Abstract
Summary: Many bacteria export extracellular polysaccharides (EPS) and capsular polysaccharides (CPS). These polymers exhibit remarkably diverse structures and play important roles in the biology of free-living, commensal, and pathogenic bacteria. EPS and CPS production represents a major challenge because these high-molecular-weight hydrophilic polymers must be assembled and exported in a process spanning the envelope, without compromising the essential barrier properties of the envelope. Emerging evidence points to the existence of molecular scaffolds that perform these critical polymer-trafficking functions. Two major pathways with different polymer biosynthesis strategies are involved in the assembly of most EPS/CPS: the Wzy-dependent and ATP-binding cassette (ABC) transporter-dependent pathways. They converge in an outer membrane export step mediated by a member of the outer membrane auxiliary (OMA) protein family. OMA proteins form outer membrane efflux channels for the polymers, and here we propose the revised name outer membrane polysaccharide export (OPX) proteins. Proteins in the polysaccharide copolymerase (PCP) family have been implicated in several aspects of polymer biogenesis, but there is unequivocal evidence for some systems that PCP and OPX proteins interact to form a trans-envelope scaffold for polymer export. Understanding of the precise functions of the OPX and PCP proteins has been advanced by recent findings from biochemistry and structural biology approaches and by parallel studies of other macromolecular trafficking events. Phylogenetic analyses reported here also contribute important new insight into the distribution, structural relationships, and function of the OPX and PCP proteins. This review is intended as an update on progress in this important area of microbial cell biology.
INTRODUCTION
Many bacteria secrete polysaccharides. Some are excreted polymers that retain only limited association with the cell surface, and these are often referred to as extracellular polysaccharides (EPS) or slime polysaccharides. In contrast, others form a discrete surface layer (the capsule) that is intimately associated with the cell surface (Fig. 1). In the case of gram-negative bacteria, the precise linkage between capsular polysaccharides (CPS) and the outer membrane is not always known, but the processes involved in the biosynthesis and export of CPS and EPS are indistinguishable. EPS and CPS play many different roles in the biology of microorganisms and are frequently essential virulence determinants in pathogens of plants, livestock, and humans. The production of EPS and CPS represents a major challenge in that they are hydrophilic high-molecular-weight polymers with molecular masses of 105 to 106 Da (or greater) that must be assembled in a process spanning the inner membrane. The nascent polymer must traverse the periplasm containing the peptidoglycan layer (an essential stress-bearing structure), and finally it must be translocated across the outer membrane without compromising the critical barrier properties. Emerging evidence points to the existence of molecular scaffolds that perform these critical polymer-trafficking functions.
FIG. 1.
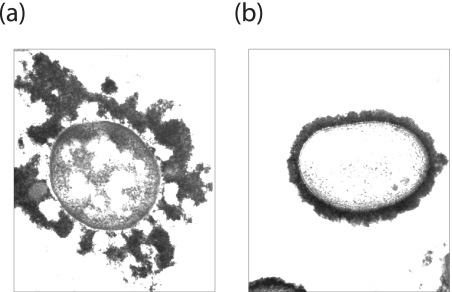
Morphology of CPS- and EPS-producing bacteria. (a) K. pneumoniae serotype K20; (b) E. coli serotype K30. The EPS and CPS in these bacteria have identical repeat-unit structures, and both forms are labeled using cationized ferritin. The E. coli isolate retains most of the polymer as CPS in a well-defined capsule structure. In contrast, the K. pneumoniae isolate has some CPS but releases substantial amounts of polymer from the surface as EPS; this “looser” association is evident in the micrograph.
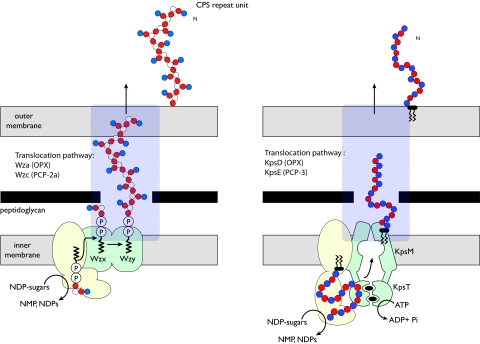
There exists a remarkable diversity of CPS and EPS structures in gram-negative bacteria, yet most are assembled by one of two primary mechanisms of biosynthesis. These processes differ fundamentally in their membrane topology and characteristic components. Both systems are found in representative serotypes of Escherichia coli, and well-studied prototypes exist in this species. These have been reviewed in detail elsewhere (115), and the reader is referred there for detailed references; only an overview of the general overall biosynthesis processes is presented here. E. coli group 1 CPS are assembled by a Wzy-dependent pathway, whereas group 2 CPS follow an ATP-binding cassette (ABC) transporter-dependent process (Fig. 2).
FIG. 2.
Overview of the Wzy-dependent (left) and ABC transporter-dependent (right) EPS biosynthesis pathways. The two biosynthesis pathways involve different components, modes of polymerization, and membrane topologies (for details, see the text). However, both involve terminal export pathways mediated by members of the PCP and OPX protein families.
Polymers in the Wzy-dependent class are built on the polyisoprenoid lipid undecaprenol diphosphate (und-PP) acceptor by glycosyltransferases located in (or at) the inner leaflet of the inner membrane. These und-PP-linked intermediates are exported across the inner membrane by the putative flippase, Wzx, and are polymerized at the periplasmic face in a reaction requiring Wzy, the putative polymerase. Wzx and Wzy are both integral membrane proteins whose exact reaction mechanisms are unknown. Their activities are largely defined by the phenotypes of wzx and wzy mutants. In E. coli, and in many other bacteria, the polymerization activity in Wzy-dependent systems is influenced by an additional component belonging to a family originally called the membrane protein auxiliary proteins (84). Subsequently these proteins have also been referred to as polysaccharide copolymerases (PCP) (75), and this designation will be used here. The assembly of group 1 CPS requires a PCP subfamily 2a (PCP-2a) representative encoded by the wzc gene (27, 117). Despite the PCP family name, the role(s) of PCP proteins seems to be more complex and is not necessarily confined to “polymerization.” Wzc plays a critical role in the translocation of CPS from the periplasm to the cell surface. It does this via critical interactions with the final component of the CPS/EPS export apparatus, a member of a family known as outer membrane auxiliary (OMA) proteins (84) encoded by the wza gene. OMA proteins were initially identified by (limited) sequence similarity before any functional data were available. With the recent structural and biochemical insight described here, we propose that these proteins should be renamed outer membrane polysaccharide export (OPX) proteins, to acknowledge their roles in CPS/EPS export. Together, Wza and Wzc comprise a molecular scaffold that spans the cell envelope and is required for the final stage of polymer export (see below).
Assembly of ABC transporter-dependent polymers (e.g., E. coli group 2 CPS) follows a fundamentally different pathway (reviewed in reference 115). The polysaccharide is polymerized at the cytoplasmic face of the inner membrane via the sequential addition of glycose residues to the nonreducing terminus of the nascent chain. The exact nature of the acceptor molecule for group 2 CPS assembly has not been unequivocally established. There is some evidence suggesting that und-PP is not involved. The exported glycan appears to be attached to a diacylglycerophosphate residue. It is possible that the terminal diacylglycerophosphate is also the acceptor for chain growth, but details of the initiation of chain growth have not yet been resolved. The polymer is exported across the inner membrane via an ABC transporter. Translocation of E. coli group 2 CPS across the periplasm requires a PCP-3 protein (KpsE) and an OPX (OMA) protein (KpsD). Existing data implicate KpsD-KpsE in an export complex analogous (but perhaps not identical) to Wza-Wzc.
While the early steps in ABC transporter-dependent and Wzy-dependent CPS/EPS biosynthesis mechanisms may vary, the involvement of members of the PCP and OPX families supports a conclusion that the later steps in export may exhibit some similarities. This is perhaps not surprising given the shared challenge of moving a large hydrophilic polymer across the periplasm and outer membrane. Since the first recognition of conserved protein families by Paulsen et al. in 1997 (84), knowledge of the structure and function of what are now called the OPX and PCP proteins has been advanced by the application of biochemistry and structural biology approaches. A significant amount of information is now available for the OPX-PCP-2a pair, which is involved in E. coli group 1 CPS assembly. Less is known of the comparable OPX-PCP-3 components for group 2 CPS systems, but conserved themes are emerging. This review is intended as an update on progress in this important area of microbial cell biology.
Wza, THE EFFLUX CHANNEL FOR E. COLI GROUP 1 CPS: THE PROTOTYPE OPX PROTEIN
Wza is essential for expression of group 1 CPS on the surface of E. coli (28, 79). It is an outer membrane lipoprotein that forms sodium dodecyl sulfate-stable multimers (28), and this resilience has been instrumental in structure-function studies. Initial electron microscopy (EM) studies, including two-dimensional (2D) crystal arrays of Wza oligomers reconstituted in lipid, demonstrated characteristic ring-shaped structures with eightfold symmetry. The images suggested Wza to be a channel-forming protein (28, 79). Subsequent single-particle analyses of Wza oligomers visualized by cryo-negative-staining EM (15.5-Å resolution), revealed a novel hollow, barrel-like structure (dimensions, 90 by 90 by 100 Å) (10). The unique features of Wza were reinforced with a 2.3-Å-resolution crystal structure (26). To date, Wza is the only OPX protein for which a high-resolution structure has been solved.
