Abstract
Mucoid strains of Pseudomonas aeruginosa that overproduce alginate are associated with chronic pulmonary disease (e.g. cystic fibrosis). Mutants defective in one of several periplasmic proteins (AlgKGX) for alginate secretion release alginate fragments due to the activity of an alginate lyase (AlgL) in the periplasm, which cleaves the newly formed polymers. However, mutants defective in Alg8 or Alg44 did not secrete polymer or alginate fragments, suggesting that both these membrane proteins have a role in the polymerization reaction. A model for the membrane topology of Alg8, a glycosyltransferase (GT), was constructed using PhoA fusions. This provided evidence for a large cytoplasmic loop containing the active domains predicted for β-GTs such as Alg8 and five transmembrane (TM) domains, one of which resembles a cleavable signal peptide. The C-terminal TM domain of Alg8 was critical for the polymerization reaction in vivo. Alanine substitution mutagenesis showed that all of the predicted active site residues in the widely spaced D, DxD, D, LxxRW motif were required for polymerization activity in vivo, and two of these substitutions also affected Alg8 protein stability. A membrane topology model for Alg44 was also constructed using PhoA fusions, and this showed a central TM domain and predicted an N-terminal TM domain that may be a membrane anchor. An N-terminal PilZ domain in Alg44 for c-di-GMP [bis-(3′,5′)-cyclic dimeric GMP] binding, which is required for alginate synthesis, was localized to the cytoplasmic loop. The long periplasmic C terminus of Alg44 contains a region similar to membrane fusion proteins (MFPs) of multi-drug efflux systems, which predicts the possibility of its interaction with another protein in this compartment. A Western blot analysis of the outer-membrane porin AlgE showed reduced AlgE levels in the alg44 mutant, whereas expression of Alg44 in trans restored AlgE within the cell. C-terminal truncations of Alg44 as small as 24 amino acids blocked alginate polymerization in vivo, indicating a critical role for the MFP domain. These studies suggest that Alg44 may act as a co-polymerase in concert with Alg8, the major GT, and that both inner-membrane proteins are required in vivo for the polymerization reaction leading to alginate production.
INTRODUCTION
Strains of Pseudomonas aeruginosa causing chronic pulmonary infections, such as in patients with cystic fibrosis (CF), often demonstrate a mucoid colony phenotype. The infecting P. aeruginosa strains undergo mucoid conversion during the course of colonization (Gilligan, 1991). The mucoid phenotype is due to the overproduction of the exopolysaccharide alginate (Jain & Ohman, 2004), which promotes persistence in the lungs and increases P. aeruginosa resistance to phagocytosis (Pier et al., 2001).
Alginate is composed of the uronic acids D-mannuronate and its C-5 epimer L-guluronate with β-1,4 linkages. All the known genes identified for alginate production except one are located in the algD operon: algD-8-44 K-E-G-X-L-I-J-F-A (Jain & Ohman, 2004). Alginate is made by many Pseudomonas species and also Azotobacter vinelandii, and the sequencing of their genomes shows the same algD operon. In the alginate biosynthesis pathway, AlgA catalyses two steps in the pathway to yield GDP-mannose (Shinabarger et al., 1991), which is then converted by the dehydrogenase activity of AlgD to GDP-mannuronic acid (Roychoudhury et al., 1989). Alg8 shares structural similarity with glycosyltransferases (GTs) of family 2, making Alg8 the probable alginate polymerase (Saxena et al., 1995). This was confirmed with an in vitro assay showing that Alg8-containing envelope fractions of mucoid P. aeruginosa could utilize GDP-mannuronate as a substrate for alginate polymerase activity (Remminghorst & Rehm, 2006a). Another protein required for alginate biosynthesis is Alg44 (Maharaj et al., 1993; Remminghorst & Rehm, 2006b). Alg44 has been shown to have a PilZ domain for the binding of bis-(3′,5′)-cyclic dimeric GMP (c-di-GMP), a cofactor that is essential for Alg44 function and alginate production (Merighi et al., 2007). The polymer undergoes further modification by periplasmic AlgG, a C-5 epimerase that converts some D-mannuronates to L-guluronates (Franklin et al., 1994), and by AlgIJF, which O-acetylates the mannuronates at the polymer level (Franklin & Ohman, 2002). AlgL is an alginate lyase that can degrade alginate polymers (Schiller et al., 1993). AlgE has been shown to be an outer-membrane porin (Rehm et al., 1994).
Mutants of mucoid P. aeruginosa strain FRD1 defective in the production of periplasmic proteins AlgK, AlgG or AlgX have a non-mucoid phenotype. However, they produce small fragments of alginate due to the action of AlgL, an alginate lyase co-expressed in the algD operon, which degrades newly formed polymers (Jain & Ohman, 1998, 2005; Jain et al., 2003; Robles-Price et al., 2004). AlgK, AlgG, AlgX and AlgL may form a periplasmic scaffold to bring newly synthesized polymers to the outer-membrane porin, AlgE, and protect the polymer from degradation by AlgL (Jain & Ohman, 2005).
Here we sought to better understand the D-mannuronate polymerization step in alginate biosynthesis, which requires the GT Alg8. GTs catalyse the transfer of a sugar moiety from an activated donor to an acceptor molecule (Campbell et al., 1997; Saxena et al., 1995). GTs can be grouped as inverting or retaining according to the stereochemistry of the linkages formed. The product of an inverting GT displays the opposite stereochemistry at the anomeric centre to that of the activated donor. In contrast, the product of a retaining GT retains the same anomeric configuration of the donor (Campbell et al., 1997). At the amino acid sequence level, GTs have very low sequence homology due to the wide diversity of acceptor molecules and sugar donors (Saxena et al., 1995). However, GTs can be placed into structural families using a computer algorithm called hydrophobic cluster analysis (HCA), which makes structural predictions based upon the secondary structure of proteins. HCA was used to demonstrate Alg8’s structural homology to family 2 (inverting) β-GTs (Campbell et al., 1997), which is one of the largest families of GTs (Campbell et al., 1997). Family 2 GTs include those involved in bacterial capsule or exopolysaccharide synthesis such as chitin synthesis by NodC in Sinorhizobium species, cellulose production by AcsAB in Gluconacetobacter xylinus (formerly Acetobacter xylinum), and hyaluronate synthesis by HasA in Streptococcus pyogenes (Campbell et al., 1997; Saxena et al., 1990, 1995).
In this study we showed that Alg8 has a membrane topology that is similar to other multi-spanning membrane family 2 (inverting) GTs in that it has a large hydrophilic region in the cytoplasm, which contains the active site. Previous predictions suggested that the active site of Alg8 has two domains (Saxena et al., 1995), and here we verified the role of the conserved active-site residues in alginate polymerization by examining the effects of alanine substitutions. In addition, given that an alg44 mutant has the same phenotype as an alg8 mutant with respect to the inability to produce alginate or alginate fragments, we investigated the membrane topology of Alg44 as well. The N-terminal PilZ domain was localized to the cytoplasm, and the C terminus of Alg44, which has homology to efflux pump proteins, was localized to the periplasm and shown to be required for polymer formation in vivo.
