Abstract
The KChIPs (K+ channel-interacting proteins) are EF hand-containing proteins required for the traffic of channel-forming Kv4 K+ subunits to the plasma membrane. KChIP1 is targeted, through N-terminal myristoylation, to intracellular vesicles that appear to be trafficking intermediates from the ER (endoplasmic reticulum) to the Golgi but differ from those underlying conventional ER–Golgi traffic. To define KChIP1 vesicles and the traffic pathway followed by Kv4/KChIP1 traffic, we examined their relationship to potential SNARE (soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptor) proteins mediating the trafficking step. To distinguish Kv4/KChIP1 from conventional constitutive traffic, we compared it to the traffic of the VSVG (vesicular-stomatitis virus G-protein). Expression of KChIP with single or triple EF hand mutations quantitatively inhibited Kv4/KChIP1 traffic to the cell surface but had no effect on VSVG traffic. KChIP1-expressing vesicles co-localized with the SNARE proteins Vti1a and VAMP7 (vesicle-associated membrane protein 7), but not with the components of two other ER–Golgi SNARE complexes. siRNA (small interfering RNA)-mediated knockdown of Vti1a or VAMP7 inhibited Kv4/KChIP1traffic to the plasma membrane in HeLa and Neuro2A cells. Vti1a and VAMP7 siRNA had no effect on VSVG traffic or that of Kv4.2 when stimulated by KChIP2, a KChIP with different intrinsic membrane targeting compared with KChIP1. The present results suggest that a SNARE complex containing VAMP7 and Vti1a defines a novel traffic pathway to the cell surface in both neuronal and non-neuronal cells.
Keywords: calcium-binding protein, K+ channel-interacting protein (KChIP), potassium channel, neuronal calcium sensor (NCS) protein, soluble N-ethylmaleimide-sensitive fusion proteinattachment protein receptor (SNARE)
Abbreviations: ARF, ADP-ribosylation factor; DMEM, Dulbecco's modified Eagle's medium; ECFP, enhanced cyan fluorescent protein; ER, endoplasmic reticulum; ERGIC, ER–Golgi intermediate compartment; EYFP, enhanced yellow fluorescent protein; FBS, foetal bovine serum; GAP, GTPase-activating protein; GFP, green fluorescent protein; KChIP, K+ channel-interacting protein; LAMP, lysosomal-associated protein; PCTV, pre-chylomicron transport vesicle; siRNA, small interfering RNA; SNARE, soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptor; TBC1D20, TBC (Tre-2/Bub2/Cdc16) domain family, member 20; TRITC, tetramethylrhodamine β-isothiocyanate; VAMP, vesicle-associated membrane protein; VSVG, vesicular-stomatitis virus G-protein
INTRODUCTION
Kv4 potassium channels were first identified as homologues of the Drosophila Shal channels and are now known to have key roles in the regulation of the excitability of neurons and cardiac myocytes. In mammals, four members of the Kv4 family (Kv4.1, 4.2, 4.3 short and 4.3 long) are known [1] and they are highly expressed in the brain and heart [2]. Kv4 potassium channels are responsible for the transient outward A-type current in neurons, and its equivalent in myocytes. In the brain, it is thought that this current is responsible for attenuating back-propagating action potentials and excitatory post-synaptic potentials [3]. The channels also limit action potentials to individual dendritic compartments [4] and abnormalities in Kv4 channel function have been implicated in the hyperexcitability underlying epilepsy [5].
When Kv4 channels are expressed alone in non-neuronal cells, their properties differ from those seen with native channels [6]. A search for interacting partners that could reconstitute native functions led to the discovery of the KChIPs (K+ channel-interacting proteins) [6], which are members of the NCS (neuronal calcium sensor) family of proteins [7,8]. The KChIPs have four EF-hand domains, of which three can potentially bind Ca2+ and two were seen to be occupied by Ca2+ in the crystal structure of KChIP1 [9,10]. Of the four KChIPs, only KChIP1 has an N-terminal myristoyl group that is required for it to bind to membranes [11] and, uniquely, allows its targeting to a distinct population of intracellular 50–100 nm diameter vesicles (referred from now onwards as KChIP1 vesicles) that partially overlap with markers of the ERGIC [ER (endoplasmic reticulum)–Golgi intermediate compartment] [12]. The KChIPs have two effects on Kv4 channels. Firstly, the KChIPs modulate the properties of the channel, particularly its inactivation kinetics [6]. Secondly, they increase current density by promoting the traffic of the Kv4 channel to the plasma membrane [6,12–15]. Kv4 proteins alone are able to exit from the ER, but then become trapped in the Golgi complex [11,12]. Co-expression of Kv4 channels with KChIPs leads to their traffic and co-localization at the plasma membrane [11,12], where the KChIPs are constitutive channel subunits [9,10].
The conventional exocytic pathway from the ER has been studied in detail by following the traffic of a temperature-sensitive mutant of the VSVG (vesicular-stomatitis virus G-protein) [16,17]. Traffic of VSVG from the ER involves budding of COPII-coated vesicles controlled by the GTPase Sar1 [18] and requires Rab1 [19]. The vesicle then passes through the intermediate compartment, the ERGIC [20] and through the Golgi complex in a COPI-dependent manner before trafficking to the plasma membrane. There is growing evidence in the literature that this single pathway is not responsible for all traffic from the ER to the Golgi. In yeast, there is evidence that both COPII- and COPI-coated vesicles can bud from the ER [21]. Proteins bound to the membrane via a GPI (glycosylphosphatidylinositol) anchor are found in vesicles distinct from those carrying standard marker proteins [22,23]. In mammalian cells, procollagen is too large to fit into the 60–80 nm diameter COPII vesicles. Although procollagen appears to require COPII for exit from the ER, it is not found in COPII-coated vesicles, and has no requirement for COPI [24,25]. Also, several proteins known to affect the transport of the marker VSVG do not affect procollagen transport and vice versa [25]. In the intestine, distinct triacylglycerol transport vesicles, PCTVs (pre-chylomicron transport vesicles) can bud from the ER in a COPII-independent manner, and are lacking in COPI, but need COPII later in the pathway to fuse with the Golgi [26]. Rab1-independent traffic from the ER has been suggested to occur for some cargos in non-neuronal [27] and neuronal cell types [28]. It has been shown that the traffic of Kv4 channels, with associated KChIP1, also occurs by an alternate pathway that is COPII-independent [12]. KChIP1 vesicles seem to be key intermediates in ER to Golgi traffic but the nature of this pathway and the KChIP1 vesicles involved in traffic are unknown.
Membrane traffic steps can be defined by the nature of the SNARE (soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptor) complex that underlies the fusion of vesicles with their target membrane. SNAREs have been defined as Q or R SNAREs and, based on sequence classification of SNARE subfamilies, active complexes appear to have the composition QaQbQcR [29]. Three such complexes have been implicated in ER to Golgi traffic. From analysis of VSVG traffic, the SNARE complex for the step between ER and the ERGIC consists of Sec22b, membrin, rBet1 and syntaxin 5 [30,31]. The complex of GOS28, Ykt6, rBet1 and syntaxin 5 is thought to be involved in the fusion of intermediates with the cis-Golgi [32]. Finally, a third complex, comprising VAMP7 (vesicle-associated membrane protein 7), Vti1a, rBet1 and syntaxin 5, has been identified in the alternative ER to Golgi trafficking pathway for PCTVs in intestinal cells [4,33]. The general significance of this latter complex in cargo traffic is unknown. We aimed to further characterize the nature of the KChIP1 vesicles and the traffic route taken by Kv4/KChIP1. The results show that the KChIP1 vesicles and Kv4/KChIP1 traffic are distinguished by a VAMP7/Vti1a-containing SNARE complex.
