants so that the distorting functions satisfy the identifiability condition (4). The confounding variable Ui was obtained as Ui ~ Uniform[2, 6].
The main CA-LME estimation results are summarized in Table 2 for the fixed effects (γ) and Table 3 for the variance components. Given are estimates based on the CALME analysis, the unadjusted analysis which is the standard LME model fitted to the observed data, and the benchmark (optimal) analysis which is the latent LME model (18) fitted to the underlying (unobserved) data (Yij, Xij, Wij). The last approach is clearly the optimal one, but these estimates would not be available in practice since (Yij, Xij) are not directly observable. However, for simulation studies they can be used as the benchmark to compare the proposed CA-LME model analysis and the unadjusted analysis. Generally, as expected, unadjusted LME estimates are biased, whereas the CA-LME estimates target the true parameters. More specifically, the fixed effects estimates in Table 2 suggest that the CA-LME estimates are close to the benchmark LME estimates (LME-benchmark, Table 2) and the bias decreases as sample size increases. Furthermore, the standard unadjusted LME estimates can be severely biased for all sample size configurations.
Table 2.
Simulation study: estimation of fixed effects for unbalanced data with 20% missing. Numbers given are (A) averages and (B) standard deviation across 1000 simulations.
| Fixed effects (γ0 = 1.5, γ1 = 2, δ1 = 0.75) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| n | CA-LME | Benchmark-LME | Unadjusted-LME | ||||||
| True | 1.5 | 2 | 0.75 | 1.5 | 2 | 0.75 | 1.5 | 2 | 0.75 |
| (A) Mean | |||||||||
| 100 | 1.4956 | 2.0044 | 0.7489 | 1.4948 | 1.9983 | 0.7504 | 1.5337 | 2.2316 | 0.8790 |
| 200 | 1.4997 | 1.9988 | 0.7523 | 1.4989 | 1.9996 | 0.7508 | 1.5395 | 2.2298 | 0.8830 |
| 400 | 1.4982 | 2.0003 | 0.7480 | 1.4991 | 2.0006 | 0.7501 | 1.5401 | 2.2334 | 0.8802 |
| 800 | 1.4997 | 1.9985 | 0.7478 | 1.4996 | 1.9984 | 0.7491 | 1.5395 | 2.2304 | 0.8802 |
|
| |||||||||
| (B) Standard deviation | |||||||||
| 100 | 0.1156 | 0.1244 | 0.1726 | 0.0972 | 0.1081 | 0.1519 | 0.1165 | 0.1517 | 0.1716 |
| 200 | 0.0861 | 0.0825 | 0.1217 | 0.0716 | 0.0739 | 0.1105 | 0.0870 | 0.1010 | 0.1209 |
| 400 | 0.0595 | 0.0575 | 0.0845 | 0.0503 | 0.0490 | 0.0770 | 0.0602 | 0.0712 | 0.0854 |
| 800 | 0.0434 | 0.0424 | 0.0611 | 0.0363 | 0.0363 | 0.0550 | 0.0438 | 0.0518 | 0.0617 |
Table 3.
Simulation study: estimation of variance components for unbalanced data with 20% missing. Numbers given are (A) averages and (B) standard deviation across 1000 simulations.