METHODS
Bacterial strains, plasmids and media
Bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa and Escherichia coli strains were grown at 37 °C in L-broth (Sigma) plus the appropriate antibiotics. For uronic acid assays, P. aeruginosa was grown in a defined medium that promotes alginate production (MAP), which contained 100 mM monosodium glutamate, 7.5 mM NaH2PO4, 16.8 mM K2HPO4, and 10 mM MgSO4. Triparental matings were used to mobilize plasmids from E. coli to P. aeruginosa using the conjugative helper pRK2013 (Jain & Ohman, 1998). Transconjugants were selected using a 1: 1 mixture of Pseudomonas Isolation Agar (PIA) (Difco) and L-agar supplemented with the appropriate antibiotics. Antibiotic concentrations (per ml) used were as follows: ampicillin at 100 μg, carbenicillin at 150 μg, and kanamycin at 30 μg.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or phenotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | φ80dlacZΔM15Δ (lacZYA–argF) endA1 recA1 | Stratagene |
| P. aeruginosa | ||
| FRD1 | Cystic fibrosis isolate, Alg+ | Ohman & Chakrabarty (1981) |
| FRD2 | algT18, Alg− | DeVries & Ohman (1994) |
| FRD1108 | alg8Δ:: GmR (non-polar), Alg− | This study |
| FRD1144 | alg44:: GmR (non-polar), Alg− | This study |
| FRD1200 | algG:: GmR (non-polar), Alg− | Jain et al. (2003) |
| PAO1 | Wild-type, non-mucoid | This laboratory |
| Plasmids | ||
| pEX100T | oriV(ColE1) bla (CbR/ApR) sacB oriT | Schweizer (1992) |
| pPHO7 | P15A oriV (TcR) ‘phoA (2.6 kb XbaI–XhoI fragment) | Gutierrez & Devedjian (1989) |
| pMF54 | ColE1-SF broad-host replicon ApR oriT(RK2) Ptrc lacIQ | Franklin et al. (1994) |
| pRK2013 | ColE1 Tra(RK2)+ KmR | Jain & Ohman (1998) |
| pSJ12 | Source of GmR non-polar cassette (0.7 kb) | Jain & Ohman (1998) |
| pSJ44 | pMF54 expressing alg44 algK (3.8 kb EcoRI fragment) | This study |
| pSJ184 | pEX100T with Δalg8:: GmR alg44 | This study |
| pSJ199 | pMF54 expressing alg8 alg44 (3.6 kb XhoI–KpnI fragment) | This study |
| pSJ220 | pEX100T with alg44:: GmR algK | This study |
| pSJ340 | pMF54 expressing alg8(1-346)-phoA | This study |
| pLO11 | pMF54 expressing alg8(1-417)-phoA | This study |
| pLO12 | pMF54 expressing alg8(1-393)-phoA | This study |
| pLO17 | pMF54 expressing alg8(1-377)-phoA | This study |
| pLO19 | pMF54 expressing alg8(1-34)-phoA | This study |
| pLO22 | pMF54 expressing alg8(1-245)-phoA | This study |
| pLO25 | pMF54 expressing alg8(1-474)-phoA | This study |
| pLO28 | pMF54 expressing alg8(1-494)-phoA | This study |
| pLO37 | pMF54 expressing alg8(D136A) | This study |
| pLO39 | pMF54 expressing alg8(D191A) | This study |
| pLO42 | pMF54 expressing alg8(D299A) | This study |
| pLO44 | pMF54 expressing alg8(L339A) | This study |
| pLO63 | pMF54 expressing alg8(V158A) | This study |
| pLO71 | pMF54 expressing alg8(R342A) | This study |
| pLO77 | pMF54 expressing alg8(W343A) | This study |
| pLO32 | pMF54 expressing alg44(1-35)-phoA | This study |
| pLO33 | pMF54 expressing alg44(1-139)-phoA | This study |
| pLO34 | pMF54 expressing alg44(1-250)-phoA | This study |
| pLO51 | pMF54 expressing alg44(1-389)-phoA | This study |
| pLO66 | pMF54 expressing alg44(1-364-stop) | This study |
| pSJ295 | pMF54 expressing alg44(1-244)-phoA | This study |
| pSJ303 | pMF54 expressing alg44(1-142)-phoA | This study |
Abbreviations: Alg+, mucoid; Alg−, non-mucoid; ApR, ampicillin resistance; CbR, carbenicillin resistance; GmR, gentamicin resistance (non-polar); KmR, kanamycin resistance; TcR, tetracycline resistance. phoA, promoterless alkaline phosphatase gene.
DNA and sequence manipulations
Pfu Turbo (Stratagene) was used to amplify DNA and the QIAEX II Gel Extraction kit was used to purify inserts for plasmid constructs. The Qiagen QIAprep spin Miniprep kit was used to purify plasmid DNA. All primers used were custom synthesized by Operon Technology. The sequences of Alg8 and Alg44 proteins were available at the P. aeruginosa PAO1 genome project website (http://www.pseudomonas.com). A Kyte–Doolittle plot of hydrophilicity was performed using Lasergene analysis software by DNASTAR. Predictions for transmembrane domains were performed using TMpred at the ExPASy Molecular Biology Server (http://www.expasy.org). Homology searches were performed using the BLAST network.
Construction of alg8 and alg44 non-polar mutants
To construct an alg8 mutant of mucoid strain FRD1, a 3.8 kb XhoI–KpnI fragment containing alg8–alg44 was cloned into pBluescript II KS(–); the SphI–BglII fragment within alg8 was replaced with a non-polar gentamicin-resistance (GmR) cartridge from pSJ12, and then it was cloned into the SmaI site of the suicide gene replacement vector pEX100T to form pSJ184. To construct an alg44 mutant, a 3.8 kb EcoRI fragment containing alg44–algK was cloned into pBluescript II KS(–); the GmR cartridge was inserted into the unique BclI site within alg44, and the EcoRI fragment was then cloned into pEX100T to form pSJ220. Suicide plasmids pSJ184 and pSJ220 were introduced into mucoid FRD1 by triparental mating with selection for bla (CbR) to obtain merodiploids. Colonies were grown without selection to promote spontaneous recombination, and then selected for GmR and sucrose resistance (i.e. loss of vector-encoded sacB) and screened for CbS (i.e. loss of vector-encoded bla). One of the non-mucoid Δalg8:: Gm mutants obtained, FRD1108, was verified by PCR for the Δalg8:: Gm allele and shown to be complemented in trans to mucoid by pSJ199 expressing alg8. One of the non-mucoid alg44:: Gm mutants obtained, FRD1144, was verified by PCR for the alg44:: Gm insertion and shown to be complemented in trans to mucoid by pSJ44 expressing alg44.
Construction of alg8′-phoA and alg44′-phoA translational fusions
For Alg8-PhoA constructions, PCR reactions were performed using a 5′ primer designed to include 9 bp upstream of the predicted ATG start codon of alg8 and to form a unique restriction site BspHI; the 3′ primers were designed to match various regions within the predicted hydrophilic domains of Alg8 and to form the unique restriction site XbaI. For Alg44-PhoA constructions, PCRs used a 5′ primer designed to include the predicted ATG start codon of alg44 and to form a unique restriction site NcoI; the 3′ primers were designed to match various regions within the predicted hydrophilic domains of Alg44 and to form the unique restriction site XbaI. The BspHI–XbaI fragments of alg8 and NcoI–XbaI fragments of alg44 were cloned into the compatible NcoI and XbaI sites of the broad-host-range expression vector pMF54 (Franklin et al., 1994), to place the genes in the correct orientation relative to a trc promoter. An XbaI–XhoI gene cassette from pPHO7 (Gutierrez & Devedjian, 1989), containing a phoA allele encoding alkaline phosphatase without its signal sequence or ribosome-binding site, was then fused in-frame downstream of each individual alg8′ or alg44′ construct in the XbaI–XhoI sites. To confirm that alg8′-phoA and alg44′-phoA constructs were in-frame, the plasmids were sequenced at ATGC, Inc. or the VCU Nucleic Acids Core Laboratory (Richmond, VA). Genes for the fusion proteins were under LacIQ control and were induced with 1 mM IPTG.