METHODS
Cell culture and transfection
HeLa human cervical carcinoma cells were grown in DMEM (Dulbecco's modified Eagle's medium; Gibco) with 5% FBS (foetal bovine serum; Gibco), 1% non-essential amino acids (Gibco), and 1% penicillin-streptomycin (10000 units/ml penicillin and 10000 μg/ml streptomycin; Gibco) and kept at 37 °C in a 5% CO2 atmosphere. Neuro2A cells were cultured in DMEM with 10% FBS and 1% penicillin/streptomycin. For transfection, cells were grown in 24 well plates, with glass coverslips for imaging studies. Cells were plated at ∼4×105 cells per well, in 1 ml supplemented medium, and incubated for 5–24 h before transfection. For the transfection reaction mixture, 3 μl of GeneJuice (Novagen) per μg of DNA was incubated for 5 min at room temperature with OptiMEM serum-free media (Gibco) to give a total reaction volume of 100 μl. Plasmid DNA (1 μg) was then added and the mixture incubated for a further 15 min, before being added drop-wise to the cells. Cells were then left for 24–72 h post-transfection.
Plasmids
Plasmids encoding KChIP1, KChIP1–EYFP (enhanced yellow fluorescent protein), KChIP1EF2–4–EYFP, KChIP1EF2–4–dsRed, KChIP2–ECFP (enhanced cyan fluorescent protein), ARF1 (ADP-ribosylation factor 1)–EYFP and Kv4.2 have been described previously [11,12,34,35]. The KChIP1EF3–EYFP plasmid bearing the mutations D135A and G140A was prepared in this laboratory by Dr Burcu Hasdemir. The ts045 VSVG–GFP (green fluorescent protein) plasmid [17] was provided by John Presley (National Institutes of Health, Bethesda, MD, U.S.A.). The plasmid encoding the Rab1 GAP (GTPase-activating protein) TBC1D20 [TBC (Tre-2/Bub2/Cdc16) domain family, member 20] was a gift from Francis Barr (School of Cancer Studies, University of Liverpool, U.K.). The EYFP–Golgi plasmid encoding EYFP linked to the N-terminus of β1,4-galactosyltransferase for trans- and medial-Golgi targeting was obtained from Clontech (Mountain View, CA, U.S.A.).
Immunocytochemistry
After transfection, cells on coverslips were gently washed three times in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 2 mM NaH2PO4, pH 7.4). Cells were then fixed in PBS with 4% formaldehyde for 30 min at room temperature (22 °C). Coverslips for imaging were then washed again before being air dried, and mounted as described below. For immunofluorescence, cells were washed in PBS after fixation, then permeabilized in PBT [PBS plus 0.1% (v/v) Triton X-100 and 0.3% (v/v) BSA] for 30 min. Cells were then incubated with the primary antibody overnight at 4 °C. Antibodies were diluted in PBS with 0.3% BSA. These included mouse monoclonal antibodies to membrin, VAMP7 and GOSR1 (GOS28), obtained from Abcam (Cambridge, U.K.) a mouse monoclonal antibody to Sec22L1 (Sec22b) from Abnova (Taipei, Taiwan) and a mouse monoclonal anti-Vti1a antibody from BD Biosciences (San Jose, CA, U.S.A.). A chicken anti-Ykt6 antibody was obtained from J. C. Hay (Division of Biological Sciences, University of Montana, Missoula, MT, U.S.A.) [36]. A rabbit anti-TI-VAMP (VAMP7) antibody was a gift from Thierry Galli (Institut Jacques Monod, Paris, France). Rabbit polyclonal antibodies against syntaxin 7 or 8 were obtained from Synaptic Systems (Goettingen, Germany). Anti-LAMP1 (lysosomal-associated protein 1) was a rabbit polyclonal obtained from Affinity Bioreagents (Golden, CO, U.S.A.). The primary antibody was removed, cells were washed three times in PBT, and the secondary antibody was added for 1 h at room temperature. Secondary antibodies were diluted in PBS with BSA, at 1:80 dilution for both anti-mouse and anti-rabbit IgG conjugated to TRITC (tetramethylrhodamine β-isothiocyanate; Sigma, St. Louis, MO, U.S.A.) or 1:250 for Alexa Fluor® 488 and Alexa Fluor® 594-linked secondary antibodies obtained from Molecular Probes/Invitrogen (Carlsbad, CA, U.S.A.). Cells were washed again in PBT, and then dried, before mounting using ProLong Gold Antifade (Invitrogen).
Confocal microscopy
Transfected cells were viewed using a Leica TCS-SP2-AOBS confocal microscope (Leica, Germany), using a 66.86 μm pinhole and a 63× water-immersion objective with a 1.2 numerical aperture. For EYFP constructs, an excitation of 488 nm was used, and an emission range of 500–540 nm was recorded. For TRITC-tagged secondary antibody signals, a 543 nm laser was used for excitation, and emission at 570–680 nm was recorded. GFP was imaged using excitation at 488 nm and light collection at 500–550 nm. ECFP was imaged using 405nm excitation and light collections between 500–550 nm. HcRed was imaged by excitation at 494 nm and light collected at 610–700 nm. Alexa Fluor® 488 was excited at 488 nm, and emissions were recorded from 500–550 nm. Alexa Fluor® 594 was excited at 594 nm, and emissions were recorded from 605–670 nm.
siRNA (small interfering RNA) knockdown
Expression of SNARE proteins was inhibited using siRNA oligonucleotide duplexes. Three duplexes were obtained against SYBL1, the gene for VAMP7 (Ambion, U.K., siRNA ID numbers 241467, 241468 and 241469) and three against Vti1a (Ambion, U.K., siRNA ID numbers 37855, 37946 and 38033). A Silence negative control siRNA#1 (Ambion, AM4611) was also obtained. All siRNAs were resuspended in nuclease-free water, to give a final concentration of 100 nM when used for transfection. Cells were transfected with untagged Kv4.2, with or without KChIP1–EYFP and siRNA or VSVG–GFP and siRNA. After 72 h, the cells were fixed and images were obtained of both KChIP1–EYFP and VSVG–GFP or Kv4.2 detected by immunofluorescent staining using an anti-Kv4.2 antibody (Exalpha Biologicals, Shirley, MA, U.S.A.) at 1:500 dilution followed by a TRITC-tagged secondary antibody. Using the Leica LCD Lite software (Leica), regions of interest were drawn around the perimeter of the cell and within the cell, to give a region of interest at the cell periphery, including the plasma membrane that was 1 μm wide. The edge of the cell could easily be identified based on either background fluorescence or that due to VSVG/KV4.2. The total level of fluorescence was recorded for each region for both channels and the percentage of fluorescence at the plasma membrane was calculated. For Kv4.2 fluorescence, the background level of fluorescence at the cell periphery in the absence of co-expression of KChIP1 (typically 10% of the total [12]) was determined for each experiment and subtracted from values for cells expressing KChIP1. The resulting values were then divided by the mean from each experiment of the percentage of Kv4.2 at the plasma membrane in the presence of KChIP1. These data are shown as the normalized value for Kv4.2 traffic. Values for VSVG were similarly normalized to the control values. All results are expressed as means±S.E.M. and statistical analyses (Student's unpaired t test) were carried out.