| Variance components: σ2 = 0.25, D11 = 0.563, D22 = 1, D12 = 0.375 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n/10 | CA-LME | Benchmark-LME | Unadjusted-LME | |||||||||
| σ2 | D11 | D22 | D12 | σ2 | D11 | D22 | D12 | σ2 | D11 | D22 | D12 | |
| (A) Mean | ||||||||||||
| 10 | 0.236 | 0.589 | 1.008 | 0.359 | 0.250 | 0.569 | 1.010 | 0.382 | 0.289 | 0.920 | 0.182 | 1.735 |
| 20 | 0.237 | 0.584 | 0.986 | 0.356 | 0.250 | 0.562 | 1.006 | 0.376 | 0.289 | 0.910 | 0.175 | 1.730 |
| 40 | 0.240 | 0.566 | 0.981 | 0.360 | 0.250 | 0.563 | 1.002 | 0.375 | 0.289 | 0.907 | 0.175 | 1.729 |
| 80 | 0.242 | 0.561 | 0.985 | 0.367 | 0.250 | 0.563 | 0.998 | 0.376 | 0.289 | 0.912 | 0.175 | 1.719 |
|
| ||||||||||||
| (B) Standard deviation | ||||||||||||
| 10 | 0.033 | 0.190 | 0.226 | 0.112 | 0.021 | 0.144 | 0.145 | 0.103 | 0.035 | 0.226 | 0.145 | 0.350 |
| 20 | 0.023 | 0.125 | 0.157 | 0.078 | 0.015 | 0.097 | 0.103 | 0.071 | 0.025 | 0.159 | 0.096 | 0.252 |
| 40 | 0.014 | 0.086 | 0.108 | 0.057 | 0.010 | 0.069 | 0.073 | 0.052 | 0.018 | 0.114 | 0.070 | 0.181 |
| 80 | 0.010 | 0.064 | 0.073 | 0.039 | 0.007 | 0.050 | 0.049 | 0.035 | 0.012 | 0.085 | 0.049 | 0.124 |
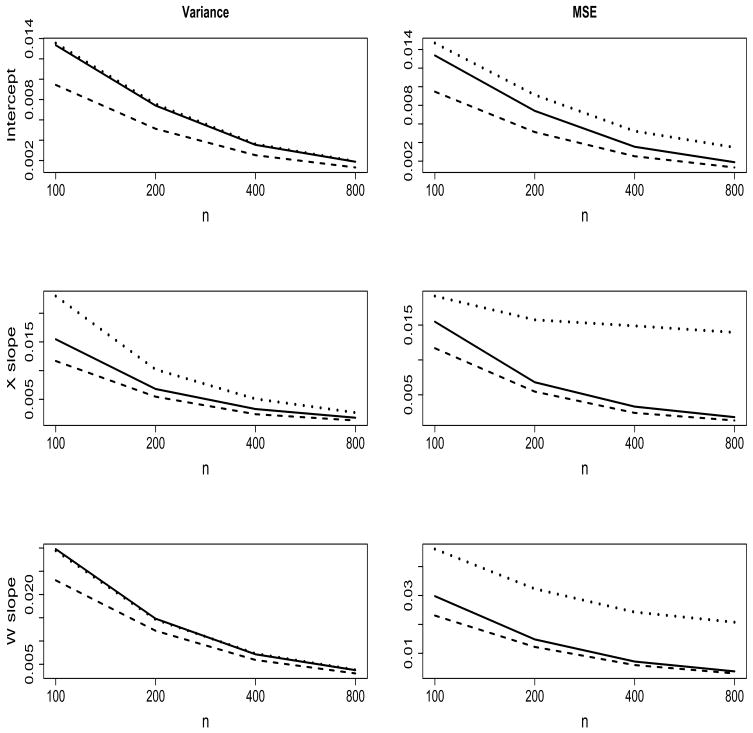
Similarly, the unadjusted LME estimates of the true variance components (σ2 and D) are generally off-target and the absolute biases are many folds larger relative to the CA-LME and benchmark estimates (Table 3). Also, similar to the fixed effects parameter estimates, the variance components estimates for CA-LME track closely the benchmark estimates. In evaluating the performance of the proposed estimators, we also examined the variance and mean square error (MSE) in estimating the fixed effects and variance components. From the fixed effects results displayed in Figure 2, we make the following observations regarding this specific simulation study. (1) The variability in the proposed CA-LME estimators is much lower in the estimation of γ1 (X-slope) and is similar for γ0 (intercept) and η1 (W-slope) compared to the unadjusted LME analysis. (2) Lower variance combined with the small bias (Table 2) result in substantially reduced MSE for the CA-LME estimators, where the estimated MSE tracks the optimal MSE closely as n increases. These results also hold for the covariate-adjusted variance components (results not shown).
Figure 2.
Estimated variance and mean square error (MSE) for estimation of fixed effects coefficients, specifically for the intercept (γ0), X slope (γ1) and W slope (δ1), corresponding to the benchmark (dashed line), CA-LME (solid line) and unadjusted LME (dotted line) estimates. Due the large MSE for the unadjusted LME estimates of the X slope, we plotted 1/4× MSE (of the unadjusted estimates) so that it would be on a similar order as the CA-LME and benchmark MSEs (column 2, row 2).