Enzyme assays
Alkaline phosphatase (PhoA) activities of each Alg8′-PhoA fusion in E. coli were visually screened for blue colonies on L-agar containing ampicillin and the chromogenic substrate XP (5-bromo-4-chloro-3-indolyl phosphate). To measure PhoA activity in P. aeruginosa, overnight cultures were diluted in L-broth with carbenicillin and grown to an OD600 of 0.4–0.6. A 1 ml sample of culture was removed and the remaining culture was induced for 2 h by adding 1 mM IPTG. An aliquot (100–200 μl) was removed from uninduced and induced cultures and added to 1 M Tris/HCl (pH 8.0) and permeabilized by adding 20 μl 0.1 % SDS and 40 μl chloroform (Haardt & Bremer, 1996). The cells were incubated for 5 min at 37 °C, vortexing periodically. Then 200 μl of the PhoA substrate p-nitrophenyl phosphate (Sigma) was added and incubated at 37 °C until a yellow colour developed. The reaction was stopped by adding 200 μl 1 M K2HPO4 (Brickman & Beckwith, 1975). The hydrolysis of the substrate was measured spectrophotometrically in Miller units. Protein concentrations were determined using a Bradford assay.
Site-directed mutagenesis
Site-directed mutagenesis of alg8 was performed using the method based on the QuikChange mutagenesis kit (Stratagene). First, primers upstream and downstream of alg8 corresponding to the putative start and last codon were designed to incorporate the unique restriction sites BspHI and XbaI, respectively. DNA from FRD1 was PCR amplified with Pfu Turbo and gel extracted using the QIAEX II Gel Extraction kit. After gel extraction, the PCR fragment was blunt ligated into pUC19 that was digested with SmaI to create pLO26. This plasmid was used as template for all mutagenic experiments. Primers used for site-directed mutagenesis were designed to be directly complementary to each other, with the mutation to be made in the middle of the primer. After PCR with the mutagenic primers, the product was digested with DpnI for 2 h and transformed into E. coli DH5α. The plasmid was reisolated using the Qiagen QIAprep spin Miniprep kit and digested with BspHI and XbaI. The band was excised from a gel, restriction enzyme digested and then ligated into the NcoI and XbaI sites of pMF54. Each clone was sequenced to confirm the presence of the mutation. The phoA-containing XbaI–XhoI gene cassette from pPHO7 was then cloned downstream of the inserts to provide a tag for immunodetection.
Western blot analysis
To visualize the Alg8′-PhoA and Alg44′-PhoA fusion proteins, or AlgE, overnight cultures were diluted into L-broth, and plasmids were maintained by the addition of ampicillin for E. coli or carbencillin for P. aeruginosa. The cultures were incubated for 2 h at 37 °C under aerobic conditions. Plasmids expressing genes from the trc promoter were induced by adding 1 mM IPTG and allowed to continue growing for 2 h at 37 °C. Aliquots of 1 ml were removed and centrifuged. The pellets were resuspended in 2× Laemmli sample buffer and heated at 100 °C for 5 min. Aliquots from each sample (20 μl for E. coli and 100 μl for P. aeruginosa) were loaded and electrophoretically separated by SDS-PAGE analysis with a 12 % separating gel. The proteins were transferred to a nitrocellulose membrane (Bio-Rad) overnight. The membrane was blocked for 2 h in TBS [100 mM Tris/HCl (Boucher et al., 1996), 0.9 % NaCl] containing 0.1 % Tween and 1 % BSA. The membrane was probed with rabbit antiserum, against alkaline phosphatase (Rockland) or recombinant AlgE, at a concentration of 1: 10 000. The antibody–antigen complexes were detected using horseradish peroxidase-conjugated goat anti-rabbit antibodies (Sigma). The bands were visualized using Super Signal West Pico chemiluminescent substrate and exposed to X-ray film (Pierce).
Detection of alginate fragments
Alginate is composed of two uronic acid residues, and thus the amount of alginate or alginate fragments in culture supernatants was determined by measuring the concentration of uronic acids. Briefly, overnight cultures were diluted into 20 ml MAP broth, and then cultures were incubated with maximum aeration for 24 h. Cultures were centrifuged (1 h at 6000 r.p.m.), and samples of the supernatant were directly assayed for uronic acids using a carbazole-spectrophotometric method (Knutson & Jeanes, 1968). MAP growth medium was used instead of L-broth to lower non-specific background reactions. Alginic acid from Macrocystis pyrifera (Sigma) was used as the standard. To detect alginate fragments, produced by some non-mucoid mutants of FRD1, culture supernatants were assayed for uronic acids before and after exhaustive dialysis against saline for 24 h. In cultures producing alginate fragments, the uronic acids were lost by dialysis treatment. To detect intracellular uronic acids, cell pellets were resuspended in 10 mM Tris/HCl (pH 7.4), lysed by sonication, centrifuged and assayed for uronic acids as described above. Culture supernatants and sonicates of strain FRD2, which produces no alginate due to a mutation in the regulator encoded by algT, was used to determine non-specific background levels in the uronic acid assay.
RESULTS
Alg8 and Alg44 play roles in polymer formation
The Alg8− and Alg44− mutants constructed here in the mucoid strain FRD1 (i.e. FRD1108 and FRD1144, respectively) were non-mucoid as expected. However, unlike an ΔalgG mutant, neither released degraded alginate, which would be detected as small (dialysable) uronic acids (Table 2). As Alg8 is the GT for alginate, utilizing GDP-mannuronate as substrate to synthesize the polymer (Remminghorst & Rehm, 2006a), an alg8 mutant would not be expected to produce alginate or alginate fragments. Interestingly, the same phenotype was observed with the Alg44− mutant, suggesting that it also plays a role in polymerization. Others have also observed this phenotype (Remminghorst & Rehm, 2006b). Thus, Alg44 may be acting as a co-polymerase by mechanisms not yet understood. To better understand the roles of Alg8 and Alg44 in alginate polymer formation, their membrane topology and other characteristics were examined.
Table 2.
Effect of mutations in the algD operon on the production of alginate polymer and alginate fragments
| Strain | Mutation | Alginate (mucoid) | Alginate fragments (μg ml−1)* |
|---|---|---|---|
| FRD1 | Wild-type | + | 0 |
| FRD1200 | ΔalgG | − | 1200 |
| FRD1108 | Δalg8 | − | 0 |
| FRD1144 | alg44 | − | 0 |
Secretion of alginate fragments was determined as previously described (Jain & Ohman, 2005) by the demonstration of uronic acids in culture supernatants of non-mucoid strains that are lost by exhaustive dialysis.
Construction of a topological model for Alg8 by PhoA fusion analysis
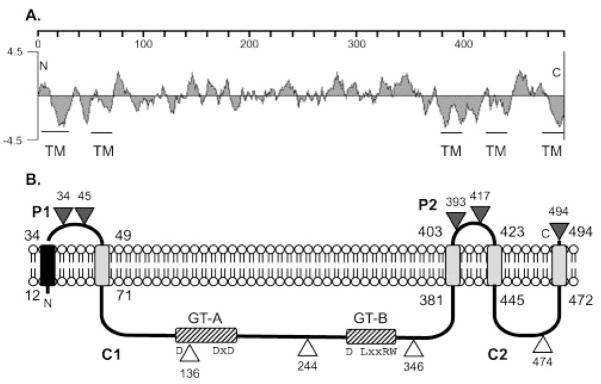
The predicted Alg8 (PA3541) coding sequence in P. aeruginosa was available through the annotated genome project (http://www.pseudomonas.com), and it encodes 494 amino acids. A hydrophilicity plot of the sequence showed several hydrophobic regions suggesting the presence of transmembrane (TM) domains (Fig. 1A). An entry for Alg8 in the SWISS-PROT database (http://www.expa-sy.org) predicted that Alg8 had four TM domains at residues 12–34, 49–71, 381–403 and 423–445. Analysis of Alg8 using TMpred predicted an additional TM domain at the C terminus: 472–494. The first TM has the characteristics of a typical signal peptide (Pugsley, 1989), ending at residue 29 with Ala-x-Ala, which is the recognition sequence for signal peptidase. A model for the membrane topology of Alg8, with a large cytoplasmic region containing the predicted enzymic domains (see below), is shown in Fig. 1B. Others have predicted a similar membrane topology for Alg8 from P. aeruginosa (Remminghorst & Rehm, 2006a) and A. vinelandii (Mejia-Ruiz et al., 1997).