Real-time PCR
After 72 h post-transfection, mRNA was extracted from HeLa cells using TRIzol reagent (Invitrogen, Paisley, U.K.) according to the manufacturer's instructions. mRNA was resuspended in 30 μl RNase-free water (Sigma, Gillingham, U.K.), and stored at −80 °C. cDNA was produced using ImProm-II reverse transcriptase (Promega) following the manufacturer's instructions, and using Oligo (dT)15 primers (Promega). The reaction proceeded with 5 min annealing at 25 °C, 60 min at 42 °C and 15 min of heat inactivation at 70 °C. For real-time PCR, cDNA was diluted into a total volume of 15 μl, 7.5 μl SYBR Green PCR master mix (Applied Biosystems, Warrington, U.K.) was used with 1 μl primer mix (50 mM each of forward and reverse primers). Each primer and cDNA combination was repeated in triplicate. Experiments were run on a BioRad iQ5 thermal cycler. After checking each run had completed correctly, the threshold cycle (Ct) was recorded, and averaged for each primer and cDNA set. The relative levels of mRNA for each primer and for each cDNA were calculated by comparison to results with primers for β-actin, using the 2−ΔΔCt method [37].
RESULTS
KChIP1 EF-hand mutants distinguish between Kv4/KChIP1 and VSVG traffic to the plasma membrane
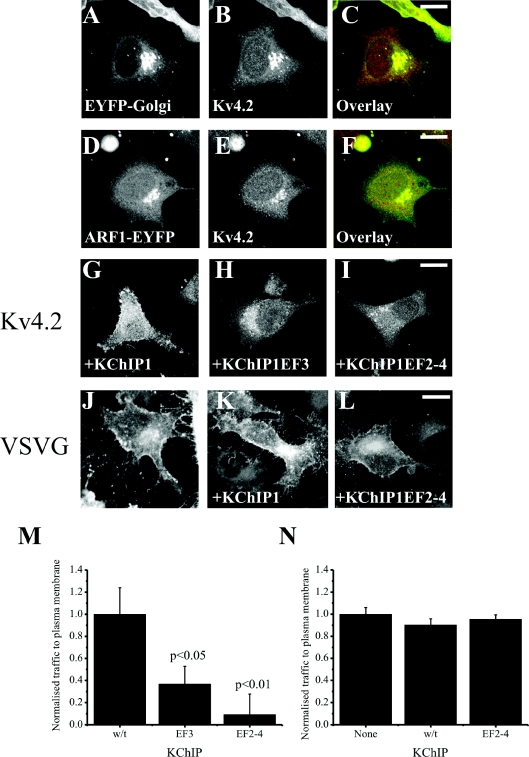
When expressed alone in HeLa cells, Kv4.2 channels do not effectively reach the plasma membrane and remain trapped intracellularly [12]. The intracellular Kv4.2 was highly co-localized with two Golgi complex markers EYFP–Golgi (Figures 1A–1C) or ARF1–EYFP (Figures 1D–1F), indicating retention in the Golgi complex. In comparison, co-expression with KChIP1 results in traffic of Kv4.2 to the plasma membrane (Figure 1G), where it co-localizes with KChIP1 [12]. Mutations of either a single EF hand (EF hand 3 in KChIP1EF3) or all three potentially functional EF hands (in KChIP1EF2–4) to disrupt co-ordination of divalent ions prevented the stimulatory effect on traffic of Kv4.2 (Figures 1H and 1I) and instead Kv4.2 was found on punctate intracellular structures that co-localized with the KChIPs (results not shown) as seen previously for the triple mutant [12]. To compare this traffic pathway to the conventional pathway followed by VSVG, we examined the effect of the inhibitory triple EF hand mutant on traffic of VSVG–GFP to the plasma membrane. Neither wild-type nor the mutant KChIP1 had any effect on the levels of VSVG–GFP that reached the plasma membrane (Figures 1J–1L). These conclusions were confirmed by quantification of the level of trafficking of proteins (Figures 1M and 1N) in assays in which fluorescence in peripheral regions of interest were determined [12] and normalized in comparison with control cells to provide a value for relative traffic to the plasma membrane for each experiment. The KChIP1 EF hand mutations appeared, therefore, to specifically impair the traffic of Kv4.2/KChIP1.
Figure 1. Effects of KChIP1 and KChIP1 with EF hand mutations on traffic of Kv4.2 and VSVG to the plasma membrane.
HeLa cells were transfected to co-express Kv4.2 and EYFP–Golgi (A–C) or ARF1–EYFP (D–F) or with wild-type KChIP1-EYFP (G), with the single EF hand mutant KChIP1EF3-EYFP (H) or with the triple EF-hand mutant KCHIP1EF2–4-EYFP (I) and Kv4.2 localization was determined by using anti-Kv4.2 immunostaining. HeLa cells were transfected to express VSVG–GFP alone (J), or with wild-type KChIP1 (K) or the triple EF-hand mutant KCHIP1EF2–4-HcRed (L) and VSVG–GFP localization was determined by confocal imaging. The scale bars represent 10 μm. Normalized traffic of Kv4.2 (M) or VSVG–GFP (N) in the presence of various KChIPs was quantified and the results shown as means±S.E.M. from 20 cells for each condition. w/t, wild-type.
Kv4.2/KChIP1 traffic requires Rab1
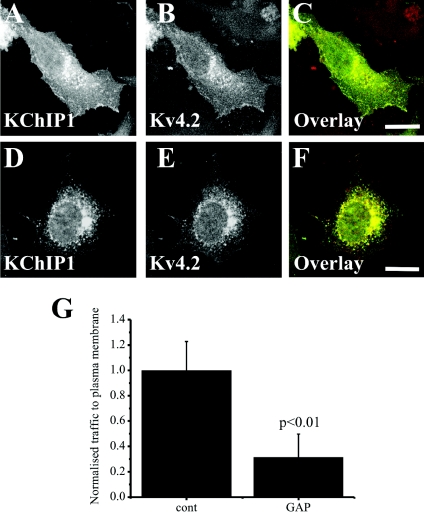
Traffic of VSVG out of the ER requires the function of Rab1 [38]. It has been suggested, however, that some cargo traffic can be independent of Rab1 function [27,28]. We tested, therefore, whether traffic of Kv4.2/KChIP1 was Rab1-dependent. The approach used was to co-express the identified Rab1 GAP TBC1D20 [38]. Since this converts Rab1 into the inactive GDP-loaded form, it acts as an effective and specific inhibitor of Rab1 function. As described previously [38], we found that co-expression of TBC1D20 inhibited traffic of VSVG–EGFP (results not shown) and also effectively inhibited traffic to the plasma membrane of Kv4.2 which remained intracellular but still co-localized with KChIP1 (Figure 2). These results indicate a common Rab1 requirement for the conventional and the KChIP1-dependent trafficking pathways.