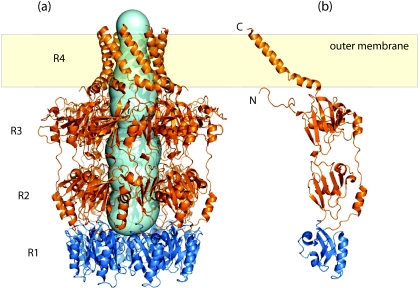
The octameric structure consists of a stack of four eightfold-symmetric rings (R1 to R4) (Fig. 3a). It has a large internal cavity (∼15 000 Å3) that is closed at its base (R1), located in the periplasm, but is open to the external environment via a narrow “neck” (R4; 17-Å diameter). The monomers have four corresponding domains (Fig. 3b), with each ring being formed by the contributions of the eight monomers. The overall shapes and sizes of R1 to R3 are consistent with the cryo-EM data (20), but R4 is destabilized (and is not visible) under the pH conditions of the original EM experiments (R. C. Ford, A. L. Brunkan-LaMontagne, R. F. Collins, B. R. Clarke, R. Harris, J. H. Naismith, and C. Whitfield, submitted for publication). At the base, R1 and R2 contain the polysaccharide export sequence (PES) (Pfam 02563) motif (Fig. 3b). This is the only region of primary sequence similarity shared by OPX proteins (see below). The motif has been identified by sequence similarity alone, and to date there is no definitive information concerning its functional significance. R2 and R3 have similar folds, probably reflecting gene duplication, and they create an internal cavity with a diameter of 25 to 30 Å. R4 provides the most striking feature of Wza; it represents the first example of an outer-membrane-spanning region (or protein) composed of amphipathic α-helices. The extreme C terminus of Wza is exposed at the cell surface, confirming that the barrel does indeed span the outer membrane (26). The oligomer is stabilized in the outer membrane via a combination of the R4 barrel (Ford et al., submitted) and the N-terminal acyl chains that insert into the inner leaflet of the outer membrane (79). The internal cavity of the oligomer is largely polar, and close homologs show little conservation in the residues that line the structure. It is proposed that a water-filled channel would ensure that the polar amino acid side chains and the hydroxyl groups of the polymer make hydrogen bonds, essentially lubricating the channel (26). This would explain the observed lack of specificity of representative OPX channels for a particular polysaccharide repeat-unit structure (91). The smallest internal diameter (R4; 17 Å) is still sufficient to accommodate a polymer in an extended conformation. However, the structure of isolated Wza octamers does not explain how it interacts with the main elements of the biosynthesis machinery in the inner membrane. Insight into that process comes from the finding that Wza interacts with the inner membrane PCP-2a protein, Wzc (20, 79).
FIG. 3.
Structure of Wza from E. coli K30, the prototype OPX protein. (a) Wza forms an octameric complex composed of four ring domains (R1 to R4), with the R4 domain spanning the outer membrane. The internal cavity of the Wza complex was calculated using the online Caver tool (http://loschmidt.chemi.muni.cz/caver/) and is indicated as a pale blue surface. (b) A single Wza monomer (Protein Data Bank accession no. 2J58). The R1 domain (dark blue) contains the majority of the OPX-specific PES motif and likely represents the minimal structural unit of the PES motif. Structure diagrams were generated using Pymol (DeLano Scientific LLC).
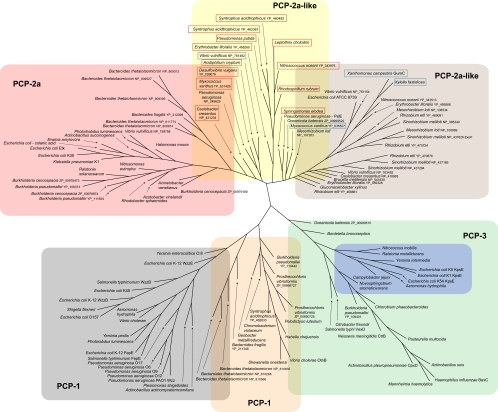
STRUCTURES AND PHYLOGENY OF OPX PROTEINS
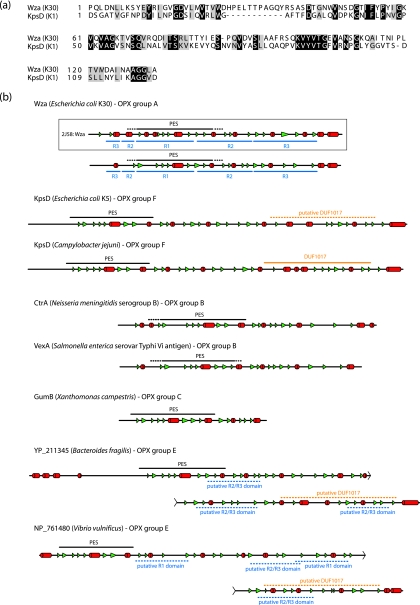
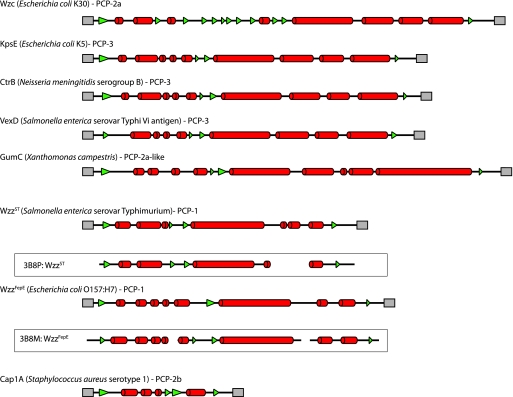
In the assembly of E. coli group 2 CPS, KpsD and KpsE are both essential for export, and mutants in the corresponding genes accumulate polysaccharide in the periplasm (12, 85, 102). The localization of KpsD in the outer membrane is dependent on coexpression of the cognate PCP-3 protein, KpsE, and ongoing CPS synthesis (3). In early studies, the absence of KpsE led to mislocalization and a periplasmic assignment for KpsD (102). KpsD contains the PES motif characteristic of OPX proteins (Fig. 4a), but other than this, the amount of overall primary sequence similarity shared with Wza is insignificant. KpsD is a larger protein (58.3 kDa for mature processed KpsD, compared to the 39.6 kDa for mature WzaK30), but there are notable similarities in the predicted secondary structures of Wza and KpsD (Fig. 4b). Like that of Wza, the C terminus of KpsD is predicted to be α-helical. KpsD unequivocally spans the outer membrane, but unlike Wza, KpsD is not a lipoprotein. In its isolated form, KpsD does not form oligomers larger than dimers (72). It is conceivable that a higher oligomerization state exists in vivo, and this may be dependent on the association with KpsE. Nevertheless, while KpsD may act as the CPS efflux channel across the membrane, its overall structure may differ from that of Wza. This will be resolved only with a solved structure.
FIG. 4.
Predicted secondary structures of OPX proteins indicate a significant degree of structural similarity between the various homologs. (a) Alignment of the primary sequences of the PES domains from Wza and E. coli KpsD (an OPX protein from the ABC transporter-dependent group 2 CPS system). (b) Predicted secondary structures from Wza and other OPX representatives. The secondary structures were assigned based on the agreement of at least three out of four structure prediction algorithms (Prof [82], PSIPred [13, 44], SSPro [16], and jPRED [18]). α-Helical structures are indicated as red cylinders, and β-structures are depicted by green arrowheads. The OPX-specific PES motifs (as detected by a conserved-domain search [65]) are shown as a black line above the secondary structural elements, and the dashed sections of these lines represent regions where no significant sequence alignment to the canonical motif could be found. The DUF1017 motif in the KpsD protein of C. jejuni was identified in a motif search. The “putative DUF1017” domain (hatched orange lines) in the E. coli KpsD protein was assigned based on similar predicted secondary structure. Several regions in the “long” OPX from B. fragilis (YP_211345) showed sequence similarity to the R2 and R3 domains from Wza. These regions were designated “putative R2/R3 domains” (hatched blue lines) if their predicted secondary structure matched the corresponding domains in the Wza prototype. The secondary structure for the solved crystal structure of Wza (bounded by a box) has been included as a measure of the accuracy of the predicted secondary structure assignments. In all cases, only the mature proteins are shown; signal sequences have been removed.
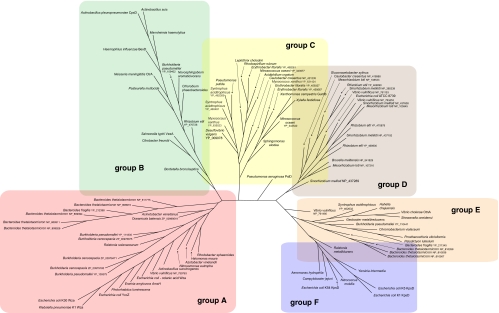
The conserved PES motif provides a foundation for phylogenetic analysis of OPX proteins from the databases of sequenced genomes (Table 1; Fig. 5). In this analysis, the relevant genomic contexts were scanned to ensure the presence of a CPS/EPS gene cluster, and characteristic predicted gene products allowed the assignment of a Wzy-dependent or ABC transporter-dependent pathway. Six clusters of OPX proteins were identified (groups A to F) (Fig. 5). In each case, the groups are homogeneous with respect to the biosynthesis pathway (i.e., ABC transporter dependent or Wzy dependent). The prototype OPX, Wza from E. coli K30, is identified in a cluster of proteins arbitrarily designated group A. The group A OPX proteins are all predicted to be lipoproteins, based on a signal peptidase II cleavage site, and fall within a size range of 261 to 449 residues. The closest relative to WzaK30 is Wza from Klebsiella pneumoniae, and this is not surprising since we have previously reported that their sequences are highly conserved and that their distribution reflects relatively recent horizontal gene transfer events (90). Also found in this group is the YccZ protein, which is a functional homolog of WzaK30 found in a genetic locus present in most E. coli strains. We have referred to this as the “22-min locus” (28). It has also been referred to as the gfc (group 4 capsule) locus (101). Recent studies have illustrated a role for this locus in group 4 CPS assembly (also Wzy dependent and essentially a variant of group 1) and in regulation of the heat shock response in E. coli (48, 86). The locus contains four other genes, designated ymcABCD, that are duplicated elsewhere on the chromosome in yjbEFGH (31). These genes may also function in CPS assembly/export, although their precise functions are currently unknown. Another close homolog, Wza from the colanic acid EPS system of E. coli, is also a member of OPX group A. The colanic acid Wza homolog can functionally replace its homolog in the K30 biosynthesis pathway (91). Colanic acid is found in many E. coli isolates but not in those with group 1 CPS (47); it is essentially a highly regulated version of a group 1 CPS that is activated by environmental cues, mostly signals outside the host. Colanic acid is important in biofilm formation and protection against desiccation (reviewed in reference 115). Although not generally considered to be a virulence determinant, colanic acid has been implicated in participating in survival against gastric fluids (64). OPX homologs in group A are not limited to Enterobacteriaceae but are also found in other gammaproteobacteria, alpha- and betaproteobacteria, and members of the family Bacteroidaceae.
TABLE 1.