Fig. 1.
(A) A hydrophilicity plot of Alg8 and a TMpred analysis suggest five TM domains as indicated. The scale shows the residue numbers in Alg8. (B) Model for the orientation of Alg8 in the inner membrane based on the activities of in-frame Alg8′-PhoA fusions. Triangles show the fusion joints to PhoA, with the number showing the terminal Alg8 residue; black indicates positive activity (periplasmic) and white indicates negative activity (cytoplasmic). P1 and P2 are predicted periplasmic loops, and C1 and C2 are predicted cytoplasmic loops. The black rectangle in the membrane represents a TM domain predicted to be a cleavable signal peptide, and the grey rectangles represent other TM domains. The locations of GT domains A and B in cytoplasmic loop C1 are indicated with hatched rectangles. The widely spaced D, DxD, D, (L/Q)xxRW motif, highly conserved in the active site of family 2 GTs, is shown.
We tested the above model of Alg8 membrane topology using a PhoA protein fusion analysis. The in-frame fusion of PhoA to a protein only produces PhoA activity if it is delivered to the periplasmic compartment (Manoil et al., 1988). Plasmids expressing alg8′-phoA fusions were constructed with the fusion joints in the hydrophilic regions immediately following amino acids 34, 45, 136, 244, 346, 393, 417, 474 and at the terminal residue, 494 (shown as triangles in Fig. 1B). These were transformed into E. coli and the transformants screened for PhoA activity on L-agar containing the chromogenic substrate XP. When PhoA was fused to the last encoded amino acid of Alg8, dark blue colonies were observed on the indicator plates (L-agar+XP), indicating that the plasmid indeed expressed the Alg8′-PhoA protein and that the C terminus was exposed to the periplasm as predicted (Fig. 1B). Likewise, the PhoA fusions in the predicted periplasmic loops P1 and P2 produced blue colonies, thus verifying their periplasmic locations. Also, PhoA fusions located in the predicted cytoplasmic loops C1 and C2 were inactive, giving rise to a white colony phenotype as expected. Although the cytoplasmic loops were not confirmed with positive LacZ protein fusions, the results with in-frame, inactive PhoA fusions were completely consistent with the computational model, which placed the GT active sites of loop C1 in the cytoplasm as predicted. Each alg8′-phoA fusion plasmid was also conjugated into P. aeruginosa FRD1 and PAO1, but the secretion of alginate or pigments by these strains interfered with the blue/white screening test. Thus, the activity of each Alg8′-PhoA in P. aeruginosa FRD1 was determined by testing cell lysates for the hydrolysis of the PhoA substrate p-nitrophenyl phosphate. The results confirmed what was seen in E. coli using the XP plate test, where periplasmic Alg8-PhoA fusions showed high activity and cytoplasmic fusions were at approximately background levels (data not shown). Overall, these experimental data were consistent with the model (Fig. 1B) for the membrane topology of Alg8.
Tests of Alg8′-PhoA constructs for restoration of alginate production
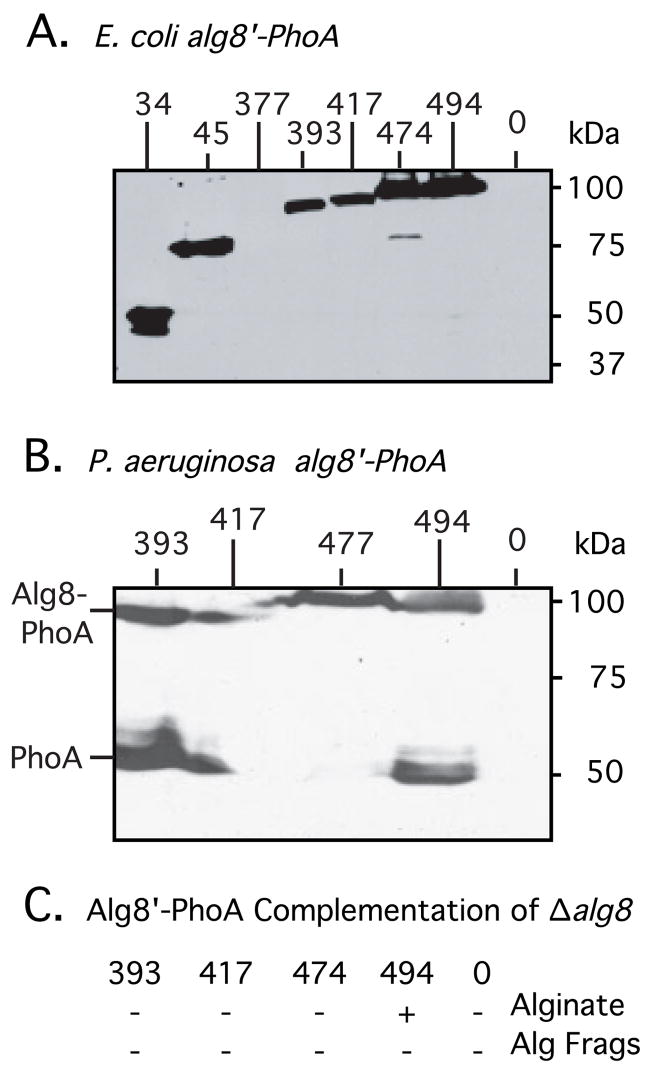
The Alg8′-PhoA fusions tested above were then used to determine the effects of C-terminal truncations of Alg8 on alginate biosynthesis in P. aeruginosa. First, we confirmed that expression of the fusion proteins resulted in stable proteins. An attempt to derive an antibody to Alg8 was unsuccessful, and so the Alg8-PhoA proteins were detected by Western blot analysis using an anti-PhoA antibody. All the strains described above carrying the Alg8′-PhoA constructs were expressed in E. coli, and the expected sizes of fusion proteins were observed (Fig. 2A). However, one Alg8-PhoA fusion at amino acid 377, not included in the analysis above, produced a protein that was difficult to detect (Fig. 2A, lane 3); the fusion joint would be in close proximity to the membrane, which may have caused its instability.
Fig. 2.
To demonstrate the expression of alg8′-phoA constructs in E. coli (A) and in P. aeruginosa FRD1108 (B), cultures were induced with IPTG, and total cell proteins were examined by Western blot analysis using anti-PhoA antibodies. Lanes are marked with the terminal amino acid in Alg8 before the PhoA fusion. Induced cultures expressed a band corresponding to the approximate size of the predicted fusion protein in E. coli. Lanes marked 0 were negative controls and contain proteins from bacteria with pLO4, which contains the unfused phoA gene cassette used to create PhoA fusions. Longer exposure was required to observe an Alg8(377)-PhoA protein of correct size encoded by pLO17, indicating that the protein was not stable. (C) Ability of Alg8-PhoA fusions, designated by their last Alg8 amino acid, to complement FRD1108 Δalg8 for alginate production or for alginate fragments (Alg Frags) is indicated.
The cytoplasmic loop C1 contained the putative active site of Alg8, and so we asked whether this alone might be sufficient for alginate production. To test this, plasmids expressing Alg8′-PhoA with the cytoplasmic loop C1 still intact were conjugated into the Δalg8 mutant FRD1108 and induced with 1 mM IPTG. A Western blot analysis of the fusion proteins in FRD1108 using anti-PhoA confirmed that the proteins were being expressed in P. aeruginosa, although variable amounts of degradation were observed resulting in a stable PhoA (48 kDa) band (Fig. 2B). Interestingly, only the plasmid expressing full-length Alg8(494 residues)-PhoA (by pLO28) was able to complement the Δalg8 mutation and restore alginate production (Fig. 2C). Even the Alg8′-PhoA fusion at residue 474, which encoded the entire protein except for the last 20 amino acids, was unable to restore the mucoid phenotype to FRD1108. We then investigated the possibility that alginate polymers were being synthesized by Alg8′-PhoA fusions, but were subject to AlgL lyase degradation. We tested for the presence of alginate fragments in 24 h culture supernatants of FRD1108(alg8′-phoA) strains; however, none was detected (Fig. 2C). Sonicates of the cell pellets were also examined for intracellular accumulation of alginate oligomers, but uronic acids were not detected above background. Although it is possible that Alg8′-PhoA fusions misfolded, these results suggest that the C-terminal TM domain of Alg8 is necessary in vivo for polymer production, even though it is well separated from the predicted catalytic domain.