Figure 2. Inhibition of Rab1 function prevents traffic of Kv4.2/KChIP1 to the plasma membrane.
HeLa cells were co-transfected to express both KChIP1–EYFP and Kv4.2 and immunostained with anti-Kv4.2 so that localization of KChIP1 (A) and Kv4.2 (B) could be visualized. Alternatively, the cells were additionally co-transfected to express the Rab1 GAP TBC1D20 (D–F). The colour overlays (C and F) show co-expressed proteins in green (KChIP1–EYFP) and red (Kv4.2), with co-localization appearing in yellow. The scale bars represent 10 μm. Normalized traffic of Kv4.2 in the absence (cont) or presence (GAP) of TBC1D20 was quantified and the data shown as means±S.E.M. from 20 cells for each condition (G).
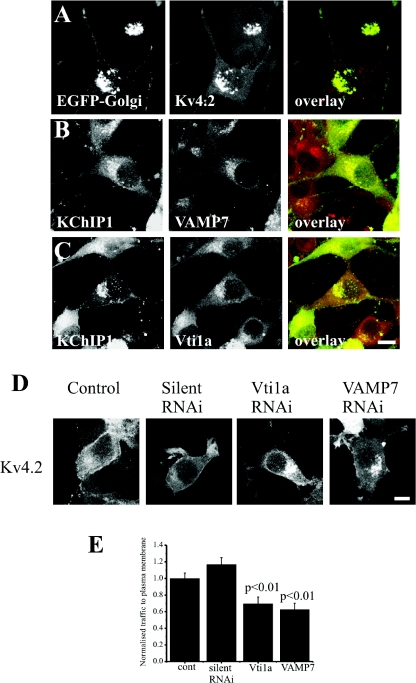
KChIP1 vesicles co-localize with the SNARE proteins VAMP7 and Vti1a
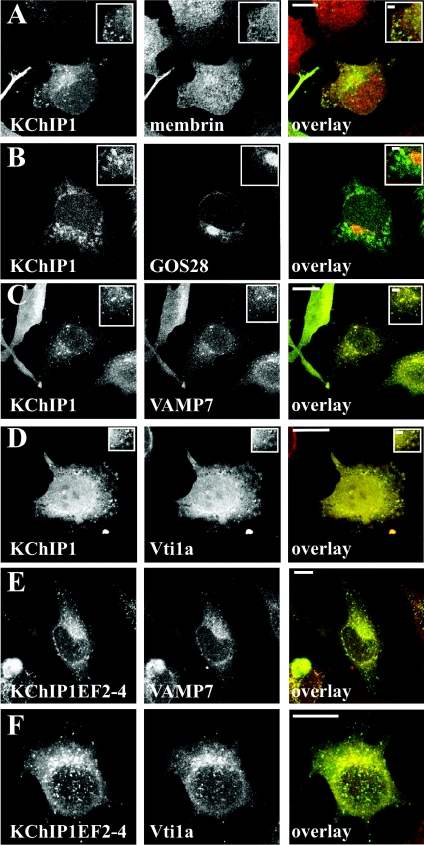
When KChIP1 is expressed alone in HeLa cells, it is present on punctate vesicular structures. However, the identity of these vesicles is unclear. The best overlap with other intracellular markers, albeit partial, was seen with the ERGIC marker ERGIC-53, and the KChIP1 vesicles were suggested to be intermediates in ER to Golgi trafficking [11,12]. To further characterize these vesicles, we aimed to determine which SNARE proteins were associated with them. HeLa cells expressing KChIP1–EYFP were stained with antibodies recognizing SNAREs that are present in only one of the three complexes implicated in ER to Golgi traffic. Membrin, a component of the SNARE complex in the conventional VSVG traffic pathway [31], did not show any significant co-localization with the KChIP1–EYFP-labelled vesicles (Figure 3A). There was also no co-localization of KChIP–EYFP with GOS28 (Figure 3B), a component of a SNARE complex in ERGIC to cis-Golgi traffic [32]. Similarly, no co-localization was seen with KChIP1 for Sec22b or Ykt6 (results not shown). In contrast, VAMP7 and Vti1a, two SNAREs implicated in traffic of pre-chylomicron transport vesicles from ER to Golgi [33], showed extensive co-localization with KChIP1-vesicles (Figures 3C and 3D). Using a polyclonal antiserum against VAMP7 [39], similar results were found (results not shown). Furthermore, the EF hand mutants of KChIP1, which also labelled punctate vesicular structures, were also co-localized with VAMP7 and Vti1a as shown for the triple EF hand mutant (Figures 3E and 3F).
Figure 3. Co-localization of KChIP1 with VAMP7 and Vti1a.
HeLa cells were transfected to express KChIP1–EYFP or KChIP1EF2–4–EYFP as indicated, fixed and immunostained with mouse monoclonal antibodies specific for membrin (A), GOS28 (B), VAMP7 (C and E) or Vti1a (D and F). The images show the localization of the KChIP1 and the indicated SNARE proteins and the colour overlays show KChIP1–EYFP in green and anti-SNARE staining in red with co-localization appearing in yellow. The scale bars represent 10 μm in the main Figures and 2 μm in the enlarged inserts.
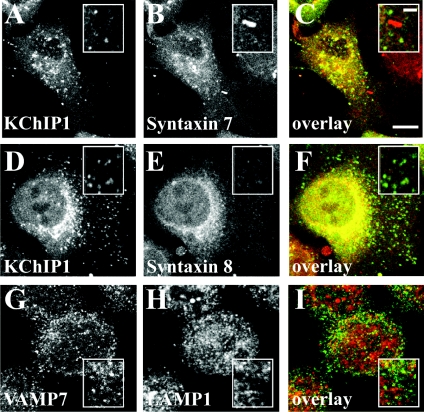
Since VAMP7 has also been shown to be part of a SNARE complex with syntaxins 7 and 8 in late endocytic traffic [40], we examined possible co-localization of KChIP1 vesicles with these proteins. Despite some overlapping localization in the perinuclear region of the cells, puncta labelled by KChIP were clearly distinct from those labelled by anti-syntaxin 7 (Figures 4A–4C) or anti-syntaxin 8 (Figures 4D–4F). We also examined potential overlap with lysosomes. Over-expressed VAMP7 has been found associated with lysosomes [41], but we saw little overlap of staining for endogenous VAMP7 with that for the lysosomal marker LAMP1. The results indicate that the VAMP7-positive KChIP vesicles are not of late endosomal/lysosomal origin.
Figure 4. Lack of co-localization of KChIP1 with syntaxin 7 and 8 or VAMP7 with LAMP1.