OPX and PCP proteins from gram-negative EPS and CPS assembly systems
| Group and organism | Serotype or strain | Classification | ABC or Wzy | OPX proteins
|
PCP proteins
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Accession no. | Length (residues) | Signal sequencea | % Identity (similarity) to PES K30 | Name | Accession no. | Length (residues) | Transmembrane segments | Walker box A | Walker box B | Canonical Walker motifsb | DxDc | Y 574c | R 614c | No. of Ys in tail (of 20) | RK-rich region | PCP | ||||
| A | |||||||||||||||||||||
| Acinetobacter venetianus | RAG-1 | Gamma-proteobacteria | Wzy | Wza | CAB57195 | 366 | SPII | 27 (71) | Wzc | CAB57193 | 726 | 24-41, 437-455 | GPAPEVGKS | HIIID | N* | Y | Y | Y | 5 | + | PCP-2a |
| Actinobacillus succinogenes | 130Z | Gamma-proteobacteria | Wzy | ZP_00732118 | 388 | SPII | 62 (91) | ZP_00732117 | 703 | 31-48, 424-442 | GVSKGVGRH | AVIVT | N | Y | N | Y | 2 | ? | PCP-2a | ||
| Azotobacter vinelandii | AvOP | Gamma-proteobacteria | ZP_00415107 | 347 | SPII* | 50 (80) | ZP_00418290 | 734 | 37-55, 446-465 | GPGPQAGKS | LVILD | Y | Y | N | Y | 5 | ? | PCP-2a | |||
| Bacteroides fragilis | NCTC9343 | Bacteroidetes | YP_212398 | 262 | SPII | 19 (59) | YP_212399 | 801 | 41-56, 515-534 | STISGEGKT | YIVLD | N* | N | N | N | 7 | − | PCP-2a | |||
| Bacteroides thetaiotaomicron | VPI-5482 | Bacteroidetes | NP_808973 | 261 | SPII | 25 (61) | NP_808 974 | 779 | 18-40, 493-514 | SFNEGAGKT | YIIVD | N* | Y | N | N | 7 | − | PCP-2a | |||
| Bacteroides thetaiotaomicron | VPI-5482 | Bacteroidetes | Wzy | NP_809311 | 264 | SPII | 23 (67) | NP_809312, NP_809313 | 381, 429 | 24-49, 127-145 | STVSGEGKS | YVILD | N* | N | N | N | 5 | − | PCP-2a | ||
| Bacteroides thetaiotaomicron | VPI-5482 | Bacteroidetes | Wzy | NP_809526 | 317 | SPII* | 26 (65) | NP_809527 | 812 | 23-50, 513-532 | STVSGEGKS | YVILD | N* | N | N | N | 6 | − | PCP-2a | ||
| Bacteroides thetaiotaomicron | VPI-5482 | Bacteroidetes | NP_809394 | 266 | SPII | 23 (69) | NP_809395 | 812 | 28-49, 512-531 | STVSGEGKS | YVILD | N* | N | N | N | 5 | − | PCP-2a | |||
| Bacteroides thetaiotaomicron | VPI-5482 | Bacteroidetes | Wzy | NP_811775 | 265 | SPII | 24 (57) | NP_811774 | 787 | 12-37, 490-510 | SFNIGAGKT | YIIVD | N* | Y | N | N | 1 | − | PCP-2a | ||
| Burkholderia cenocepacia | PC184 | Betaproteobacteria | Wzy | ZP_00979185 | 368 | SPII | 42 (78) | ZP_00979180 | 716 | 24-42, 438-454 | SAAPAQGKS | MVIVD | Y | N | N | N | 0 | − | PCP-2a | ||
| Burkholderia cenocepacia | PC184 | Betaproteobacteria | ZP_00978471 | 417 | SPI | 40 (72) | ZP_00978473 | 752 | 38-54, 453-470 | GPAPGAGKS | MVVID | Y | Y | N | N | 0 | ? | PCP-2a | |||
| Burkholderia cenocepacia | PC184 | Betaproteobacteria | Wzy | ZP_00978875 | 392 | SPII | 44 (70) | ZP_00978874 | 741 | 42-60, 459-475 | GPTPGIGKS | VVLID | Y | Y | N | ? | 3 | ? | PCP-2a | ||
| Burkholderia pseudomallei | K96243 | Betaproteobacteria | Wzy | YP_111835 | 391 | SPII | 45 (69) | YP_111834 | 739 | 43-61, 460-479 | GPTPGIGKS | MVIVD | Y | Y | N | ? | 3 | ? | PCP-2a | ||
| Burkholderia pseudomallei | K96243 | Betaproteobacteria | Wzy | YP_109375 | 400 | SPII* | 42 (73) | YP_109373 | 746 | 42-57, 457-472 | GPAPGVGKS | AVVID | Y | Y | N | N | 5 | + | PCP-2a | ||
| Erwinia amylovora | Gamma-proteobacteria | Wzy | AmsH | CAA54880 | 377 | SPII | 76 (93) | AmsA | CAA54882 | 726 | 38-54, 430-449 | GASPGIGKT | LVLID | Y | Y | Y | Y | 7 | + | PCP-2a | |
| Escherichia coli (colanic acid) | K-12 | Gamma-proteobacteria | Wzy | Wza | P0A930 | 379 | SPII | 70 (94) | Wzc | P76387 | 720 | 45-61, 437-455 | GVSPSIGKT | LVLID | Y | Y | Y | Y | 6 | + | PCP-2a |
| Escherichia coli (22 min) | K-12 | Gamma-proteobacteria | Wzy | YccZ | P75881 | 379 | SPII | 78 (97) | Etk | P38134 | 726 | 37-55, 430-449 | GATPDSGKT | LVIVD | Y | Y | Y | Y | 7 | + | PCP-2a |
| Escherichia coli | K30 | Gamma-proteobacteria | Wzy | Wza | AAD21562 | 379 | SPII | Wzc | AAD21564 | 721 | 40-59, 435-457 | GASPSAGKT | LIIID | Y | Y | Y | Y | 7 | + | PCP-2a | |
| Halomonas maura | S-30 | Gamma-proteobacteria | Wzy | EpsA | AAX14034 | 376 | SPII | 53 (83) | EpsC | AAX14036 | 729 | 37-56, 444-461 | GPSPGIGKS | LVIID | Y | Y | N | Y | 6 | + | PCP-2a |
| Klebsiella pneumoniae | K1 | Gamma-proteobacteria | Wzy | Wza | AAV27322 | 377 | SPII | 92 (100) | Wzc | AAV27324 | 717 | 38-58, 432-451 | GPSPEIGKT | LVLVD | Y | Y | Y | N | 6 | + | PCP-2a |
| Nitrosomonas eutropha | C71 | Betaproteobacteria | Wzy | ZP_00669712 | 405 | SPII | 55 (84) | ZP_00669713 | 747 | 37-55, 457-475 | GPSPEAGKT | LIIID | Y | Y | N | N | 5 | + | PCP-2a | ||
| Oceanicola batsensis | HTCC2597 | Alpha-proteobacteria | ABC/Wzy | ZP_00999519 | 449 | SPII | 31 (66) | ZP_00999515, ZP_00999522 | 416, 708 | 51-68, 399-415; 40-61, 432-450 | SSVPGEGKT | YIIID | N* | N | N | N | 6 | ? | PCP3, PCP-2a | ||
| Photorhabdus luminescens | W14 | Gamma-proteobacteria | ORF39 | AAO18064 | 381 | SPII | 77 (95) | AAO18066 | 715 | 37-54, 430-448 | GSAPELGKS | YVLVD | Y | Y | N | Y | 6 | + | PCP-2a | ||
| Ralstonia solanacearum | UW551 | Betaproteobacteria | Wzy | EpsA | ZP_00945865 | 377 | SPII | 41 (72) | EpsB | ZP_00945863 | 750 | 35-54, 453-470 | GPTPGVGKS | LVLVD | Y | Y | Y | N | 4 | ? | PCP-2a |
| Rhodobacter sphaeroides | ATCC17025 | Alpha-proteobacteria | ZP_00912185 | 356 | SPII | 48 (83) | ZP_00912187 | 730 | 39-49, 443-458 | SSAPEAGKS | LTIFD | Y | Y | N | ? | 7 | ? | PCP-2a | |||
| Vibrio vulnificus | CMCP6 | Gamma-proteobacteria | NP_759763 | 378 | SPII | 68 (92) | NP_759758 | 727 | 35-54, 435-453 | GPAPGIGKS | LVIID | Y | Y | Y | Y | 6 | + | PCP-2a | |||
| B | |||||||||||||||||||||
| Actinobacillus pleuropneumoniae | J45 | Gamma-proteobacteria | ABC | CpxD | AAB64442 | 394 | SPII | 29 (62) | CpxC | AAB64443 | 385 | 30-49, 359-377 | PCP-3 | ||||||||
| Actinobacillus suis | SO4-K1 | Gamma-proteobacteria | ABC | Wza | AAO65490 | 393 | SPII | 30 (62) | Wzf | AAO65489 | 379 | 29-48, 358-376 | PCP-3 | ||||||||
| Bordetella bronchiseptica | RB50 | Betaproteobacteria | ABC | Wza | NP_889462 | 365 | SPII | 33 (62) | WcbD | NP_889465 | 389 | 37-54, 373-389 | PCP-3 | ||||||||
| Burkholderia pseudomallei | K96243 | Betaproteobacteria | ABC | WcbC | YP_109402 | 387 | SPII | 27 (67) | WcbD | YP_109401 | 382 | 35-56, 366-382 | PCP-3 | ||||||||
| Chlorobium phaeobacteroides | DSM 266 | Bacteroidetes | ABC | ZP_00527694 | 363 | SPII* | 32 (66) | ZP_00527693 | 399 | 40-60, 373-391 | PCP-3 | ||||||||||
| Citrobacter freundii | OU7004 | Gamma-proteobacteria | ABC | VexA | AAK14185 | 355 | SPII | 22 (54) | VexD | AAK14188 | 434 | 83-103, 408-428 | PCP-3 | ||||||||
| Haemophilus influenzae | serotype b | Gamma-proteobacteria | ABC | BexD | CAA38730 | 394 | SPII | 26 (60) | BexC | CAA38731 | 377 | 31-48, 357-375 | PCP-3 | ||||||||
| Mannheimia hemolytica | A1 | Gamma-proteobacteria | ABC | CpxD | AAF08243 | 394 | SPII | 26 (61) | CpxC | AAF08242 | 367 | 16-38, 347-365 | PCP-3 | ||||||||