Identification of critical residues for Alg8 activity in vivo
A powerful sequence comparison method called hydrophobic cluster analysis (HCA) has been used to discover putative GTs. An HCA analysis of the database of available protein sequences led to the identification of Alg8 as a probable β-GT, which included the prediction of several critical active-site residues (Saxena et al., 1995). A region in Alg8 called GT domain A contains the two conserved aspartic acids D133 and D190, with D188–D190 in the often-seen DxD motif. Alg8 also contains the GT domain B with a conserved D296 and an LxxRW motif (residues 336–340), although the latter is typically seen as a QxxRW motif. An in vitro analysis of other GTs has shown the requirement of these conserved amino acids for GT activity (Garinot-Schneider et al., 2000; Keenleyside et al., 2001; Saxena & Brown, 1997). A comparison of the Alg8 sequence to its most distant Alg8 homologue, found in A. vinelandii, showed not only the presence of these conserved residues, but also that the overall sequences of GT domains A and B were well conserved (data not shown).
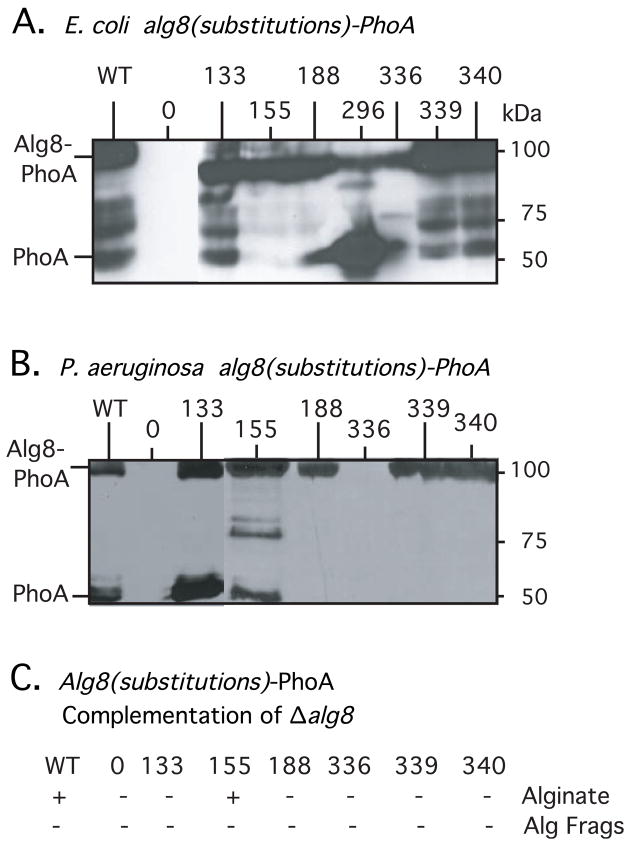
To test the prediction of Saxena et al. (1995) that the critical conserved residues in Alg8 were important for GT function, each codon was changed to encode alanine by site-directed mutagenesis in the full-length alg8(1-494)-phoA construct. Because each Alg8 protein was tagged with PhoA, it was possible to test for their expression and stability in bacterial cells. A Western blot analysis showed that the mutant Alg8-PhoA proteins were all being expressed in E. coli (Fig. 3A). However, compared to wild-type Alg8-PhoA (Fig. 3A, lane 1), the bands of Alg8-PhoA with D296A or L336A substitutions were much less intense, suggesting increased protein degradation. A Western blot analysis of mutant proteins D133A, V155A, D188A, L336A, R339A and W340A in P. aeruginosa produced similar results, and also showed that the L336A substitution resulted in an unstable Alg8-PhoA protein (Fig. 3B). We were unable to maintain the plasmid encoding Alg8-PhoA with the D296A change in FRD1108, suggesting that the unstable protein produced had deleterious effects on growth. Alg8-PhoA constructs with the D133A substitution also showed a more prominent PhoA band (48 kDa) compared to wild-type, suggesting some protein instability was caused by this alteration, too.
Fig. 3.
To demonstrate the expression in E. coli (A) and in P. aeruginosa FRD1108 (B) of alg8-phoA constructs with substitutions in GT domains A and B, cultures were induced with IPTG, and total cell proteins were examined by a Western blot analysis using anti-PhoA antibodies. Lanes are marked with the terminal amino acid in Alg8 before the PhoA fusion. Induced cultures expressed a band corresponding to the approximate size of the predicted fusion protein in E. coli. Lanes marked 0 were negative controls and contain proteins from bacteria with pLO4, which contains the unfused phoA gene cassette used to create PhoA fusions. All mutant fusion proteins displayed some degree of instability, but the constructs with the D296A and L336A substitutions displayed a high level of instability such that the most prominent band detected corresponded to the size of PhoA (48 kDa). (C) Ability of Alg8(with substitutions)-PhoA fusions to complement FRD1108 Δalg8 for alginate production or for fragmented alginate (Alg Frags). Constructs are designated by their terminal amino acid in Alg8 before the PhoA fusion.
When the Alg8-PhoA constructs with the D133A, D188A, L336A, R339A and W340A substitutions were tested for the ability to complement the Δalg8 mutation in FRD1108, none but the wild-type Alg8-PhoA (pLO28) restored the mucoid phenotype, suggesting that these conserved amino acids were necessary for the GT activity of Alg8 (Fig. 3C). As a positive control, an Alg8-PhoA with an irrelevant substitution (V155A in pLO81) was tested, and it also showed complementation to the mucoid phenotype (Fig. 3C). To check the possibility that polymer was being synthesized by mutant Alg8-PhoAs but degraded by the AlgL lyase, we tested culture supernatants for fragments of degraded alginate, but none was detected (Fig. 3C). Overall, this characterization verified the residues predicted to be critical for Alg8 function, and suggested that L336 and D296 also play a structural role in maintaining the stability of the protein.
Construction of a topological model for Alg44 by PhoA fusion analysis
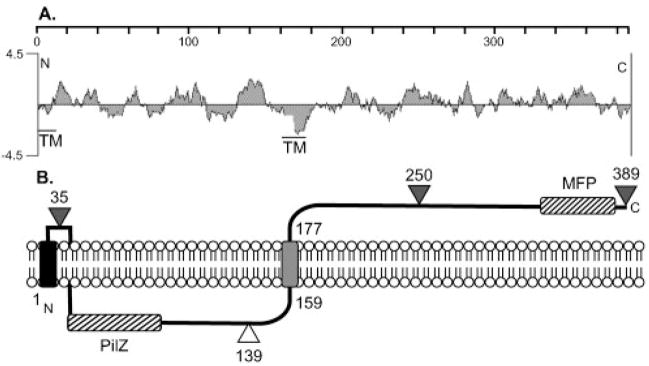
The predicted Alg44 (PA3542) coding sequence was obtained from the P. aeruginosa annotated genome project (http://www.pseudomonas.com). A hydrophilicity plot of the 389 amino acid Alg44 protein shows it to be mostly hydrophilic, with one strong hydrophobic area in the middle of the protein (Fig. 4A). The transmembrane prediction program TMpred predicted that this hydrophobic area (residues 159–177) was a TM domain. TMpred also predicted the potential for a TM domain between residues 42 and 62, but this is unlikely considering its weak hydrophobicity and the presence of a c-di-GMP-binding PilZ domain (residues 7–104), which would have a cytoplasmic localization (Merighi et al., 2007). SignalP predicts that Alg44 does not contain a cleavable signal sequence.