HeLa cells were transfected to express KChIP1–EYFP, fixed and immunostained with mouse monoclonal antibodies specific for syntaxin 7 (A–C) or syntaxin 8 (D–F). The images show that KChIP1 punctae are distinct from those stained by anti-syntaxin 7 or 8. In cells immunostained with anti-VAMP 7 and anti-LAMP1 (G–I), there was little overlap of the stained punctae. The colour overlays show KChIP1–EYFP or anti-VAMP7 in green and anti-syntaxin 7, 8 or anti-LAMP1 in red, with co-localization appearing in yellow. The scale bars represent 10 μm in the main Figures and 2 μm in the enlarged inserts.
Functional involvement of VAMP7 and Vti1a in traffic of Kv4.2/KChIP1
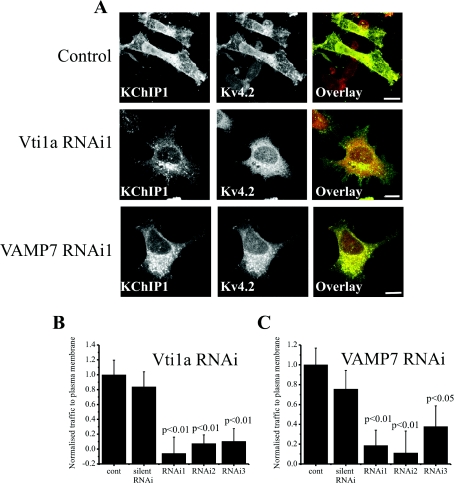
The co-localization data suggested a potential role for VAMP7 and Vti1a in the unconventional traffic route taken by Kv4.2/KChIP1. In order to test this possibility, we used RNA silencing to examine the effect of knockdown of these SNAREs on Kv4.2/KChIP1 traffic. Three distinct siRNAs were tested for both Vti1a and VAMP7 and their effect on the localization of KChIP1–EYFP and Kv4.2 was examined. Cells treated with the siRNAs were compared with cells in the absence of siRNA and also with cells in the presence of a silent siRNA. In control cells, KChIP1 and Kv4.2 were co-localized and found at the plasma membrane (Figure 5A). Treatment with Vti1a or VAMP7 siRNA blocked traffic of Kv4.2 channels to the plasma membrane and KChIP1–EYFP was also found to be intracellular (Figure 5A). The proportion of Kv4.2 present at the plasma membrane was determined and expressed as a normalized value for plasma membrane traffic. The silent siRNA had no effect on the amount of Kv4.2 traffic but all three Vtila and all three VAMP7 siRNAs had a significant inhibitory effect (Figures 5B and 5C). Further studies concentrated on use of the most inhibitory Vti1a or VAMP7 siRNA.
Figure 5. VAMP7 and Vti1a siRNA specifically inhibits traffic of Kv4.2/KChIP1 to the plasma membrane in HeLa cells.
(A) HeLa cells were co-transfected to express both KChIP1–EYFP and Kv4.2 and immunostained with anti-Kv4.2 so that localization of KChIP1 and Kv4.2 could be visualized. Control cells are shown as well as cells co-transfected with VAMP7 or Vtila siRNA as indicated. The colour overlays show co-expressed proteins in green (KChIP1–EYFP) and red (Kv4.2) with co-localization appearing in yellow. The scale bars represent 10 μm. Normalized traffic of Kv4.2 in control cells and cells co-transfected with Vti1a (B) or VAMP7 (C) siRNAs was quantified and the results shown as means±S.E.M. from 20 (Vti1A) or 25 (VAMP7) cells for each condition. cont, control; RNAi, RNA interference.
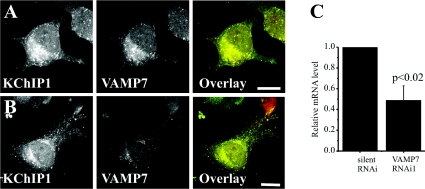
In order to determine whether siRNA treatment affected the KChIP1 vesicles, staining for VAMP7 was carried out on cells transfected to express KChIP1–EYFP. As seen above, VAMP7 co-localized with KChIP1 in control cells (Figure 6A). In cells treated with VAMP7 siRNA, the levels of VAMP7 protein appeared to be significantly reduced and the effectiveness of the siRNA knockdown of VAMP7 was confirmed by real-time PCR to measure relative mRNA levels (Figure 6C). Despite the knockdown, the KChIP1 vesicles did not show any obvious alterations in distribution or morphology and still showed co-localization with low levels of VAMP7 (Figure 6B).
Figure 6. VAMP7 knockdown does not affect the morphology or distribution of KChIP1 vesicles.
HeLa cells were transfected to express KChIP1–EYFP, fixed and immunostained with anti-VAMP7 in the absence (A) or presence (B) of VAMP7 siRNA so that localization of KChIP1 and VAMP7 could be visualized. The colour overlays show co-expressed proteins in green (KChIP1–EYFP) and red (VAMP7) with co-localization appearing in yellow. The scale bars represent 10 μm. (C) For quantitative PCR, mRNA was prepared from HeLa cells transfected in the presence of silent or VAMP7 siRNA and relative mRNA levels determined using real-time PCR. RNAi, RNA interference.
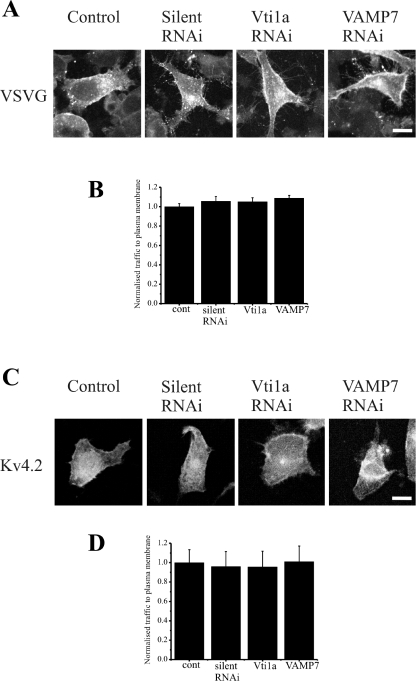
To determine whether the ability of the Vti1a and VAMP7 siRNAs to inhibit Kv4.2 traffic was specific, we tested the effect of the selected siRNAs on VSVG traffic to the plasma membrane. The appearance of VSVG–GFP at the plasma membrane was apparent in control cells and also in cells treated with control siRNA or Vti1a and VAMP7 siRNAs (Figure 7A). Quantification of VSVG at the plasma membrane indicated that the siRNAs were without effect (Figure 7B). These findings support the conclusion that Vti1a and VAMP7 are specifically involved in an alternative traffic pathway that is distinct from the conventional pathway from the ER followed by VSVG.
Figure 7. VAMP7 and Vti1a siRNA do not inhibit traffic of VSVG or Kv4.2/KChIP2 to the plasma membrane.
HeLa cells were transfected to express VSVG–GFP and imaged. Control cells are shown as well as cells co-transfected with silent, VAMP7 or Vtila siRNA as indicated (A). Normalized traffic of VSVG in control cells (cont) and cells co-transfected with silent, Vti1a or VAMP7 siRNAs was quantified and the results are shown from 20 cells for each condition (B). HeLa cells were transfected to express KChIP2 and Kv4.2 and immunostained with anti-Kv4.2 to allow imaging of Kv4.2 localization. Control cells are shown as well as cells co-transfected with silent, VAMP7 or Vtila siRNA as indicated (C). Normalized traffic of Kv4.2 in control cells and cells co-transfected with silent, Vti1a or VAMP7 siRNAs was quantified and the results shown as means±S.E.M. from 30 cells for each condition (D). The scale bars represent 10 μm. RNAi, RNA interference.