The general pattern of results described above for 20% missing data is similar to the cases with 10%, 30% and 40% missing data. The implementation of the binning algorithm require specification of the number of bins H. There are some practical restrictions on the choice of the number of bins, H. From the practical point of view, H should be chosen such that there are enough points to fit the corresponding mixed effects models in each bin. From a theoretical point of view, H → ∞and m0/(H log(m0)) → ∞. This means that the number of bins and the number of points within each bin both increase with sample size. Thus, we performed a sensitivity analysis to analyze the effects of the number of bins on the proposed estimators. For the current longitudinal data setting, our study suggests that given the minimal requirement that there are enough data to fit the LME model within each bin, the CA-LME estimates are fairly robust to the bin choice H. This was also reported in the case of linear regression models for cross-sectional data (ªentürk and Müller 2006). For the simulation results reported, the average number of bins were 10, 19, 29 and 45 corresponding to sample size n = 100, 200, 400 and 800, respectively. For each of these sample sizes, Monte Carlo simulation suggests that the correponding range of bins where the estimates are robust are (8, 14), (10, 21), (20, 33) and (30, 54), respectively. For example, Table 4 illustrates the robustness of the proposed estimators for n = 200 for the number of bins ranging from 10 to 21. Given are the bias and MSE for each parameter of the model.
Table 4.
Effects of the number of bins H on estimation (γ0 = 1.5, γ1 = 2.0, δ = 0.75) Results given are averages over simulation runs.
| # bin H | Estimates | MSE | ||||
|---|---|---|---|---|---|---|
| 10 | 1.4967 | 1.9988 | 0.7497 | 0.0060 | 0.0083 | 0.0110 |
| 13 | 1.4938 | 1.9879 | 0.7514 | 0.0053 | 0.0073 | 0.0125 |
| 16 | 1.4992 | 1.9975 | 0.7565 | 0.0075 | 0.0070 | 0.0120 |
| 19 | 1.5007 | 1.9880 | 0.7481 | 0.0066 | 0.0066 | 0.0137 |
| 21 | 1.5052 | 2.0048 | 0.7681 | 0.0069 | 0.0059 | 0.0127 |
Next, to assess the subject-specific estimators {γ̂0i, γ̂1i}, we define the following average residual sum of squares error (ARSSE) quantities of prediction error,
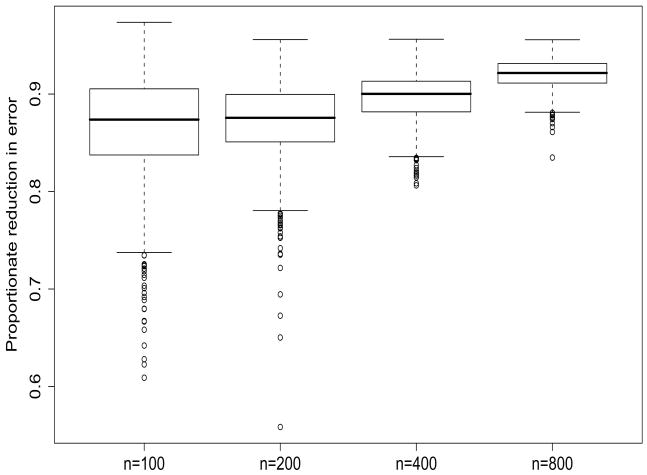
where is the population fitted values and is the fitted values with the addition of subject-specific random effects. We examine the proportionate reduction in error (PRE) due to the addition of the random effects, namely PRE = (ARSSE1 − ARSSE2)/ARSSE1. For example, with n = 200 subjects, the minimum, mean, and maximum PRE among the 1000 simulation replications are 0.56, 0.87, and 0.96 with a standard deviation of 0.04. Thus, the addition of the CA-LME predicted subject-specific effects reduce, on average, about 87% reduction in prediction error. Note that this is a substantial reduction, especially given that we have demonstrated that the bias in the population estimates (γ̂0, γ̂1, δ̂1) is very small (Table 2). The distribution of the PREs, for each sample size (20% missing), are summarized in Figure 3. The results show appreciable reduction in error for all sample sizes and, as expected, this improves with sample size.
Figure 3.
Proportionate reduction in error (PRE) due to the inclusion of estimated subject-specific effects. Displayed are results for 1000 Monte Carlo data sets (20% missing data) for each sample size n.
Finally, we note that proposed method can be implemented in standard statistical software with LME model routines, including SAS, R or Splus since they involve fitting LME models after binning by U. The computational cost is modest as it involves mainly fitting the H LME models.