Fig. 4.
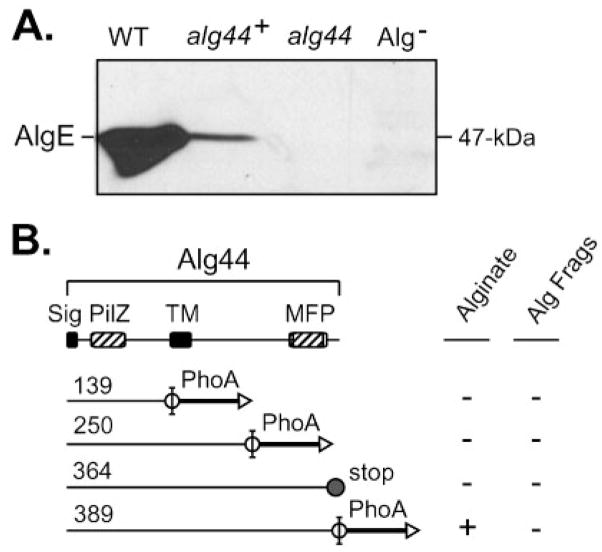
(A) A hydrophilicity plot of Alg44 and a TMpred analysis suggests a central TM domain as indicated. A putative TM is also indicated at the N terminus. The scale shows the residue numbers in Alg44. (B) Model for the orientation of Alg44 in the inner membrane based on the results of Alg44′-PhoA fusions. Triangles show the fusion joints to an in-frame PhoA, with black indicating positive activity (periplasmic) and white indicating negative activity (cytoplasmic). A TM functioning as a putative membrane anchor (e.g. uncleaved signal peptide) is indicated on the N terminus as a black rectangle in the membrane, and the grey rectangle represents the central TM domain. The locations of the PilZ domain in the cytoplasmic loop and a MFP domain in the periplasmic terminus are indicated with hatched rectangles.
To experimentally determine the membrane topology of Alg44, alg44-phoA fusions were constructed. E. coli expressing an Alg44′-PhoA with the fusion joint at amino acid 35 was positive for PhoA activity when screened on indicator plates (L-agar+XP). Thus, amino acids 1–35 of Alg44 allowed for membrane translocation of PhoA even though its sequence is predictably weak as a signal peptide and does not contain the Ala-x-Ala recognition site for signal peptidase. If uncleaved, the signal is likely to serve as an N-terminal membrane anchor (Pugsley, 1989). E. coli expressing an Alg44′-PhoA, fused to the last amino acid at 389, was positive for PhoA activity on XP agar as indicated by blue colonies. A positive result was also observed for bacterial colonies expressing PhoA fused to N250 (Fig. 4B). As predicted by the model, Alg44′-PhoA fused at T139 was negative for PhoA activity when screened on XP agar (Fig. 4B). A Western blot analysis using anti-PhoA showed that these Alg44′-PhoA constructs were expressed (data not shown). Thus, these data suggest that Alg44 contains a putative N-terminal membrane anchor, a central TM region, and a long C terminus located in the periplasm (Fig. 4B).
Alg44 shares homology with efflux proteins
A computer search for homologies and predicted domains showed that the cytoplasmic region of Alg44 contains a PilZ domain at the N terminus (labelled PilZ in Fig. 4). PilZ domains function as receptors for c-di-GMP, which can act as a cofactor to regulate the activity of the protein (Romling et al., 2005). Alg44’s PilZ domain has recently been shown to bind c-di-GMP, and that this is critical for alginate biosynthesis (Merighi et al., 2007). In addition to PilZ in the N terminus, the C terminus of Alg44 (residues 220–364) shares homology with proteins in multi-drug efflux pumps, and in particular those known as membrane fusion proteins (labelled MFP in Fig. 4). MFPs have been suggested to function as adaptor proteins, connecting the efflux pump located in the cytoplasmic membrane to the outer-membrane pore (Elkins & Nikaido, 2003; Tikhonova et al., 2002). This suggested the possibility of an interaction between the periplasmic C terminus of Alg44 and the outer-membrane protein AlgE, the putative alginate porin through which alginate is secreted to the extracellular space. A Western blot analysis with antibodies raised to AlgE showed that AlgE (54 kDa) was present in mucoid strain FRD1, and absent as expected in FRD2, which does not express the algD operon (Fig. 5A). Interestingly, AlgE was generally below detection in the Δalg44 non-polar mutant FRD1144 (Fig. 5A). Conjugation of pLO51 encoding full-length Alg44-PhoA into FRD1144 resulted in a restored mucoid phenotype. The production of alginate in L-broth culture by FRD1144(pLO51) was determined to be ~35 % of wild-type FRD1. This partial complementation of FRD1144 by Alg44-PhoA also resulted in a partial restoration of AlgE as well, as determined by Western blot analysis (Fig. 5A).
Fig. 5.
(A) A Western blot analysis was used to test for the presence of outer-membrane porin, AlgE. AlgE was detected in the wild-type mucoid strain P. aeruginosa FRD1 (WT, lane 1), but was difficult to detect in the alg44 mutant, FRD1044 (alg44, lane 3). Expression of full-length Alg44(1-389)-PhoA (from pLO51) in FRD1044 restored AlgE (alg44+, lane 2) and alginate production. The negative control strain FRD2, which does not express the alginate operon, showed no AlgE band (Alg−, lane 4). (B) Diagram showing the ability of Alg44′-PhoA fusions to complement the alg44 mutation for alginate production (i.e. a mucoid phenotype) or for production of alginate fragments (Alg Frags). Constructs are designated by the number of the last Alg44 amino acid before the PhoA fusion, or in one case by the introduction of a stop codon in Alg44(1-364-stop). FRD1044 expressing the full-length Alg44(1-389)-PhoA fusion protein (from pLO51) produced approximately 490 μg alginate per ml culture supernatant, which was ~35 % of that produced by wild-type strain FRD1 under the same conditions.
Given that the C terminus of Alg44 may be involved in polymer transport across the periplasm to AlgE, we tested the possibility that some C-terminal deletions might still allow for polymer production, although newly formed polymers would be subjected to degradation by AlgL. A complementation analysis was performed using the various Alg44′-PhoA constructs with truncated C termini in the Δalg44 mutant FRD1144. Of the Alg44′-PhoA constructs, only full-length Alg44(1-389)-PhoA was able to restore the mucoid phenotype (Fig. 5B). Also, no detectable secreted alginate fragments or intracellular uronic acids were found when FRD1144 expressed Alg44(1-139)-PhoA (pLO33) or Alg44(1-250)-PhoA (pLO34), which were missing the MFP domain. A Western blot analysis confirmed that the Alg44′-PhoA proteins were produced and stable in FRD1144 (data not shown). Another plasmid (pLO66) was constructed in which a stop codon was introduced in Alg44 so as to truncate only the last 25 residues, and the PhoA fusion was not included in case it might interfere with Alg44’s terminal MFP function (e.g. potential interaction with AlgE). However, even FRD1144(pLO66) expressing Alg44′ (1-364) remained non-mucoid and did not release alginate fragments into the supernatant (Fig. 5B). These data suggest that the extreme terminus of Alg44 plays a role in Alg8-mediated alginate polymer formation.