KChIP1 is the only one of the four KChIPs that is myristoylated, and the only one that targets to the pool of intracellular vesicles [11,13,34]. KChIP2, in contrast, is palmitoylated and targets to the plasma membrane when expressed alone [15,34]. This raised the possibility that its stimulation of Kv4 traffic may involve interaction with the channel in a distinct cellular compartment and via a distinct cellular mechanism to that used by KChIP1. To test this possibility, the effect of Vti1a and VAMP7 siRNAs on KChIP2-stimulated traffic of Kv4.2 was examined. No effect of the siRNAs was detected (Figures 7C and 7D), consistent with distinct pathways being used by KChIP1 and KChIP2 and again confirming the lack of a general disruptive effect of the siRNAs on cellular function.
The studies so far were all carried out in HeLa cells, in which KChIPs and Kv4 channels are not normally expressed. To test the validity of the findings in a neuronal background, Neuro2A neuroblastoma cells were used. In this cell line, Kv4.2 expressed alone again remained intracellular and co-localized with a Golgi complex marker (Figure 8A), and required co-expression of a KChIP for effective trafficking to the plasma membrane as seen in HeLa cells. KChIP1 vesicles had a more perinuclear distribution in Neuro2A cells compared with HeLa cells, but showed co-localization with both Vti1a and VAMP7 (Figures 8B and 8C). Treatment of Neuro2A cells with Vti1a and VAMP7 siRNAs specifically inhibited traffic of Kv4.2 channels to the plasma membrane and instead these remained, in part, in an intracellular compartment (Figures 8D and 8E) suggesting that the Kv4.2/KChIP1 traffic also involves Vti1a and VAMP7 in neuronal cells.
Figure 8. VAMP7 and Vti1a siRNA specifically inhibit traffic of Kv4.2/KChIP1 to the plasma membrane in Neuro2A cells.
(A) Neuro2A cells were transfected to co-express Kv4.2 and EYFP–Golgi. (B and C) Neuro2a cells were transfected to express KChIP1–EYFP, fixed and immunostained with anti-Vti1a or anti-VAMP7 as indicated. (D) Neuro2a cells were co-transfected to express both KChIP1-EYFP and Kv4.2 and immunostained with anti-Kv4.2 so that localization of KChIP1 and Kv4.2 could be visualized. Control cells are shown as well as cells co-transfected with silent, VAMP7 or Vtila siRNA as indicated. (E) Normalized traffic of Kv4.2 in control cells (cont) and cells co-transfected with siRNAs was quantified and the results shown as means±S.E.M. from 20 cells for each condition. The scale bars represent 10 μm. RNAi, RNA interference.
DISCUSSION
The results presented in the present study support a role for a VAMP7/Vti1a-containing SNARE complex in a novel traffic pathway that is used by KChIP1 in its stimulation of Kv4 traffic to the plasma membrane, but not by the conventional pathway followed by the VSVG protein. This pathway appears to exist in both non-neuronal (HeLa) and neuronal-derived (Neuro2A) cell lines. Where in the secretory pathway are the KChIP1 vesicles placed? KChIP1 is associated with 50–100 nm vesicles, and we have previously concluded that the KChIP1 vesicles are an intermediate in ER to Golgi traffic based on their partial co-localization with ERGIC-53 and the finding that an expressed N-terminal fragment of KChIP1 blocked Kv4.2 traffic and became fully co-localized with an enlarged ERGIC compartment [11]. They were not COPII-coated nor was COPII function crucial for Kv4.2/KChIP1 traffic, although COPI function was [12]. This pathway is likely to be distinct from the other non-conventional ER to Golgi route, such as the well studied route for procollagen, as this is COPII-dependent and COPI-independent [25]. We now show that the VAMP-7-positive KChIP1 vesicles do not overlap with endosomal or lysosomal markers, consistent with the interpretation that they lie in the early exocytic pathway and with previous results suggesting a role for VAMP/Vti1a in ER to Golgi traffic of PCTVs [33].
VAMP7, which is otherwise known as TI-VAMP [42], appears to be involved in multiple SNARE complexes in various traffic steps in different cell types. It has been shown to be involved in endosomal/lysosomal traffic where it can function in at least two distinct SNARE complexes [40,41,43]. Despite a focus on VAMP7 in endosomal traffic, VAMP7 is also involved in transport of apical vesicles to the plasma membrane in epithelial cells [39]. In addition, it is clear that many of the vesicular VAMP7 structures identified by immunofluorescence do not overlap with endosomal/lysosomal markers. In PC12 cells, VAMP7 overlaps to only a low extent with endosomal and lysosomal markers, and it was noted that in HeLa cells the endogenous VAMP7 does not co-localize with the lysosomal protein LAMP1 [44]. We have now confirmed the latter observation in the present study. These findings suggest that VAMP7 may associate with a variety of other SNARE partners to function in multiple membrane traffic steps which may in part be cell type-specific [42]. It is noteworthy and potentially relevant to the current study that, in neurons, VAMP7 is involved in axonal and dendritic outgrowth [45] and is associated with as yet unidentified vesicular structures present in axons and dendrites [44]. In PC12 cells, the VAMP7-labelled vesicles seen by immuno-EM [44] are very similar in size and morphology to those labelled by KChIP1 in HeLa cells [12].
The specificity of siRNA on Kv4.2/KChIP1 traffic and lack of general disruption of the secretory pathway was demonstrated by the lack of effect on VSVG traffic to the plasma membrane. In addition, Vti1a and VAMP7 siRNAs had no effect on Kv4.2 traffic stimulated by KChIP2. This latter result is not surprising given the differing intrinsic localization of KChIP2 compared with KChIP1. Since KChIP2 effectively targets to the plasma membrane due to its palmitoylation even in the absence of Kv4.2 [15,34], it would be unlikely to encounter Kv4.2 in the same compartment as KChIP1, but may do so at the level of the Golgi where Kv4.2 accumulates when expressed alone and where many palmitoyltransferases are located [46]. Interestingly, KChIP2 is expressed at the highest level in the heart [47] where it is the major KChIP and is indispensable for normal Kv4 function [48] and VAMP7 is apparently not expressed in the heart [49].
SNARE complexes suggested to mediate ER to Golgi traffic all contain syntaxin 5 and rBet1, but three distinct complexes have been indentified based on the identity of the other two SNAREs. KChIP1/Kv4.2 traffic appears to involve Vti1a and VAMP7 based on co-localization and functional siRNA data. Vti1a has been shown to interact with syntaxin 5 [50], and both Vti1a and VAMP7 have been found to be quantitatively recovered in a SNARE complex with syntaxin 5 and rBet1, which was implicated in ER to Golgi traffic of PCTVs [33]. Since these organelles are expressed in a tissue-specific manner, it was possible that the Vti1a/VAMP7-dependent ER to Golgi traffic could have been an enterocyte-specific process. The present results further highlight the distinctive nature of Kv4.2/KChIP1 traffic and now suggest that Vti1a/VAMP7 define a non-conventional pathway that may be used for the traffic of a range of cargo proteins in neuronal and non-neuronal cell types.