7 Discussion
As described in Section 3.1, the varying coefficient model can be applied to study the relationship between the observed variables Ỹ and X̃ at varying levels of U, although this is not the objective of inference for CAR (or the CA-LME) model. Thus, the main distinction (difference) between the two approaches of analyzing the data with VCMs and with CAR is that in the latter approach, U is viewed as a “nuisance” parameter. Hence in the CAR analysis, the result is a model that is free from or adjusted for the effects of U (by removing its effect). The object of inference in the CAR/CA-LME model is the relationship between X and Y (not directely observed). Thus, if one were interested in inference for the relationship between Ỹ and X̃ and how this relationship/effect is modified by U, the VCM is the preferred/suitable modelling approach and CAR would not be suitable for this specific aim. Also, we emphasize that in the VCM analysis, U is an important part of the modelling where one of the main goals is to recover (understand) the effects of U via the varying coefficient functions, i.e. the βr(·)’s.
Although VCMs and CAR models both serve useful, but different, purposes (as described above) the applicability of CAR can seem limited, when viewed from a latent variable modelling perspective. From a latent variable modeling perspective, it is true that not being able to check the linearity of the underlying model is a limitation that it shares with other latent variable models. However, the applicability and advantage of CAR is in (biomedical) applications where the adjustments (for U) is due to general measurement errors induced by U, therefore, U is nuissance (so that the relationship of interest is between X and Y). The most common example of this in literature is the adjustment for U via division (which originally motivated the CAR method). That is, the motivation for the treatment of U as a nuisance variable and, hence, for our covariate adjusted approach comes from studies that involve adjustment via division by body configuration measures (such as body mass index or body surface area). An example is the 2002 study of Kaysen et al. [18] where albumin turnover and protein catabolic rate were among the variables that were adjusted for body surface area via division. After the adjustment, researchers proceed to analyze the linear regression relation between the adjusted variables. Here the assumption is that the effects of U on the variables are removed by the division. Thus, in such cases, CAR has an advantage over the simple adjustment via division, in that the distortion effects of U are in fact unknown (a priori) and CAR provides a way to model this uncertainty.
Also, we point out here that for the particular simulation set-up considered in Section 6, it was observed that the variance of CA-LME estimators are lower than or approximately equal to those of the unadjusted estimators. Even though we know that the unadjusted estimators do not target the underlying regression parameters (bias does not decrease with increasing n), since the variance formulas are not derived for CA-LME estimators, one cannot claim that their variance will always be larger compared to the variance of the CA-LME estimators generally. Also, unlike other typical studies where the bias and variance trade-off of two estimators can be studied via a tuning parameter, like the bandwidth, the current setting does not lend itself to such a simple analysis. With respect to this issue, there are (at least) two key considerations that explains the complicated bias-variance relationship. First, the effect of the distortion functions on the variance of both estimators are different, which makes it difficult to predict which variance will be larger in a given set-up (i.e. generally). Second, since the unadjusted estimators do not target the underlying regression parameters, they are estimating a different quantity. Hence, the estimators can potentially have very different variances. In our study, we have two estimators that have completely different forms due to the distortion and they target two different quantities.
Finally, while estimation of the varying coefficient functions (i.e. the β(·)’s) are a natural by-product of the proposed estimation procedure, estimation of φ(·) and ψ(·) is not. Although this can be viewed as an advantage of the covariate adjusted estimation approach, i.e. that without having to know the form of distortions, or without targeting these distortion functions, the underlying relationship can be targeted directly; these functions can provide information graphically. Formulating an alternative estimation algorithm to also target the distortion functions is a challenge for further research.
Acknowledgments
We are grateful to two reviewers and an Associate Editor for many detailed suggestions which substantially improved the paper. Support for this work includes the National Institute of Health (NIH) grants UL1RR024922, RL1AG032119 and RL1AG032115, National Institute of Child Health and Human Development grant HD036071, NIEHS grant P01-ES011269-06 and grant UL1 RR024146 from the National Center for Research Resources, a component of NIH.
Appendix A: Covariate-Adjusted Variance Components
Denote the vector of distorted and undistorted random effects for the underlying LME model (15) by , where and . We express the covariance matrix of these random effects, D ≡ Var(γi), as
where . The observable varying coefficient model is (16) where the random varying coefficients at Ui = u is denoted by with and . Direct calculation yields,
where
and . Therefore, the average of the variance components estimates from each bin, namely , can be adjusted to obtain the covariate-adjusted variance components estimators. From the above expression for D̃(u), the required adjustment coefficients involve
Similar to Section 3.3, it can be shown that and λrs = (γrγs)−1E{βr(U)βs(U)}. The estimators for the adjustment coefficients are:
They are sample moments corresponding to theoretical moments {λ0, λrr, λr, λrs}.