| Neisseria meningitidis | Serogroup A | Betaproteobacteria | ABC | CtrA | NP_283045 | 387 | SPII | 28 (62) | CtrB | NP_283044 | 387 | 39-57, 368-386 | PCP-3 | ||||||||
| Novosphingobium aromaticivorans | DSM 12444 | Alpha-proteobacteria | ABC | ABD25200 | 392 | SPII | 29 (56) | ABD25193 | 365 | 17-35, 343-362 | PCP-3 | ||||||||||
| Pasteurella multocida | M1404 | Gamma-proteobacteria | ABC | CexD | AAF67275 | 393 | SPII | 30 (61) | CexC | AAF67274 | 370 | 22-41, 350-368 | PCP-3 | ||||||||
| Rhizobium etli | CFN 42 | Alpha-proteobacteria | Wzy | PssN | YP_470726 | 403 | SPII | 30 (62) | PssP | YP_470724 | 758 | 41-59, 452-465 | SSLPGEGKS | YIIVD | N | Y | N | Y | 6 | ? | PCP-2a-like |
| Salmonella enterica serovar Typhi | Gamma-proteobacteria | ABC | VexA | D14156 | 355 | SPII | 22 (59) | VexD | D14156 | 434 | 92-109, 416-434 | PCP-3 | |||||||||
| C | |||||||||||||||||||||
| Acidiphilium cryptum | JF-5 | Alpha-proteobacteria | Wzy | YP_001236072 | 213 | SPI | 28 (59) | YP_001236073, YP_001236074 | 493, 309 | 24-39, 408-428, 470-488 | SPRDGDGKS | IVVVD | Y | Y | N | N | 0 | − | PCP-2a-like | ||
| Caulobacter crescentus | CB15 | Alpha-proteobacteria | Wzy | HfsD | NP_421235 | 239 | SPII | 28 (58) | HfsA, HfsB | NP_421234, NP_421233 | 501, 233 | 41-59, 454-471 | AARRGEGTS | VIVVD | N | Y | N | N | 0 | − | PCP-2a-like |
| Desulfovibrio vulgaris | Hildenborough | Delta-proteobacteria | Wzy | YP_009078 | 268 | SPII | 25 (62) | YP_009079, YP_009080 | 500, 372 | 39-57, 405-424, 440-450, 468-483 | SSVMGEGKT | YVIID | N* | Y | N | N | 3 | ? | PCP-2a-like | ||
| Erythrobacter litoralis | HTCC2594 | Alpha-proteobacteria | Wzy | YP_459567 | 239 | SPII | 32 (60) | YP_459566 | 739 | 61-80, 467-487 | STRPAEGKS | HVIID | N* | Y | N | N | 5 | − | PCP-2a-like | ||
| Erythrobacter litoralis | HTCC2594 | Alpha-proteobacteria | Wzy | YP_459327 | 237 | SPI | 27 (65) | YP_459328 | 719 | 53-75, 455-471 | SSQQSEGKS | IVIFD | Y | Y | N | N | 7 | ? | PCP-2a-like | ||
| Erythrobacter litoralis | HTCC2594 | Alpha-proteobacteria | YP_458261 | 206 | SPII | 33 (56) | YP_458260, YP_458259 | 507, 341 | 21-40, 416-434, 485-500 | SPHPDEGKT | IVIFD | Y | Y | N | N | 2 | ? | PCP-2a-like | |||
| Leptothrix cholodnii | SP-6 | Betaproteobacteria | YP_001792528 | 213 | SPII | 25 (64) | YP_001792527, YP_001792526 | 517, 305 | 21-40, 427-446, 493-512 | SAMPEEGKT | IIVFD | Y | Y | N | N | 4 | + | PCP-2a-like | |||
| Myxococcus xanthus | DK 1622 | Delta-proteobacteria | YP_631424 | 190 | SPII | 32 (69) | YP_631426, YP_631427 | 465, 231 | 15-33, 391-409 | SAMPGEGKT | EVYVD | N* | Y | N | N | 2 | − | PCP-2a-like | |||
| Myxococcus xanthus | DK 1622 | Delta-proteobacteria | Wzy | YP_635523 | 219 | SPI | 32 (61) | YP_635527 | 507 | 51-72, 461-479 | PCP-2a-like | ||||||||||
| Nitrosococcus oceani | ATCC 19707 | Gamma-proteobacteria | YP_343540 | 243 | SPII | 29 (60) | YP_343513 | 753 | 68-83, 467-483 | STSKGEGKS | HVIVD | N* | Y | N | N | 4 | − | PCP-2a-like | |||
| Nitrosococcus oceani | ATCC 19707 | Gamma-proteobacteria | Wzy | YP_343977 | 208 | SPII | 26 (59) | YP_343976, YP_343975 | 533, 297 | 29-45, 444-460, 505-524 | SALSGEGKT | IIIID | Y | Y | N | N | 6 | + | PCP-2a-like | ||
| Pseudomonas aeruginosa | PAO1 | Gamma-proteobacteria | Wzy | PslD | NP_250924 | 256 | SPII | 27 (63) | PslE | NP_250925 | 662 | 24-39, 459-481 | EATYNPGRI | VVLLD | N | N | N | N | 1 | − | PCP-2a-like |
| Pseudomonas putida | W619 | Gamma-proteobacteria | Wzy | YP_001749781 | 185 | SPI | 25 (61) | YP_001749779, YP_001749778 | 522, 268 | 21-39, 427-444, 486-513 | SPTPEAGKT | ICIFD | N | Y | N | ? | 2 | ? | PCP-2a-like | ||
| Rhodospirillum rubrum | ATCC 11170 | Alpha-proteobacteria | YP_428199 | 209 | SPII | 29 (58) | YP_428200, YP_428201 | 532, 335 | 22-39, 415-435, 475-494 | SSMPGEGKT | LILID | Y | Y | N | ? | 3 | + | ||||
| Sphingomonas elodea | ATCC31461 | Alpha-proteobacteria | GelD | AAO85845 | 300 | SPI | 32 (61) | GelC, GelE | AAO85846, AAO85847 | 532, 335 | 22-39, 415-435, 475-494 | SSMPGEGKT | LILID | Y | Y | N | ? | 3 | + | PCP-2a-like | |
| Syntrophus aciditrophicus | SB | Delta-proteobacteria | YP_462359 | 188 | SPI | 29 (59) | YP_462361, YP_462362 | 448, 235 | 15-33, 375-397 | AASTGVGCS | ITIFD | N | N | N | N | 1 | − | PCP-2a-like | |||
| Syntrophus aciditrophicus | SB | Delta-proteobacteria | Wzy | YP_460481 | 238 | SPI* | 29 (69) | YP_460482, YP_460483 | 514, 275 | 22-42, 441-459 | SANPGEGKT | FIIFD | Y | Y | N | N | 0 | ? | PCP-2a-like | ||
| Xanthomonas campestris | pv. campestris | Gamma-proteobacteria | Wzy | GumB | AAL28080 | 232 | SPII | 29 (62) | GumC | AAA86371 | 525, 289 | 28-43, 424-439, 494-514 | SALPGEGKT | YIFFD | N | Y | N | N | 1 | + | PCP-2a-like |
| Xylella fastidiosa | Temecula 1 | Gamma-proteobacteria | Wzy | GumB | AAO29243 | 217 | SPII* | 27 (62) | AAO29242 | 449 | 13-33, 422-439 | PCP-2a-like | |||||||||
| 467 | 38-53, 437-455 | PCP-2a-like | |||||||||||||||||||
| D | |||||||||||||||||||||
| Brucella melitensis | 16 M | Alpha-proteobacteria | Wzy | NP_541829 | 392 | SPI* | 30 (59) | NP_541830 | 729 | 29-45, 434-450 | SLSEGDGKG | IILVN | N | N | N | N | 0 | − | PCP-2a-like | ||
| Caulobacter crescentus | CB15 | Alpha-proteobacteria | Wzy | NP_418988 | 186 | SPI | 28 (65) | NP_418983 | 739 | 36-53, 449-471 | SSLPGEGKT | IVLLD | Y | Y | N | N | 9 | ? | PCP-2a-like | ||
| Escherichia coli | ATCC 8739 | Gamma-proteobacteria | Wzy | YP_001726124 | 185 | SPII | 35 (64) | YP_001726125 | 710 | 32-49, 425-446 | AKQGEGRSL | RIIVD | N | Y | N | Y | 0 | − | PCP-2a-like | ||
| Gluconacetobacter xylinus | BPR2001 | Alpha-proteobacteria | Wzy | AceH | BAB88849 | 214 | SPII* | 27 (61) | AceD | BAB88845 | 736 | 36-53, 448-464 | SAGPEEGKS | LIIID | Y | Y | N | ? | 2 | ? | PCP-2a-like |
| Mesorhizobium loti | MAFF303099 | Alpha-proteobacteria | Wzy | NP_108533 | 172 | SPII* | 31 (65) | NP_108534 | 760 | 25-43, 437-456 | VSPEGDEAA | LVVVE | N | N | N | N | 0 | − | PCP-2a-like | ||
| Mesorhizobium loti | MAFF303099 | Alpha-proteobacteria | Wzy | NP_105945 | 419 | SPI | 27 (61) | NP_105969 | 795 | 41-56, 471-488 | SVLPGEGKS | YIVVD | N* | Y | N | Y | 3 | ? | PCP-2a-like | ||
| Mesorhizobium loti | MAFF303099 | Alpha-proteobacteria | Wzy | NP_107216 | 448 | SPI | 23 (50) | NP_107203 | 734 | 54-69, 444-465 | SSILDDGKT | VVIIN | N | Y | N | N | 1 | − | PCP-2a-like | ||
| Rhizobium etli | CFN 42 | Alpha-proteobacteria | Wzy | YP_469090 | 191 | SPII | 30 (62) | YP_469091 | 724 | 27-41, 435-455 | ISPTGDNGS | LVVVE | N | N | ? | N | 0 | − | PCP-2a-like | ||
| Rhizobium etli | CFN 42 | Alpha-proteobacteria | Wzy | ExoF1 | YP_468900 | 438 | SPI | 24 (56) | YP_468881 | 735 | 44-64, 452-470 | SAHSGEGKS | VVIFD | Y | Y | N | N | 2 | ? | PCP-2a-like | |
| Rhizobium etli | CFN 42 | Alpha-proteobacteria | Wzy | ExoF2 | YP_470878 | 459 | SPI | 24 (53) | YP_470876 | 555 | 11-28, 285-301 | GVSPGSGAS | ITIFD | N | N | N | N | 0 | ? | PCP-2a-like | |