DISCUSSION
We have recently proposed that in mucoid P. aeruginosa, the periplasmic proteins AlgKGXL form a scaffold that guides alginate polymers through the periplasm to AlgE, the outer-membrane porin (Jain & Ohman, 2005). When either AlgK, G or X is absent from the cell, new polymers are subjected to degradation by AlgL, an alginate lyase encoded by the algD operon (Schiller et al., 1993). Such degraded polymers are readily found in the culture supernatant as dialysable uronic acids (Jain & Ohman, 1998, 2005; Jain et al., 2003; Robles-Price et al., 2004). When AlgL is absent, polymer cannot escape the periplasm, suggesting a role for AlgL in polymer transport as well as its alginate-lyase activity (Jain & Ohman, 2005). Thus, the detection of secreted alginate fragments (i.e. dialysable uronic acids) can be used to determine if polymer synthesis has occurred in vivo even though the non-mucoid phenotype is presented. Alg8 is the GT for alginate biosynthesis, and the lack of secreted alginate fragments in an Alg8− mutant’s culture supernatant was verified here. However, it was intriguing that an Alg44− mutant does not secrete alginate fragments, suggesting that it also has a role in alginate polymer formation. Thus, this study examined both Alg8 and Alg44 to better understand the mechanism of alginate polymerization, which presumably involves the transport of newly synthesized oligouronates across the cytoplasmic membrane to the growing chains.
Alg8 is known to have topological similarity to family 2 GTs involved in the formation of polysaccharides such as cellulose, chitin and hyaluronate (Saxena et al., 1995), suggesting that Alg8 plays a similar role in alginate biosynthesis. The GTs that show structural likeness to Alg8 have been demonstrated or predicted to be multi-spanning membrane proteins (Barny et al., 1996; Saxena et al., 1990). Here we investigated the membrane topology of Alg8 using translational fusions with PhoA to predict periplasmic and cytoplasmic regions of the protein. The results predicted that Alg8 contains an N-terminal TM that resembles a cleavable signal sequence and four other TM regions, which results in a large cytoplasmic loop containing the active domains of β-GTs. The enzymes that share structural topology with Alg8 are responsible for the synthesis of large polymers and contain active sites located in the cytoplasm. In vitro activity assays have been used to demonstrate the activity of GTs such as cellulose synthase in Gluconacetobacter xylinus and Cps3S in Streptococcus pneumoniae using their respective UDP-linked sugars (Forsee et al., 2000; Saxena et al., 2001). Unfortunately, the substrate of Alg8, GDP-mannuronate, is not commercially available. However, radio-labelled GDP-mannuronate was recently synthesized to show in vitro that Alg8-containing membranes of P. aeruginosa can use GDP-mannuronate for polymer production, thus confirming the role of Alg8 in alginate polymer formation (Remminghorst & Rehm, 2006a).
Alg8 is classified as a family 2 GT member, which is one of the largest groups of GTs (Campbell et al., 1997). Domain A is found in all β-GTs, and is characterized by just two conserved aspartic acid residues, with one often in a DxD motif. This forms two sites, one of which is probably occupied by an NDP-sugar and the other by the acceptor molecule. Domain B, when present as it is in Alg8, may permit transfer of multiple NDP-linked sugars to the acceptor molecule (Saxena et al., 1995). The nature of the acceptor molecule for Alg8 is currently unknown but it could be a membrane lipid. Thus, one might predict that the cytoplasmic loop C1 of Alg8, if still membrane anchored, would be sufficient to bind GDP-mannuronate and allow for polymerization and alginate production in vivo. However, our studies here showed that only full-length Alg8 could restore the mucoid phenotype. Even a deletion of the last 21 amino acids, removing the terminal TM domain, prevented the synthesis of polymer or alginate fragments. Similar results were also seen with terminal deletions of NodC, a structurally homologous β GT from the same family as Alg8 (Barny et al., 1996). The role played by the C-terminal membrane anchor in Alg8 is unclear, but appears to be critical. It may be necessary for some protein–protein interaction with other proteins involved in alginate biosynthesis.
Alanine substitution mutagenesis was used to validate the importance of the predicted conserved aspartates and the LxxRW motif in Alg8. Using an alg8 mutant for complementation analysis, all of the substitutions made in domains A and B blocked both the production of polymer (i.e. the mucoid phenotype) and also the ability to release alginate fragments, indicating that no polymer synthesis occurred. Most of these mutant Alg8 proteins were relatively stable in E. coli and P. aeruginosa except for those with the D296A and L336A changes. The most extensive degradation was observed with the D296A change, and this construct proved difficult to even maintain in P. aeruginosa. Thus, although D296 and L336 are predicted to be involved in substrate binding and catalysis of Alg8, they appear to be involved in the stability of the protein as well. However, substitutions in the structurally aligned amino acids in cellulose synthase were not reported to affect the stability of those mutant proteins (Saxena et al., 2001). It was also interesting that domain B of Alg8 does not have a true ‘QxxRW’ motif, but instead has a leucine instead of the glutamine. The LxxRW motif is also present in the Alg8 proteins found in other bacterial species that have the alginate biosynthesis operon. A leucine is found bound to the guanine base of GDP in the active site of the Saccharomyces cerevisiae α-1,2-mannosyl-transferase Kre2p/Mnt1p, a retaining GT (Lobsanov et al., 2004), suggesting that the presence of Q or L in this motif may depend upon the nucleotide diphosphate of the activated sugar.
The PhoA fusion analysis of Alg44 suggests a central TM domain plus an N-terminal TM (e.g. uncleaved signal peptide) that may form a membrane anchor. The end of the long periplasmic C terminus of Alg44 has similarity to some MFPs of multi-drug efflux systems that export various substances out of the cell. These three-component systems typically consist of a pump located in the cytoplasmic membrane, a periplasmic MFP, and an outer-membrane channel (Johnson & Church, 1999). The membrane pumps located in the cytoplasmic membrane usually belong to the resistance-nodulation division family (RND) or the major facilitator superfamily (MFS) (Johnson & Church, 1999; Nehme et al., 2004). Multi-drug efflux systems are associated with conferring broad-range antibiotic resistance to various Gram-negative bacteria (Elkins & Nikaido, 2003; Tikhonova et al., 2002). MFPs have been suggested to function as an adaptor, connecting the cognate efflux pump located in the cytoplasmic membrane to the outer-membrane pore (Elkins & Nikaido, 2003; Tikhonova et al., 2002). Although full-length Alg44-PhoA could complement an alg44 mutation to restore alginate production, an Alg44′-PhoA truncating the MFP domain could not restore alginate production or any polymer formation, as indicated by the absence of alginate fragments in the culture supernatants.
An interesting phenotype of the Δalg44 mutation in FRD1144 was the loss of a stable AlgE porin in the cell, but this defect was complemented by a functional Alg44-PhoA in trans. Even a truncation of the C-terminal 24 amino acids from Alg44 resulted in a failure to complement the alg44 mutation and even blocked the ability of Alg8 to synthesize new alginate polymers that could be subjected to AlgL degradation. It is tempting to speculate that a protein–protein interaction takes place between the C terminus of Alg44 and AlgE or AlgK to help stabilize AlgE. Obtaining direct evidence for such interactions is in progress. Also, interactions with Alg44’s C terminus in the periplasm may send a conformational signal to its N terminus that affects Alg8’s ability to polymerize D-mannuronate. Perhaps this signalling mechanism acts to control the binding of c-di-GMP to Alg44 in the cytoplasm. Such possibilities are currently under study.
Acknowledgments
We gratefully acknowledge the VCU Nucleic Acids Core Facility for assistance in the DNA sequence analysis. We thank Michael Harwich (this laboratory) for providing antibodies to AlgE. This work was supported by Veterans Administrations Medical Research Funds (D. E. O.) and in part by Public Health Service grants AI-19146 (D. E. O.) from the National Institute of Allergy and Infectious Diseases, and by a grant from the Cystic Fibrosis Foundation (D. E. O.).