Acknowledgments
We are grateful to Francis Barr, Burcu Hasdemir, Jesse Hay, Thierry Galli and John Presley for the gift of reagents.
FUNDING
This work was supported by a Wellcome Trust Prize Studentship to S. E. F.
References
- 1.Birnbaum S. G., Varga A. W., Yuan L.-L., Anderson A. E., Sweatt J. D., Schrader L. A. Structure and function of Kv4-family transient potassium channels. Physiol. Rev. 2004;84:803–833. doi: 10.1152/physrev.00039.2003. [DOI] [PubMed] [Google Scholar]
- 2.Isbrandt D., Leicher T., Waldschutz R., Zhu X., Luhmann U., Michel U., Sauter K., Pongs O. Gene structures and expression profiles of three human KCND (Kv4) potassium channels mediating A-type currents I(TO) and I(SA) Genomics. 2000;64:144–154. doi: 10.1006/geno.2000.6117. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman D. A., Magee J. C., Colbert C. M., Johnston D. K+ channel regulation of signal propagation in dendrites of hippocampal pyramidal neurons. Nature. 1997;387:869–875. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- 4.Cai X., Liang C. W., Muralidharan S., Kao J. P., Tang C. M., Thompson S. M. Unique roles of SK and Kv4.2 potassium channels in dendritic integration. Neuron. 2004;44:351–364. doi: 10.1016/j.neuron.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 5.Bernard C., Anderson A., Becker A., Poolos N. P., Beck H., Johnston D. Acquired dendritic channelopathy in temporal lobe epilepsy. Science. 2004;305:532–535. doi: 10.1126/science.1097065. [DOI] [PubMed] [Google Scholar]
- 6.An W. F., Bowlby M. R., Bett M., Cao J., Ling H. P., Mendoza G., Hinson J. W., Mattsson K. I., Strassle B. W., Trimmer J. S., Rhodes K. J. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- 7.Burgoyne R. D., Weiss J. L. The neuronal calcium sensor family of Ca2+-binding proteins. Biochem. J. 2001;353:1–12. [PMC free article] [PubMed] [Google Scholar]
- 8.Burgoyne R. D. Neuronal calcium sensor proteins: generating diversity in neuronal Ca(2+) signalling. Nat. Rev. Neurosci. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pioletti M., Findeisen F., Hura G. L., Minor D. L. Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer. Nat. Struct. Mol. Biol. 2006;13:987–995. doi: 10.1038/nsmb1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H., Yan Y., Liu Q., Huang Y., Shen Y., Chen L., Chen Y., Yang Q., Hao Q., Wang K., Chai J. Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits. Nat. Neurosci. 2007;10:32–39. doi: 10.1038/nn1822. [DOI] [PubMed] [Google Scholar]
- 11.O'Callaghan D. W., Hasdemir B., Leighton M., Burgoyne R. D. Residues within the myristoylation motif determine intracellular targeting of the neuronal Ca2+ sensor protein KChIP1 to post-ER transport vesicles and traffic of Kv4 K+ channels. J. Cell Sci. 2003;116:4833–4845. doi: 10.1242/jcs.00803. [DOI] [PubMed] [Google Scholar]
- 12.Hasdemir B., Fitzgerald D. J., Prior I. A., Tepikin A. V., Burgoyne R. D. Traffic of Kv4 K+ channels mediated by KChIP1 is via a novel post-ER vesicular pathway. J. Cell Biol. 2005;171:459–469. doi: 10.1083/jcb.200506005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibata R., Misonou H., Campomanes C. R., Anderson A. E., Schrader L. A., Doliveira L. C., Carroll K. I., Sweatt J. D., Rhodes K. J., Trimmer J. S. A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. J. Biol. Chem. 2003;278:36445–36454. doi: 10.1074/jbc.M306142200. [DOI] [PubMed] [Google Scholar]
- 14.Bahring R., Dannenberg J., Peters H. C., Leicher T., Pongs O., Isbrandt D. Conserved Kv4 N-terminal domain critical for effects of Kv channel interacting protein 2.2 on channel expression and gating. J. Biol. Chem. 2001;276:23888–23894. doi: 10.1074/jbc.M101320200. [DOI] [PubMed] [Google Scholar]
- 15.Takimoto K., Yang E.-K., Conforti L. Palmitoylation of KChIP splicing variants is required for efficient cell surface expression of Kv4.3 channels. J. Biol. Chem. 2002;277:26904–26911. doi: 10.1074/jbc.M203651200. [DOI] [PubMed] [Google Scholar]
- 16.Beckers C. J. M., Balch W. E. Calcium and GTP: essential components in vesicular trafficking between the endoplasmic reticulum and Golgi apparatus. J. Cell Biol. 1989;108:1245–1256. doi: 10.1083/jcb.108.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J. M., Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 18.Kuge O., Dascher C., Orci L., Rowe T., Amherdt M., Plutner H., Ravazzola M., Tanigawa G., Rothman J. E., Balch W. E. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J. Cell Biol. 1994;125:51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurkan C., Koulov A. V., Balch W. E. An evolutionary perspective on eukaryotic membrane trafficking. Adv. Exp. Med. Biol. 2007;607:73–83. doi: 10.1007/978-0-387-74021-8_6. [DOI] [PubMed] [Google Scholar]
- 20.Bannykh S. I., Balch W. E. Membrane dynamics at the endoplasmic reticulum-Golgi interface. J. Cell Biol. 1997;138:1–4. doi: 10.1083/jcb.138.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bednarek S. Y., Ravazzola M., Hosobuchi M., Amherdt M., Perrelet A., Schekman R., Orci L. COPI- and COPII-coated vesicles bud directly from the endoplasmic reticulum in yeast. Cell. 1995;29:1183–1196. doi: 10.1016/0092-8674(95)90144-2. [DOI] [PubMed] [Google Scholar]
- 22.Takida S., Maeda Y., Kinoshita T. Mammalian GPI-anchored proteins require p24 proteins for their efficient transport from the ER to the plasma membrane. Biochem. J. 2008;409:555–562. doi: 10.1042/BJ20070234. [DOI] [PubMed] [Google Scholar]
- 23.Muniz M., Morsomme P., Riezman H. Protein sorting upon exit from the endoplasmic reticulum. Cell. 2001;104:313–320. doi: 10.1016/s0092-8674(01)00215-x. [DOI] [PubMed] [Google Scholar]
- 24.Stephens D. J., Pepperkok R. Imaging of procollagen transport reveals COPI-dependent cargo sorting during ER-to-Golgi transport in mammalian cells. J. Cell Sci. 2002;115:1149–1160. doi: 10.1242/jcs.115.6.1149. [DOI] [PubMed] [Google Scholar]
- 25.Starkuviene V., Pepperkok R. Differential requirements for ts-O45-G and procollagen biosynthetic transport. Traffic. 2007;8:1035–1051. doi: 10.1111/j.1600-0854.2007.00582.x. [DOI] [PubMed] [Google Scholar]
- 26.Siddiqi S. A., Gorelick F. S., Mahan J. T., Mansbach C. M. COPII proteins are required for Golgi fusion but not for endoplasmic reticulum budding of the pre-chylomicron transport vesicle. J. Cell Sci. 2003;116:415–427. doi: 10.1242/jcs.00215. [DOI] [PubMed] [Google Scholar]