| Sinorhizobium meliloti | 1021 | Alpha-proteobacteria | Wzy | NP_385339 | 191 | SPII | 29 (65) | NP_385340 | 712 | 28-40, 437-456 | VSPGGDEGS | LVLIE | N | N | N | N | 0 | − | PCP-2a-like | ||
| Sinorhizobium meliloti | 1021 | Alpha-proteobacteria | Wzy | ExoF1 | NP_437608 | 421 | SPI | 27 (64) | ExoP | NP_437626 | 786 | 41-62, 463-477 | SALPDEGKS | YVVVD | N* | Y | N | N | 4 | ? | PCP-2a-like |
| Sinorhizobium meliloti | 1021 | Alpha-proteobacteria | Wzy | ExoF2 | NP_437192 | 455 | SPI | 23 (57) | ExoP2 | NP_437189 | 632 | 38-56, 361-375 | SATPDDGKS | IIIVD | Y | Y | N | N | 1 | − | PCP-2a-like |
| Sinorhizobium meliloti | 1021 | Alpha-proteobacteria | Wzy | ExoF3 | NP_437289 | 416 | SPI | 28 (62) | NP_437284 | 662 | 45-62, 335-355 | SCVHNIANA | FIVLH | N | N | N | N | 0 | − | PCP-2a-like | |
| Vibrio vulnificus | CMCP6 | Gamma-proteobacteria | Wzy | NP_761153 | 174 | SPI* | 27 (60) | NP_761154 | 723 | 31-48, 430-449 | ASCEGEGAS | RVVVN | N | Y | N | Y | 0 | − | PCP-2a-like | ||
| Vibrio vulnificus | CMCP6 | Gamma-proteobacteria | Wzy | NP_763453 | 163 | SPI* | 34 (63) | NP_763452 | 726 | 31-47, 449-467 | SAIPEEGKT | RIIID | N* | Y | N | N | 3 | ? | PCP-2a-like | ||
| E | |||||||||||||||||||||
| Bacteroides fragilis | NCTC9343 | Bacteroidetes | YP_211345 | 849 | SPI | 23 (65) | YP_211346 | 370 | 46-63, 348-367 | PCP-1 | |||||||||||
| Bacteroides thetaiotaomicron | VPI-5482 | Bacteroidetes | Wzy | NP_810636 | 789 | SPI* | 24 (61) | NP_810635 | 382 | 44-57, 341-361 | PCP-1 | ||||||||||
| Bacteroides thetaiotaomicron | VPI-5482 | Bacteroidetes | Wzy | NP_810269 | 789 | SPI* | 24 (61) | NP_810268 | 379 | 44-57, 340-359 | PCP-1 | ||||||||||
| Bacteroides thetaiotaomicron | VPI-5482 | Bacteroidetes | Wzy | NP_810567 | 789 | SPI* | 24 (61) | NP_810566 | 365 | 35-49, 331-350 | PCP-1 | ||||||||||
| Burkholderia pseudomallei | K96243 | Betaproteobacteria | Wzy | YP_110441 | 850 | SPI | 27 (61) | YP_110442 | 405 | 39-61, 352-372 | PCP-1 | ||||||||||
| Chromobacterium violaceum | ATCC 12472 | Betaproteobacteria | AAQ58449 | 630 | SPI | 20 (61) | AAQ58448 | 401 | 44-61, 364-381 | PCP-1 | |||||||||||
| Geobacter metallireducens | GS-15 | Delta-proteobacteria | Wzy | ABB32412 | 833 | SPI* | 27 (64) | ABB32411 | 409 | 43-59, 361-379 | PCP-1 | ||||||||||
| Hahella chejuensis | KCTC 2396 | Gamma-proteobacteria | Wzy | YP_433622 | 990 | SPI* | 21 (60) | YP_433623 | 317 | 35-52, 295-314 | PCP-1 | ||||||||||
| Pelodictyon luteolum | DSM 273 | Bacteroidetes | ABB24737 | 576 | SPI* | 23 (60) | ABB24736 | 343 | 43-59, 305-321 | PCP-1 | |||||||||||
| Prosthecochloris vibrioformis | DSM 265 | Bacteroidetes | ZP_00660724 | 576 | SPI* | 23 (61) | ZP_00660725, ZP_00660727 | 343, 370 | 49-65, 311-327; 50-66, 336-354 | PCP-1, PCP-1 | |||||||||||
| Shewanella oneidensis | MR-1 | Gamma-proteobacteria | Wzy | NP_718749 | 921 | SPI | 20 (65) | NP_718747 | 329 | 51-67, 310-326 | PCP-1 | ||||||||||
| Syntrophus aciditrophicus | SB | Delta-proteobacteria | Wzy | YP_462632 | 1048 | SPI | 26 (59) | YP_462633 | 342 | 41-59, 303-323 | PCP-1 | ||||||||||
| Vibrio cholerae | O139 | Gamma-proteobacteria | OtnA | CAA62140 | 911 | SPI | 24 (65) | OtnB | CAA62141 | 335 | 50-67, 306-325 | PCP-1 | |||||||||
| Vibrio vulnificus | CMCP6 | Gamma-proteobacteria | Wzy | NP_761480 | 699 | SPI | 16 (50) | NP_761492 | 476 | 20-37, 420-437 | PCP-1 | ||||||||||
| F | |||||||||||||||||||||
| Aeromonas hydrophila | PPD134/91 | Gamma-proteobacteia | ABC | KpsD | AAM22568 | 546 | SPI | 26 (59) | KpsE | AAM22567 | 397 | 51-67, 378-394 | PCP-3 | ||||||||
| Campylobacter jejuni | NCTC 11168 | Epsilon-proteobacteria | ABC | KpsD | CAB73868 | 552 | SPI | 25 (62) | KpsE | CAB73869 | 372 | 20-42, 350-368 | PCP-3 | ||||||||
| Escherichia coli | K1 | Gamma-proteobacteria | ABC | KpsD | AAA21682 | 558 | SPI | 26 (57) | KpsE | AAB51624 | 346 | 8-17, 324-344 | PCP-3 | ||||||||
| Escherichia coli | K5 | Gamma-proteobacteria | ABC | KpsD | CAA52656 | 558 | SPI | 26 (57) | KpsE | CAA52655 | 382 | 37-54, 361-381 | PCP-3 | ||||||||
| Escherichia coli | K54 | Gamma-proteobacteria | ABC | KpsD | AAC38077 | 581 | SPI | 23 (59) | KpsE | AAC38080 | 415 | 76-93, 402-415 | PCP-3 | ||||||||
| Nitrococcus mobilis | Nb-231 | Gamma-proteobacteria | ABC | ZP_01127551 | 575 | SPI | 24 (57) | ZP_01127548 | 361 | 19-32, 345-361 | PCP-3 | ||||||||||
| Ralstonia metallidurans | CH34 | Betaproteobacteria | ABC | ZP_00596582 | 606 | SPI | 23 (56) | ZP_00596581 | 368 | 23-40, 349-368 | PCP-3 | ||||||||||
| Yersinia intermedia | ATCC 29909 | Gamma-proteobacteria | ABC | ZP_00832080 | 558 | SPI | 24 (62) | ZP_00832081 | 390 | 43-59, 365-385 | PCP-3 | ||||||||||
SPI, predicted signal peptidase I cleavage site; SPII, predicted signal peptidase II cleavage site; SPI* and SPII*, the signal peptidase cleavage site predicted based on an alternative start site. In the database accession, a cytoplasmic localization was predicted using the annotated start site.
Y, yes; N, no; N*, only minor deviations present in the hydrophobic amino acids in the Walker B motif.
Y, yes; N, no.
FIG. 5.
Phylogenetic tree showing the relationship between OPX proteins. Sequences were identified using key word and BLAST searches (2). Only the PES motif of each OPX was used in the analysis. Alignments were performed using CLUSTALW (57). Phylogenetic analysis was carried out on 100 bootstrapped data sets using the parsimony program in the PHYLIP package (30). Trees were viewed using SplitsTree (41). The tree is based on data available in September 2008.
Groups C, D, and E include additional OPX proteins from Wzy-dependent systems in a variety of plant pathogenic genera (Xanthomonas and Xylella), plant symbionts (Sinorhizobium and Mesorhizobium), free-living organisms, and human pathogens (Pseudomonas, Vibrio, and Bacteroides). There is limited biochemical information concerning polysaccharide export in most of these bacteria. However, the OPX proteins are involved in the assembly of products, including traditional EPS such as xanthan gum (114) and succinoglycan (93), as well as polymers with specialized functions, such as the holdfast adhesive that allows the stalks of Caulobacter crescentus to stick to surfaces (104). Group C proteins are among the smallest identified OPX protein (Fig. 4b) and are predicted to be a mixture of lipidated and nonlipidated representatives. PslD is an outlier located between OPX groups C and D. It is involved in the export of an EPS in Pseudomonas aeruginosa that is required for biofilm formation (14, 96). Group D OPX proteins are heterogeneous with respect to size and lipidation. Group E OPX proteins are not lipidated and include some of the largest family members; the representative from Syntrophus acidotrophicus holds the current record, with a predicted size of 1,048 residues. One group E representative is OtnA, a protein associated with the production of the O139 capsule in Vibrio cholerae (11).