Abbreviations
- c-di-GMP
bis-(3′,5′)-cyclic dimeric GMP
- GT
glycosyl-transferase
- HCA
hydrophobic cluster analysis
- MFP
membrane fusion protein
- PhoA
alkaline phosphatase
- TM
transmembrane
- XP
5-bromo-4-chloro-3-indolyl phosphate
References
- Barny MA, Schoonejans E, Economou A, Johnston AW, Downie JA. The C-terminal domain of the Rhizobium leguminosarum chitin synthase NodC is important for function and determines the orientation of the N-terminal region in the inner membrane. Mol Microbiol. 1996;19:443–453. doi: 10.1046/j.1365-2958.1996.382911.x. [DOI] [PubMed] [Google Scholar]
- Boucher JC, Martinez-Salazar J, Schurr MJ, Mudd MH, Yu H, Deretic V. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J Bacteriol. 1996;178:511–523. doi: 10.1128/jb.178.2.511-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries CA, Ohman DE. Mucoid to nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternative sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins CA, Nikaido H. Chimeric analysis of AcrA function reveals the importance of its C-terminal domain in its interaction with the AcrB multidrug efflux pump. J Bacteriol. 2003;185:5349–5356. doi: 10.1128/JB.185.18.5349-5356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsee WT, Cartee RT, Yother J. Biosynthesis of type 3 capsular polysaccharide in Streptococcus pneumoniae. Enzymatic chain release by an abortive translocation process. J Biol Chem. 2000;275:25972–25978. doi: 10.1074/jbc.M002613200. [DOI] [PubMed] [Google Scholar]
- Franklin MJ, Ohman DE. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J Bacteriol. 2002;184:3000–3007. doi: 10.1128/JB.184.11.3000-3007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Chitnis CE, Gacesa P, Sonesson A, White DC, Ohman DE. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol. 1994;176:1821–1830. doi: 10.1128/jb.176.7.1821-1830.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinot-Schneider C, Lellouch AC, Geremia RA. Identification of essential amino acid residues in the Sinorhizobium meliloti glucosyltransferase ExoM. J Biol Chem. 2000;275:31407–31413. doi: 10.1074/jbc.M004524200. [DOI] [PubMed] [Google Scholar]
- Gilligan PH. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev. 1991;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C, Devedjian JC. A plasmid facilitating in vitro construction of phoA gene fusions in Escherichia coli. Nucleic Acids Res. 1989;17:3999. doi: 10.1093/nar/17.10.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haardt M, Bremer E. Use of phoA and lacZ fusions to study the membrane topology of ProW, a component of the osmoregulated ProU transport system of Escherichia coli. J Bacteriol. 1996;178:5370–5381. doi: 10.1128/jb.178.18.5370-5381.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Ohman DE. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J Bacteriol. 1998;180:634–641. doi: 10.1128/jb.180.3.634-641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Ohman DE. Alginate biosynthesis. In: Ramos J-L, editor. Pseudomonas. New York: Kluwer Academic/Plenum Publishers; 2004. pp. 53–81. [Google Scholar]
- Jain S, Ohman DE. Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect Immun. 2005;73:6429–6436. doi: 10.1128/IAI.73.10.6429-6436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Franklin MJ, Ertesvag H, Valla S, Ohman DE. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol Microbiol. 2003;47:1123–1133. doi: 10.1046/j.1365-2958.2003.03361.x. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Church GM. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J Mol Biol. 1999;287:695–715. doi: 10.1006/jmbi.1999.2630. [DOI] [PubMed] [Google Scholar]
- Keenleyside WJ, Clarke AJ, Whitfield C. Identification of residues involved in catalytic activity of the inverting glycosyl transferase WbbE from Salmonella enterica serovar borreze. J Bacteriol. 2001;183:77–85. doi: 10.1128/JB.183.1.77-85.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson CA, Jeanes A. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal Biochem. 1968;24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- Lobsanov YD, Romero PA, Sleno B, Yu B, Yip P, Herscovics A, Howell PL. Structure of Kre2p/Mnt1p: a yeast α1,2-mannosyltransferase involved in mannoprotein biosynthesis. J Biol Chem. 2004;279:17921–17931. doi: 10.1074/jbc.M312720200. [DOI] [PubMed] [Google Scholar]
- Maharaj R, May TB, Wang SK, Chakrabarty AM. Sequence of the alg8 and alg44 genes involved in the synthesis of alginate by Pseudomonas aeruginosa. Gene. 1993;136:267–269. doi: 10.1016/0378-1119(93)90477-k. [DOI] [PubMed] [Google Scholar]
- Manoil C, Boyd D, Beckwith J. Molecular genetic analysis of membrane protein topology. Trends Genet. 1988;4:223–226. doi: 10.1016/0168-9525(88)90154-0. [DOI] [PubMed] [Google Scholar]
- Mejia-Ruiz H, Guzman J, Moreno S, Soberon-Chavez G, Espin G. The Azotobacter vinelandii alg8 and alg44 genes are essential for alginate synthesis and can be transcribed from an algD-independent promoter. Gene. 1997;199:271–277. doi: 10.1016/s0378-1119(97)00380-6. [DOI] [PubMed] [Google Scholar]
- Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- Nehme D, Li XZ, Elliot R, Poole K. Assembly of the MexAB-OprM multidrug efflux system of Pseudomonas aeruginosa: identification and characterization of mutations in mexA compromising MexA multimerization and interaction with MexB. J Bacteriol. 2004;186:2973–2983. doi: 10.1128/JB.186.10.2973-2983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman DE, Chakrabarty AM. Genetic mapping of chromosomal determinants for the production of the exopolysac-charide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981;33:142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect Immun. 2001;69:1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley AP. Early stages in the secretory pathway, chapter III. In: Pugsley AP, editor. Protein Targeting. San Diego: Academic Press; 1989. pp. 45–111. [Google Scholar]
- Rehm BH, Boheim G, Tommassen J, Winkler UK. Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J Bacteriol. 1994;176:5639–5647. doi: 10.1128/jb.176.18.5639-5647.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remminghorst U, Rehm BH. In vitro alginate polymerization and the functional role of Alg8 in alginate production by Pseudomonas aeruginosa. Appl Environ Microbiol. 2006a;72:298–305. doi: 10.1128/AEM.72.1.298-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remminghorst U, Rehm BH. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 2006b;580:3883–3888. doi: 10.1016/j.febslet.2006.05.077. [DOI] [PubMed] [Google Scholar]
- Robles-Price A, Wong TY, Sletta H, Valla S, Schiller NL. AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J Bacteriol. 2004;186:7369–7377. doi: 10.1128/JB.186.21.7369-7377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Roychoudhury S, May T, Gill J, Singh S, Feingold D, Chakrabarty A. Purification and characterization of guanosine diphospho-D-mannose dehydrogenase. A key enzyme in the biosynthesis of alginate by Pseudomonas aeruginosa. J Biol Chem. 1989;264:9380–9385. [PubMed] [Google Scholar]
- Saxena IM, Brown RM., Jr Identification of cellulose synthase(s) in higher plants: sequence analysis of processive β-glycosyltransferases with the common motif ‘D,D,D35Q(R,Q)XRW’. Cellulose. 1997;4:33–49. [Google Scholar]
- Saxena IM, Lin FC, Brown RM., Jr Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol Biol. 1990;15:673–683. doi: 10.1007/BF00016118. [DOI] [PubMed] [Google Scholar]
- Saxena IM, Brown RM, Fevre M, Geremia RA, Henrissat B. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Brown RM, Jr, Dandekar T. Structure-function characterization of cellulose synthase: relationship to other glycosyltransferases. Phytochemistry. 2001;57:1135–1148. doi: 10.1016/s0031-9422(01)00048-6. [DOI] [PubMed] [Google Scholar]
- Schiller NL, Monday SR, Boyd CM, Keen NT, Ohman DE. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J Bacteriol. 1993;175:4780–4789. doi: 10.1128/jb.175.15.4780-4789.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer HP. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- Shinabarger D, Berry A, May TB, Rothmel R, Fialho A, Chakrabarty AM. Purification and characterization of phosphomannose isomerase-guanosine diphospho-D-mannose pyrophosphorylase – a bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J Biol Chem. 1991;266:2080–2088. [PubMed] [Google Scholar]
- Tikhonova EB, Wang Q, Zgurskaya HI. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J Bacteriol. 2002;184:6499–6507. doi: 10.1128/JB.184.23.6499-6507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]