- 27.Wu G., Zhao G., He Y. Distinct pathways for the trafficking of angiotensin II and adrenergic receptors from the endoplasmic reticulum to the cell surface: Rab1-independent transport of a G protein-coupled receptor. J. Biol. Chem. 2003;278:47062–47069. doi: 10.1074/jbc.M305707200. [DOI] [PubMed] [Google Scholar]
- 28.Ye B., Zhang Y., Song W., Younger S. H., Jan L. Y., Jan Y. N. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kloepper T. H., Kienle C. N., Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell. 2007;18:3463–3471. doi: 10.1091/mbc.E07-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dascher C., Matteson J., Balch W. E. Syntaxin 5 regulates endoplasmic reticulum to Golgi transport. J. Biol. Chem. 1994;269:29363–29366. [PubMed] [Google Scholar]
- 31.Xu D., Joglekar A. P., Williams A. L., Hay J. C. Subunit structure of a mammalian ER/Golgi SNARE complex. J. Biol. Chem. 2000;275:39631–39639. doi: 10.1074/jbc.M007684200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang T., Hong W. Ykt6 forms a SNARE complex with syntaxin 5, GS28, and Bet1 and participates in a late stage in endoplasmic reticulum-Golgi transport. J. Biol. Chem. 2001;276:27480–27487. doi: 10.1074/jbc.M102786200. [DOI] [PubMed] [Google Scholar]
- 33.Siddiqi S. A., Siddiqi S., Mahan J., Peggs K., Gorelick F. S., Mansbach C. M. The identification of a novel ER to Golgi SNARE complex used by the pre-chylomicron transport vesicle. J. Biol. Chem. 2006;281:20974–20982. doi: 10.1074/jbc.M601401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venn N., Haynes L. P., Burgoyne R. D. Specific effects of KChIP3/calsenilin/DREAM but not KChIPs1, 2 and 4 on calcium signalling and regulated secretion in PC12 cells. Biochem. J. 2008;413:71–80. doi: 10.1042/BJ20080441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Handley M. T. W., Haynes L. P., Burgoyne R. D. Differential dynamics of Rab3A and Rab27A on secretory granules. J. Cell Sci. 2007;120:973–984. doi: 10.1242/jcs.03406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasegawa H., Zinsser S., Rhee Y., Vik-Mo E. O., Davanger S., Hay J. C. Mammalian ykt6 is a neuronal SNARE targeted to a specialized compartment by its profilin-like amino terminal domain. Mol. Biol. Cell. 2003;14:698–720. doi: 10.1091/mbc.E02-09-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Haas A. K., Yoshimura S., Stephens D. J., Preisinger C., Fuchs E., Barr F. A. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J. Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- 39.Galli T., Zahraoui A., Vaidyanathan V. V., Raposo G., Tian J. M., Karin M., Niemann H., Louvard D. A novel tetanus neurotoxin-insensitive vesicle-associated membrane protein in SNARE complexes of the apical plasma membrane of epithelial cells. Mol. Biol. Cell. 1998;9:1437–1448. doi: 10.1091/mbc.9.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pryor P. R., Mullock B. M., Bright N. A., Lindsay M. R., Gray S. R., Richardson S. C., Stewart A., James D. E., Piper R. C., Luzio J. P. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;5:590–595. doi: 10.1038/sj.embor.7400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Advani R. J., Yang B., Prekeris R., Lee K. C., Klumperman J., Scheller R. H. VAMP-7 mediates vesicular transport from endosomes to lysosomes. J. Cell Biol. 1999;146:765–776. doi: 10.1083/jcb.146.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossi V., Banfield D. K., Vacca M., Dietrich L. E., Ungermann C., D'Esposito M., Galli T., Filippini F. Longins and their longin domains: regulated SNAREs and multifunctional SNARE regulators. Trends Biochem. Sci. 2004;29:682–688. doi: 10.1016/j.tibs.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Ward D. M., Pevsner J., Scullion M. A., Vaughn M., Kaplan J. Syntaxin 7 and VAMP-7 are soluble N-ethylmaleimide-sensitive factor attachment protein receptors required for late endosome-lysosome and homotypic lysosome fusion in alveolar macrophages. Mol. Biol. Cell. 2000;11:2327–2333. doi: 10.1091/mbc.11.7.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coco S., Raposo G., Martinez S., Fontaine J. J., Takamori S., Zahraoui A., Jahn R., Matteoli M., Louvard D., Galli T. Subcellular localization of tetanus neurotoxin-insensitive vesicle-associated membrane protein (VAMP)/VAMP7 in neuronal cells: evidence for a novel membrane compartment. J. Neurosci. 1999;19:9803–9812. doi: 10.1523/JNEUROSCI.19-22-09803.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez-Arca S., Coco S., Mainguy G., Schenk U., Alberts P., Bouille P., Mezzina M., Prochiantz A., Matteoli M., Louvard D., Galli T. A common exocytotic mechanism mediates axonal and dendritic outgrowth. J. Neurosci. 2001;21:3830–3838. doi: 10.1523/JNEUROSCI.21-11-03830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greaves J., Salaun C., Fukata Y., Fukata M., Chamberlain L. H. Palmitoylation and membrane interactions of the neuroprotective chaperone cysteine-string protein. J. Biol. Chem. 2008;283:25014–25026. doi: 10.1074/jbc.M802140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pruunsild P., Timmusk T. Structure, alternative splicing, and expression of the human and mouse KCNIP gene family. Genomics. 2005;86:581–593. doi: 10.1016/j.ygeno.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Kuo H.-C., Cheng C.-F., Clark R. B., Lin J. J.-C., Lin J. L.-C., Hoshijima M., Nguyen-Tran V. T. B., Gu Y., Ikeda Y., Chu P.-H., et al. A defect in the Kv channel-interacting protein 2 (KChIp2) gene leads to a complete loss of Ito and confers susceptibility to ventricular tachycardia. Cell. 2001;107:801–813. doi: 10.1016/s0092-8674(01)00588-8. [DOI] [PubMed] [Google Scholar]
- 49.Advani R. J., Bae H. R., Bock J. B., Chao D. S., Doung Y. C., Prekeris R., Yoo J. S., Scheller R. H. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J. Biol. Chem. 1998;273:10317–10324. doi: 10.1074/jbc.273.17.10317. [DOI] [PubMed] [Google Scholar]
- 50.Xu Y., Wong S. H., Tang B. L., Subramaniam V. N., Zhang T., Hong W. A 29-kilodalton Golgi soluble N-ethylmaleimide-sensitive factor attachment protein receptor (Vti1-rp2) implicated in protein trafficking in the secretory pathway. J. Biol. Chem. 1998;273:21783–21789. doi: 10.1074/jbc.273.34.21783. [DOI] [PubMed] [Google Scholar]