The OPX proteins involved in ABC transporter-dependent pathways are split into two groups (B and F) (Fig. 5). The E. coli group 2 CPS OMA homologs (KpsD) are located in group F, with the highly conserved homologs from serotypes K1 and K5 being closely linked, as expected. Also in this group is the KpsD homolog from serotype K54, a group 3 CPS, which shares biosynthetic machinery with group 2 CPS systems but lacks the characteristic transcriptional thermoregulation of capsule expression seen in K1 and K5 (17). KpsD from the CPS assembly system of Campylobacter jejuni is another member of group F; the kpsD gene sits adjacent to homologs of other genes found in the E. coli group 2 locus (46). The OPX homologs in group F are all larger than Wza and other group A proteins (546 to 606 residues), and none are lipoproteins, based on the predicted signal peptidase I cleavage site. Group B OPX proteins include CtrA from Neisseria meningitidis, BexD from Haemophilus influenzae, and VexA from Salmonella enterica serovar Typhi (Vi antigen). These have well-conserved predicted secondary structures (Fig. 4b). In terms of size, they are more similar to Wza than KpsD, and they are predicted to be lipoproteins (unlike group F). Despite these differences, group B OPX proteins can replace KpsD in export of E. coli group 2 CPS providing that they are coexpressed with their cognate ABC transporter and PCP protein (103). The substrate recognition and export processes for group B and F OPX proteins therefore must function in the same manner.
Many of the OPX members in group E contain an extra motif (DUF1017) that is identified by a conserved domain search (65). The DUF1017 motif is also identified in the same manner in one group F OPX protein, the KpsD homolog from C. jejuni (Fig. 4b). However, analysis of predicted secondary structures indicate that all of the OPX proteins in groups E and F may actually contain a domain corresponding to DUF1017; the putative DUF1017 domain in E. coli KpsDK5 is illustrated in Fig. 4b. The functional significance of the DUF1017 motif has yet to be established, and it is not confined to OPX proteins. For example, YmcB of the E. coli 22-min locus contains a DUF1017 motif, but the precise role of YmcB in CPS export is currently unknown. Sequence analysis and secondary structure predictions indicate that many of the long OPX proteins in group E (e.g., Bacteroides fragilis YP_211345 [Fig. 4b]) may contain additional domains sharing similarity with R2 and R3 of WzaK30 (Fig. 4b). This is perhaps not surprising given that R2 and R3 are structurally related and share a ubiquitin fold; they are speculated to have arisen through gene duplication (26). Using the portion of the PES motif corresponding to the R1 domain of WzaK30 to search for local similarities (40), the potential for multiple R1 domains is also apparent in some of the long OPX proteins (e.g., Vibrio vulnificans NP_761480; Fig. 4b). Although the structure and function of the longer OPX proteins have not been directly addressed, it is conceivable that additional R2/R3-like domains are required in these OPX proteins to span the periplasm. An alternative intriguing possibility is that some of the long OPX proteins may represent a fusion of two Wza-like monomers.
Some bacteria possess multiple genes for OPX proteins, and members of the Bacteroidales lead the way in this respect. As an example, Bacteroides fragilis can produce eight different phase-variable CPS forms (22, 54). The ability to produce multiple polysaccharides is essential for intestinal colonization (61). One (PS-A) has important roles in the commensal lifestyle of this organism and has implications for inflammatory disorders (71). We can identify only four OPX homologs in the B. fragilis genome. Three of these are highly conserved and map within group E (only one representative is included in Fig. 5). However, this organism also makes an EPS (15), and the corresponding OPX sits in group A. The genome of Bacteroides thetaiotaomicron encodes a remarkable eight OPX homologs distributed between groups A (five representatives) and E (three representatives). The trend of multiple OPX proteins is also evident in Burkholderia species. The genome of Burkholderia pseudomallei (strain K96243) encodes four OPX proteins distributed between groups A (three representatives, with two shown) and E, whereas Burkholderia cenocepacia has three representatives that are confined to group A.
STRUCTURES AND PHYLOGENY OF PCP PROTEINS
Like Wza, Wzc is also essential for export and assembly of CPS (27, 83, 91, 117). PCP proteins were initially identified in lipopolysaccharide (LPS) O-antigen biosynthesis systems in which PCP-1 representatives (often encoded by wzz genes) are involved in the determination of chain length (89) (see below). All PCP proteins have a characteristic membrane topology in which a large periplasmic loop is flanked by two transmembrane (or membrane-associated) regions. Studies with the E. coli KpsE (PCP-3) protein suggest that the C terminus does not contain a transmembrane region but instead is anchored to the membrane via the hydrophobic face of an amphiphilic α-helix (3, 87). All PCP proteins also contain a proline- and glycine-rich domain overlapping the second transmembrane domain. The PCP-2a proteins such as Wzc contain a unique additional domain, a C-terminal cytoplasmic region that contains a tyrosine autokinase. The functional significance of the kinase domain will be discussed in more detail below. Secondary structure predictions for CPS/EPS PCP proteins (Fig. 6) illustrate the conserved features, regardless of the subfamily, the mode of biosynthesis, or whether the polysaccharide product is CPS (e.g., the E. coli and N. meningitidis capsular antigens and the S. enterica serovar Typhi Vi antigen) or EPS (e.g., Xanthomonas campestris xanthan gum). In all cases, the periplasmic domain contains extensive predicted α-helical structure. These features are evident in the PCP-1 proteins (Wzz), although the PCP-1 proteins are typically smaller than the gram-negative CPS/EPS PCP proteins. Similar features are also seen in the PCP-2b proteins involved in CPS assembly by Wzy-dependent processes in gram-positive bacteria (e.g., CapA1 from Staphylococcus aureus) (Fig. 6). In the case of Wzz, the validity of the secondary structure predictions is supported with X-ray structure data (112).
FIG. 6.
The predicted secondary structures of the periplasmic regions of PCP proteins demonstrate the high degree of α-helical content found in the different PCP families. The structures were calculated using the approach used for the OPX proteins in Fig. 4. The transmembrane regions were assigned based on the agreement of three transmembrane prediction algorithms (TMHMM [105], TMPred [39], and TMPro [33]). The secondary structures from solved crystal structures of the periplasmic domains of the Wzz homologs, WzzST from S. enterica serovar Typhimurium (Protein Data Bank accession no. 3B8P) and WzzFepE from E. coli O157:H7 (Protein Data Bank accession no. 3B8M), are bounded by black boxes.
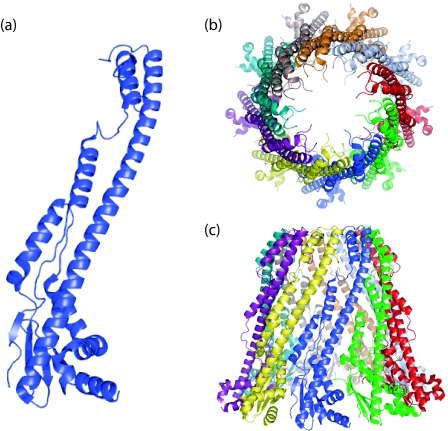
PCP-1 protomers consist of a membrane-proximal mixed α/β base domain which extends into a long (∼100-Å) α-helical hairpin (Fig. 7a). Secondary structure predictions for the periplasmic region of Wzc indicate that the majority of the β structure is located toward the first transmembrane region, with predominantly extended α-helical content toward the second. Like Wzc, the PCP-1 periplasmic domains form oligomers, and these adopt a cone-like structure in the crystal units (Fig. 7b and c). The precise physiological oligomerization states of the PCP-1 homologs are uncertain. In the three homologs for which X-ray structures are available, highly conserved protomers pack with subtle differences in the interfaces and generate different oligomerization states (five, eight, and nine protomers). Furthermore, several studies of periplasmic domains and full-length PCP-1 proteins involving various biochemical and biophysical methods have yielded various results with respect to oligomerization (67, 110, 112). In an attempt to resolve this issue, three full-length PCP-1 homologs were reconstituted in proteoliposomes to examine the oligomerization states under conditions that more accurately reflect the natural environment for these proteins. EM data indicated that all three form hexamers, and the overall conical shape and size of the solvent-exposed periplasmic domain in cryo-EM images are consistent with those predicted by the α-helical hairpin in the X-ray structure of the protomer (58). It therefore seems likely that the varying oligomerization states in the X-ray structures reflect altered crystal packing and that PCP-1 proteins in fact have a conserved quaternary (hexameric) structure. In contrast, an EM structure (∼14-Å resolution) of Wzc reveals a tetramer with extensive contact between the periplasmic domains of the monomers (Fig. 8a) (19). The overall shape of the PCP-1 homolog (58, 112) is comparable with particles of the full-length Wzc protein visualized in single-particle EM analysis (19), although the PCP-1 oligomer structures are substantially more extended than the Wzc periplasmic domain. In Wzc, the secondary structure predictions suggest more β structure in the region corresponding to the PCP-1 base domain, and this may result in a more compact overall structure, as seen in EM data (Fig. 8a).
FIG. 7.
Structures of PCP-1 proteins. The figure shows data obtained for FepE (Protein Data Bank accession no. 3B8M) from E. coli O157:H7 (112). The protomer (a) has an N-terminal α/β base domain from which a long α-helix extends. The protomers are thought to assemble into a nonamer structure that extends ∼100 Å into the periplasm (b). The oligomer is open at the top (c), generating a solvent-filled cavity with the lipid of the inner membrane at its base. The transmembrane regions of the protein were deleted from the construct used in crystallization.
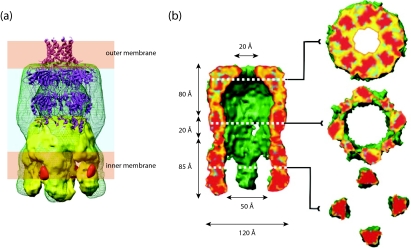
FIG. 8.
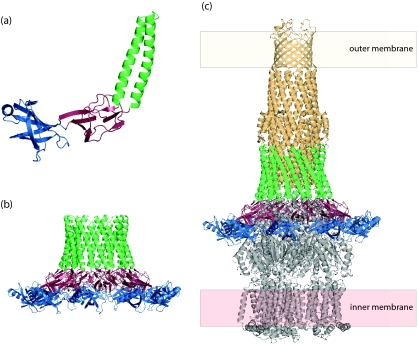
3D reconstruction of the Wza-Wzc complex from cryo-EM. (a) Proposed organization of the complex (green wire frame) in the cell envelope (20). The Wzc EM structure (yellow) and the ribbon structure of Wza are included. The orange densities are regions occupied by cytoplasmic N-terminal hexahistidine tags (labeled with nanogold). Note that the α-helical outer membrane barrel in Wza is destabilized under the pH conditions of the EM experiments (Ford et al., submitted). (b) Fifty percent of the foremost volume is removed to reveal the central cavity and the (upper) exit pore. The slices provide detail through the structure. It is expected that lipid from the cytoplasm would fill the center of the Wzc tetramer, precluding a contiguous connection from the cytoplasm to the exterior of the cell. (Reprinted from reference 20 with permission of the publisher.)