Appendix B: Proof
Technical conditions
The following assumptions are made.
C1. The covariate U is bounded below and above: −∞< a ≤ U ≤ b < ∞, for real numbers a < b. The density f(u) of U satisfies infa≤u≤b f(u) > 0, supa≤u≤b f(u) < ∞, and is uniformly Lipschitz; that is there exists a real number M such that supa≤u≤b|f(u + c) − f(u)|≤M|c| for any real number c.
C2. The needed dependence/independence structure is that U is independent of ej, Xrj is independent of ej and U; and Wsj is independent of ej for r = 1, …, p, s = 1, …, q, j = 1, …, T.
C3. For the predictors, sup |Xrij|≤B1 and sup |Wrij|≤B2 for some bounds B1, B2 ℝ+ and where sup is taken over 1 ≤ i ≤ n, 1 ≤ r ≤ p, 1 ≤ j ≤ ni. In addition for 1 ≤ r ≤ p.
C4. The functions ψ(·) and φr(·), 1 ≤ r ≤ p, are twice continuously differentiable, satisfying E{ψ(U)} = 1 E{φr(U)} = 1 and |φr(·)| > c, for some constant c > 0.
C5. Define to be the normalization of Ωi, where the row and column element of is given as , and mj1j2denotes the number of subjects observed at both times tj1 and tj2. As infj1j2 mj1j2 → ∞, in probability where the limiting p + q + 1 × p + q + 1 matrix
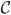 is nonsingular.
is nonsingular.
Proof of Theorem 1
We first introduce some boundedness considerations. Since Xrij and Wrij are assumed to be bounded (C3) and U has compact support (C1), X̃rij are also bounded. The matrices Ṽvk and have been assumed to be invertible previously; more precisely, assume , where 1a×b is an a ×b dimensional matrix of ones. We define and for 1 ≤ k ≤ Lv, 1 ≤ r ≤ p, 1 ≤ v ≤ H, where , is the average of the U’s in Bv. The following boundedness results for 1 ≤ r ≤ p, 1 ≤ k ≤ Lv can be obtained using Taylor expansions: ; supv,k |Δvk| = O(H−1); .
The coefficient function estimators β̂v can be expressed as
Next we expand and in the above formulation around , the average of the U’s in bin Bv. The remainder terms from this expansion, when averaged to form γ̂r, will be shown to be either or op(H−1), while the leading term will be shown to be , hence Theorem 1 follows. Details are given below.
Making use of the remainder terms Δvk and defined above, we have
where . Thus, the CA-LME estimators γ̂r becomes
where and . Let us analyze each term separately. Term P1 becomes
Note that E(P2|U,
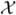 ˜, Lv,
˜, Lv,
 ) = 0 and since
is bounded uniformly in v,
) = 0 and since
is bounded uniformly in v,
Thus, and so is P3, …, Pp+q+3. With similar expansions and using the fact that Δvk, are O(H−1), . Similar considerations of conditional mean and variance can be used to show that . Thus, it follows that for 0 ≤ r ≤ p. Showing that follows closely the derivation above and therefore is omitted here.
Appendix C: Bootstrap estimate of standard errors
To obtain standard error estimates for the data analysis of section 5, the wild bootstrap is used as it is more suitable for heteroskedastic cases that commonly arise in nonparametric regression. For the covariate-adjusted regression model, the point estimators for the fixed effects are obtained by averaging heteroskedastic estimators coming from each bin given by equation (11). Bin specific estimators are indeed heteroskedastic since their variance depends on U. The wild bootstrap algorithm implemented is as follows.
Within each bin v, bin specific estimates are obtained based on the original data and the resulting residual vectors are for subject k in bin v (k = 1, …, Lv).
We multiply each residual belonging to a specific subject and repetition (i.e. each component of ) by a random variable sampled from the two-point distribution attaching masses and to the points and to obtain the wild bootstrap residual vectors , k = 1, …, Lv. (These wild bootstrap residuals approximate the variance and skewness of the residuals for each subject.)
The new responses in bin v, , are obtained from the wild bootstrap residuals; and bootstrap bin-specific estimates (11) are obtained based on the bootstrap data.
We repeat the above procedure to obtain B = 400 bootstrap samples to estimate the standard errors. (Standard error estimates in this case stabalize after B = 100 bootstrap samples.)
Contributor Information
Danh V. Nguyen, Division of Biostatistics, University of California School of Medicine Davis, California 95616, U.S.A.