The PCP-3 KpsE protein has a tendency to form dimers and higher-order oligomers (3), but structural information comparable to that for PCP-2a and PCP-1 oligomers is not yet available. Given the existing data, some similarities in the structures of PCP-1 and other family members might be anticipated. However, subtle differences are certainly expected, depending on the interacting partners determined by the biosynthesis pathway, i.e., Wzy-dependent versus ABC transporter-dependent pathway and CPS/EPS versus LPS O antigen.
To examine the diversity of PCP proteins from CPS/EPS systems, we investigated phylogenetic relationships among PCP homologs corresponding to the OPX proteins described above. A selection of known PCP-1 proteins was included for reference. The phylogenetic tree (Fig. 9) was built using the periplasmic domains as a basis for comparison (i.e., the kinase domain in PCP-2a proteins was excluded). In general, the major clusters of CPS/EPS PCP proteins follow the groupings established for the corresponding OPX proteins, perhaps reflecting their coevolution. The PCP-2a homologs associated with OPX group A cluster together, as do most of the PCP proteins linked to OPX groups C and D. Interestingly, the group C-associated PCP proteins include several representatives in which the PCP-2a protein consists of two separate polypeptides (Table 1). Examples include the HfsAB protein involved in holdfast biosynthesis in C. crescentus. One polypeptide (HfsA) contains the PCP periplasmic domain flanked by its transmembrane helices, and the other (HfsB) comprises the C-terminal kinase domain. This two-polypeptide organization is reminiscent of the situation with PCP-2b proteins from gram-positive CPS systems (35). However, the kinase domains from the PCP-2b (81) and PCP-2a (59) proteins have distinct structural and mechanistic features. The group C-associated PCP proteins are unmistakably derivatives of PCP-2a. There seems to be no obligatory requirement for the gram-negative PCP-2a proteins to take the form of a single polypeptide, as a Wzc-derivative designed as two polypeptides could operate in E. coli group 1 CPS assembly (117). In another structural variation, several group C OPX-associated PCP proteins from free-living bacteria appear to have an additional predicted transmembrane region located at the extreme C terminus (Table 1).
FIG. 9.
Phylogenetic tree showing the relationship between PCP proteins associated with the OPX proteins in Fig. 5. Sequences were cropped so that only the sequence aligning with COG3206 was used in the analysis. This includes the periplasmic domain of each PCP and the flanking transmembrane domains. Alignments were performed and trees prepared as for Fig. 5. The background colors highlighting each group of proteins follow the same pattern used for the OPX proteins. PCP proteins highlighted by a red box are those PCP-2a homologs where the protein exists as two separate polypeptides (one being the integral membrane protein and the other comprising the cytosolic kinase). The PCP proteins highlighted by a blue box are close relatives of PCP-2a proteins, but the kinase domain is absent. The tree is based on data available in September 2008.
In this analysis, the PCP-3 proteins associated with OPX groups B and F that are involved in ABC transporter-dependent CPS biosynthesis form a more unified group. This is not surprising given the high conservation of predicted secondary structures (Fig. 6). Interestingly, these proteins arise from a common branch that also gives rise to the PCP proteins associated with group E OPX representatives and the PCP-1 proteins from O-antigen biosynthesis (Fig. 9).
The group E OPX proteins and their cognate PCP proteins participate in a Wzy-dependent biosynthesis CPS/EPS system, and, like the PCP-1 proteins, they lack a kinase domain. In the case of the V. cholerae O139 polysaccharide, the same repeat-unit structure exists as a short oligosaccharide (a single repeat unit) linked to lipid A core (21, 50) and as high-molecular-weight CPS (49), a situation very similar to that for E. coli group 1 CPS (63). OtnB, the V. cholerae O139 PCP-1 protein, is required for the formation of CPS but not for the LPS-linked form of O139 antigen (11). Interestingly, when OtnB is expressed in an S. enterica serovar Typhimurium Δwzz strain, it does result in an altered distribution of short O-antigen chains, although no large modal chain lengths are produced (78). In many respects, the role of OtnB in biosynthesis of the O139 CPS is remarkably similar to the role of Wzc in E. coli (27). The appearance of CPS/EPS biosynthesis proteins in this broad PCP group is perhaps not surprising, because PCP-1 proteins are not necessarily confined to lipid A-linked polymers. The PCP-1 group includes WzzE, a protein implicated in the biosynthesis of enterobacterial common antigen (ECA). ECA is found in many genera of the Enterobacteriaceae (53). WzzE participates in the biosynthesis of both ECAPG (4), a phosphoglyceride-linked form, and ECACYC (45), an unusual cyclic molecule containing four to six ECA repeat units. In the absence of any detailed characterization of the PCP proteins associated with group E OPX proteins, we have retained the PCP-1 designation.
Three other examples in which a PCP protein lacking a kinase domain participates in a Wzy-dependent EPS system were identified. These are GumC from Xanthomonas campestris, the closely related protein from Xylella fastidiosa, and the PCP from Myxococcus xanthus. They are all associated with group C OPX proteins. From the perspective of secondary structure predictions, these PCP proteins are comparable to the membrane-embedded parts of the two-polypeptide PCP-2a proteins. They lack the accompanying kinase polypeptide, and no obvious kinase partner could be identified elsewhere on the chromosome.
MOLECULAR SCAFFOLDS SPANNING THE CELL ENVELOPE FACILITATE CPS EXPORT
The interactions between Wza and Wzc can be reconstituted using purified proteins, and the resulting 3D heterocomplex structure (∼12-Å resolution) has been obtained by EM methods (20) (Fig. 8). The only notable deviation from the structures of the individual oligomers is the staining-artifact-driven absence of the R4 domain of Wza in the EM structure (Ford et al., submitted). The dimensions of the complex (170 Å along the axis perpendicular to the plane of the membrane) are consistent with the prediction of a contiguous scaffold spanning the cell envelope. The identification of these structures may explain earlier in vivo studies that identified specific sites of membrane apposition (often referred to as Bayer junctions or Bayer bridges) where de novo CPS export occurs in conditional E. coli group 1 CPS biosynthesis mutants (7, 8). Current estimates of the thickness of the periplasm (∼200 Å) are derived from electron micrographs of frozen thin sections (70). This value is considerably larger than the ∼125 to 145 Å predicted from the cryo-EM structure (20). Interestingly, the TolC-linked efflux pumps (29) and the type III protein secretion needle complex (66) also seem to create a local narrowing of the periplasm, so this phenomenon may be widespread.
The periplasmic location of the polymerization step for Wzy-dependent CPS/EPS creates additional complexity and requires a mechanism to open up the periplasmic domain of Wza and/or Wzc to allow polymer entry into the channel. In the complex, there is a broadening of the interface between the Wza and Wzc oligomers, compared to each isolated component. This appears to reflect both an opening (or spreading) of the periplasmic domain of the Wzc oligomer and a widening of the base of the Wza octamer. This is evident when the boundaries of the individual oligomeric components are considered within the space occupied by the heterocomplex (Fig. 8b). These conformational changes generate putative portals in the side of the complex that could connect the periplasm (the site of CPS polymerization) to the cavity of the structure and consequently to the exterior of the cell (20). Computational estimates of the relative stabilities of the rings in the Wza oligomer suggest that the most likely region to “open” is R2 (Ford et al., submitted). The other rings have significantly higher ΔG values for dissociation. The necessary conformational changes could be mediated by the intermeshing of α-helices in Wza and Wzc protomers (see below). Once the polymer enters the central cavity, growth of the chain by addition of repeat units to the reducing terminus would be sufficient to drive extrusion through the channel.
The active conformation of the Wza translocon therefore appears to need the association of the Wza and Wzc oligomers, and this interdependence is evident in comparable acapsular phenotypes for wza and wzc mutants (28, 117). The mutant bacteria still produce short oligosaccharides of the CPS, but no large molecules are formed and capsule export is absent (27, 28, 117). Logically, one might expect that an export defect from null wza and wzc mutations would result in the accumulation of periplasmic polymer export intermediates. Their absence may reflect the loss of essential protein-protein interactions in a biosynthesis complex that creates a feedback system to inactivate early steps in the assembly pathway when export is defective. To date, only one mutation that uncouples polymerization and export has been isolated. This involves a Wza derivative engineered to eliminate its N-terminal acylation (79). The mutant makes aberrant Wza oligomers, and while no CPS is found on the cell surface, polysaccharide does accumulate in the periplasm. This phenotype is interpreted as the Wza derivative being able to reconstitute the essential interface between Wza and Wzc, thereby preventing the feedback process that turns down polymer synthesis in the null mutants. In this scenario, the improper outer membrane association of the mutant Wza oligomers renders them unable to complete the export process. However, as will be described below, this interpretation is complicated by the observation that Wzc may play additional roles in CPS assembly and may not be confined to export.
Do similar structures participate in the export of other CPS/EPS? Information for most systems is scant, but the reliance on KpsE for KpsD location in the outer membrane certainly argues for their interaction in E. coli isolates with group 2 CPS (72). The inferred structural similarities discussed above also favor an interpretation of shared functions. As with the group 1 CPS system, nascent group 2 CPS emerges on the cell surface at discrete sites (52). However, different studies have reported those sites to be either distributed randomly on the cell surface (52, 116) or located at the poles together with components in the biosynthesis system (72). The basis for this disparity is currently unknown. Interestingly, the polar localization of KpsDE is not an inherent property of these proteins and is lost in favor of random distribution in the absence of other components of the CPS assembly system (72). The observed cross-linking of KpsD within a complex containing cytosolic CPS biosynthesis proteins also suggests the existence of a trans-envelope assembly machine, analogous to Wza-Wzc in E. coli making group 1 CPS (72). One important difference in the systems is that kpsDE mutants still make polymer (12, 85, 102) and therefore lack the feedback regulation apparent in wza and wzc mutants in group 1 CPS systems.