Damla Şentürk, Department of Statistics, Pennsylvania State University University Park, Pennsylvania 16802, U.S.A.
Raymond J. Carroll, Department of Statistics, Texas A&M University College Station, Texas 77843, U.S.A
References
- 1.Diggle PJ, Heagerty P, Liang K-Y, Zeger SL. Analysis of Longitudinal Data. 2. Oxford University Press; Oxford: 2002. [Google Scholar]
- 2.Fitzmaurice GM, Laird NM, Ware JH. Applied Longitudinal Analylsis. John Wiley & Sons Inc; New Jersey: 2004. [Google Scholar]
- 3.Davidian M, Giltinan D. Nonlinear models for repeated measurement data. Chapman and Hall; New York: 1995. [Google Scholar]
- 4.Dawson-Hughes B. In: The role of calcium in the treatment of osteoporosis, in Osteoporosis. Marcus R, Feldman D, Kelsey J, editors. Academic Press; San Diego: 1996. pp. 1159–1168. [Google Scholar]
- 5.Wu K, Willett WC, Fuchs CS, Colditz GA, Giovannucci EL. Calcium intake and risk of colon cancer in women and men. Journal of the National Cancer Institute. 2002;94:437–46. doi: 10.1093/jnci/94.6.437. [DOI] [PubMed] [Google Scholar]
- 6.Allender PS, Cutler JA, Follmann D, Cappuccio FP, Pryer J, Elliott P. Dietary calcium and blood pressure: a meta-analysis of of randomized clinical trials. Annals of Internal Medicine. 1996;124:825–831. doi: 10.7326/0003-4819-124-9-199605010-00007. [DOI] [PubMed] [Google Scholar]
- 7.Zemel MB. Regulation of adiposity and obesity risk by dietary calcium: mechanisms and implications. Journal of the American College of Nutrition. 2002;21:146S–151S. doi: 10.1080/07315724.2002.10719212. [DOI] [PubMed] [Google Scholar]
- 8.Şentürk D, Müller HG. Covariate adjusted regression. Biometrika. 2005;92:59–74. [Google Scholar]
- 9.Carroll RJ, Ruppert D, Stefanski LA, Crainiceanu CM. Measurement Error in Nonlinear Models: A Modern Perspective. 2. Chapman and Hall CRC Press; Boca Raton: 2006. [Google Scholar]
- 10.Hwang JT. Multiplicative error-in-variables models with applications to recent data released by the U.S. Department of Energy. Journal of the American Statistical Association. 1986;81:680–688. [Google Scholar]
- 11.Iturria S, Carroll RJ, Firth D. Multiplicative measurement error estimation: estimating equations. Journal of the Royal Statistical Society, Series B. 1999;61:547–561. [Google Scholar]
- 12.Diggle PJ, Verbyla A. Nonparametric estimation of covariance structure in longitudinal data. Biometrics. 1998;54:401–415. [PubMed] [Google Scholar]
- 13.Wu H, Liang H. Backfitting random varying-coefficient models with time-dependent smoothing covariates. Scandinavian Journal of Statistics. 2004;31:3–19. [Google Scholar]
- 14.Şentürk D, Müller HG. Inference for covariate adjusted regression via varying coefficient models. Annals of Statistics. 2006;34:654–679. [Google Scholar]
- 15.Davis CS. Statistical methods for the analysis of repeated measurements. Springer-Verlag; New York: 2002. [Google Scholar]
- 16.Matkovic V. Calcium metabolism and calcium requirements during skeletal modeling and consolidation of bone mass. American Journal of Clinical Nutrition. 1991;54:245S–605S. doi: 10.1093/ajcn/54.1.245S. [DOI] [PubMed] [Google Scholar]
- 17.Heaney RP, Recker RR, Stegman MR, Moy AJ. Calcium absorption in women: relationships to calcium intake, estrogen status, and age. Journal of Bone and Mineral Research. 1989;4:469–475. doi: 10.1002/jbmr.5650040404. [DOI] [PubMed] [Google Scholar]
- 18.Kaysen GA, Dubin JA, Müller HG, Mitch WE, Rosales LM, Levin NW, the Hemo Study Group Relationship among inflammation nutrition and physiologic mechanisms establishing albumin levels in hemodialysis patients. Kidney International. 2002;61:2240–2249. doi: 10.1046/j.1523-1755.2002.00076.x. [DOI] [PubMed] [Google Scholar]