The potential conservation in PCP protomer structure raises interesting questions about the role of PCP-1 proteins in LPS O-antigen biosynthesis. From the crystal data, the α-helical hairpins may create a structure large enough to span the periplasm to interact with the outer membrane (112). However, to date, there is no evidence for an interaction partner analogous to Wza. Furthermore, PCP-1 proteins are not essential for LPS export, so the rationale for a periplasmic scaffold in the O-antigen assembly system is currently not apparent.
STRUCTURES OF DRUG EFFLUX PUMPS; IMPLICATIONS FOR THE STRUCTURE AND FUNCTION OF PCP-OPX COMPLEXES
From the available data for E. coli group I CPS, molecular recognition between Wza and Wzc homologs influences the outcome of genetic complementation experiments using even closely related heterologous proteins (91). The precise interface between Wza and Wzc is currently unknown, and, as indicated above, there appears to be significant conformational rearrangement in the interface when the two oligomers interact. How might this occur? Some clues may be provided by considering parallel information from envelope-spanning bacterial drug efflux systems.
The periplasmic extended α-helical hairpin motif seen in PCP-1 proteins is also evident in the periplasmic adaptor proteins (membrane fusion proteins) from tripartite drug efflux systems. In these systems, a periplasmic adaptor protein couples a drug pump to the outer membrane efflux channel (TolC) (Fig. 10). The outer-membrane-spanning domain of TolC is a trimeric 12-stranded β-barrel with a long (100-Å) α-helical barrel projecting across the periplasmic space (reviewed in reference 51). The entrance of the α-helical domain is closed in purified TolC protein, but in the presence of the other components, it is proposed to untwist (in an iris-like mechanism) to open the channel. This action results from engagement with a substrate-binding, energy-charged efflux pump as well as interactions with the periplasmic adaptor protein. Partial structures of two adaptors, MexA (1, 38) and AcrA (73), have been solved. These conserved proteins are normally N-terminally acylated (but not in the crystallized form). Acylation provides a means of association with the inner membrane, although membrane linkage is not essential for function. MexA and AcrA are both composed of three domains: a membrane-proximal α/β-barrel, a β-sandwich “lipoyl” domain, and an extended α-helical domain (Fig. 10a). The C terminus of the native protein is missing in the crystallized constructs and is involved in interactions with the membrane component. The α-helical domain in AcrA is slightly longer than MexA (58 versus 47 Å). The superficial similarity in the architectures of the adaptor proteins and PCP-1 representatives is apparent. Equally striking is the propensity for MexA and AcrA protomers to pack side-by-side in crystal units, despite the fact that they behave as monomers in solution. In the case of MexA, 13 molecules are present in the asymmetric unit. These take the form of two arcs (six and seven protomers, respectively) that are arranged head-to-head through association with the tops of the α-helical domains (38). In the case of AcrA, the crystal has four monomers arranged as a dimer of dimers (73). This propensity of the α-helical domains to interact may help explain the uncertainty surrounding the oligomeric status of the PCP-1 proteins. Given the overall similarities in the protomer, similar intermonomer contacts, and their interaction with a common partner (TolC), it seems unlikely that the oligomerization status differs for these adaptor protein homologs. In the accepted model, the physiological unit has nine protomers to accommodate the threefold symmetry in this system (62) (Fig. 10b). This would be sufficient for the adaptor to wrap around the base of TolC. There are obvious superficial similarities shared by the oligomers of adaptor proteins and the PCP-1 and PCP-2a proteins (Fig. 7 and 8).
FIG. 10.
The periplasmic adaptor protein MexA in the tripartite drug efflux pump has an architecture similar to that of PCP proteins. (a) Protomer of MexA (Protein Data Bank accession no. 1T5E). It has three domains, an α/β barrel (blue), a “lipoyl” domain (red), and an extended α-helical domain (green). (b) The nonameric complex of MexA is compatible with the threefold symmetry of the drug efflux pump. (c) A model of the tripartite TolC-MexA-AcrB pump. The α-helical domain of a nonameric MexA complex could accommodate the periplasmic end of the TolC efflux channel (pale orange; Protein Data Bank accession no. 1EK9). The AcrB pump (gray; Protein Data Bank accession no. 1IWG) anchors the complex in the inner membrane. The structure of the tripartite complex was generated based on the model presented by Higgins et al. (38).
The RND (resistance-nodulation-cell division) transporter, AcrB has been used to model and unravel the complex interactions involved in the tripartite drug efflux pumps (25, 77, 98, 99, 113) (Fig. 10c). AcrA is crucial to the recruitment of the TolC efflux channel, and the formation of the tripartite pump is proposed to occur via an induced-fit model (6). Existing biochemical data support an intermeshing of β hairpins in the periplasmic crown of AcrB and α helices in TolC without extensive side chain interactions. It is proposed that this creates a partially open state where two helices in TolC partially relax. In this state, a side groove is exposed in TolC, providing a binding site for AcrA; this is supported by mutagenesis and docking experiments. The interaction of AcrA then allows full channel opening (6), and the conformational flexibility of AcrA may be important in this respect (73). In summary, the opening of the pump is dependent on a dynamic process requiring contributions of all three partners, in addition (in at least some situations) to the presence of the efflux substrate (111).
The possible structural similarities between ABC transporter-dependent CPS/EPS assembly systems, tripartite drug efflux pumps, and (TolC-dependent) type 1 protein secretion systems have been recognized before (103, 108, 115). In ABC transporter-dependent EPS/CPS systems, completed polymer is presumed to be exported via the ABC transporter directly into the putative channel formed by the OPX protein. The proposed involvement of components of the polymer biosynthesis machinery and polymer biosynthesis in determining the proper location and conformation of KpsD is also a feature reminiscent of the tripartite drug efflux pumps (see above). While the Wzy-dependent pathway is reliant on polymer substrate that must access the terminal export pathway (the Wza-Wzc heterocomplex), there are examples of efflux pumps (e.g., AcrB) that pump molecules from the periplasm (77, 98, 100). A true cytoplasm-to-cell exterior route is certainly not essential. The merger of helices at the interface of TolC and the adaptor protein provides a predictive model for Wza-Wzc interactions that may underpin the conformational changes (broadening) seen in the heterocomplex.
ARE PCP PROTEINS CHAIN LENGTH REGULATORS IN EPS/CPS ASSEMBLY?
PCP-1 proteins such as Wzz unequivocally influence the chain length of O-antigen polysaccharides synthesized by Wzy-dependent pathways (reviewed in reference 89). The wild-type polymers possess a characteristic “modal” chain length, where the majority of the chains fall within a narrow size range. In the absence of PCP-1 activity, modality is lost and predominantly short oligosaccharides are produced. However, their influence is not confined to lipid A-core-linked polymers, because one representative, WzzE, regulates modality of the phosphoglyceride-linked ECAPG (4). The phenotype resulting from PCP-1 defects suggests a loss of processivity in the polymerization process, but to date there is no evidence supporting a direct physical interaction between Wzy and PCP proteins. Two models have been proposed for the establishment of modality, and these invoke the PCP-1 protein being either a molecular timer (5) or a molecular chaperone (76) acting to regulate the relationship between chain extension and chain termination (i.e., ligation of O antigen to lipid A-core) activities. Shared structural features in PCP proteins were influential in the proposal that Wzc is also involved in chain regulation of EPS/CPS, but only limited experimental evidence exists to support the hypothesis that Wzc homologs (and Wzc mutants) influence polymer chain length in E. coli K30 (91). The conserved proline- or glycine-rich domain overlapping the second transmembrane region has been targeted for mutagenesis in Sinorhizobium meliloti succinoglycan (9, 34) and Acinetobacter venetianus emulsan (23, 24). These mutations do affect polymerization to a certain extent, but none are as dramatic as Wzc null mutations. Unfortunately, without a structural guide for the mutations, it is difficult to discriminate between mutations affecting catalysis and those perturbing protein structure. As an example, many of the PCP-1 mutations that cause the most dramatic chain length phenotypes do affect structural elements of the proteins (67, 88, 112). In the case of Wza-Wzc interactions, the picture is further complicated because of confounding similarities in the phenotypes of wza and wzc mutants.
The idea that PCP proteins all modulate polymerization is hard to rationalize when the role of PCP-3 proteins in ABC transporter-dependent CPS systems is taken into consideration. In this case, the biosynthesis steps are confined to the cytosolic face of the inner membrane and there is no Wzy-like polymerase step. Polymerization is catalyzed solely by the cytosolic glycosyltransferases, and it is not clear how these could interact in any substantial way with a PCP-3 protein. This could only occur indirectly in the context of the larger, multiprotein assembly complex that has been proposed for E. coli group 2 CPS (72, 94, 108). kpsED mutants of E. coli do still make high-molecular-weight polymers that are exported to the periplasm (12, 85, 102). Any effect of a KpsE defect on polymerization is therefore likely to be subtle, but further analysis of its potential effects on modality is certainly warranted. In fact, further studies with PCP-3 proteins will be essential to establish whether all PCP proteins live up to the PCP designation.
A simple alternative explanation is that PCP proteins provide periplasmic scaffolds for different protein-protein interactions that vary according to the system. In O-antigen systems, genetic data predict interactions and/or recognition between the initiating sugar of the und-PP-linked repeat unit and the cognate Wzz, Wzx, and Wzy proteins (68, 69). A speculative model for PCP-1 function suggests that these proteins “organize” Wzy into an active complex (112), although there are currently no direct experimental data to support this. Mutations directed at the solvent-exposed parts of the oligomer had little or no effect, whereas mutations affecting the top of the periplasmic cone structure did influence chain length, suggesting that this may be a site of interaction (112). Candidates for interaction logically include Wzy, but they also include the initiating hexose-1-P transferases. It has been established that mutations in the initiating hexose-1-P transferase (WbaP) alter chain length in S. enterica serovar Typhimurium (97), so the influence of the PCP-1 protein could be mediated at the level of the flow of und-PP-linked intermediates into the system, rather than by directly affecting the putative polymer protein, Wzy. Since the functions of initiating enzymes and Wzy proteins are highly conserved in O-antigen biosynthesis and Wzy-dependent CPS/EPS biosynthesis, there are obvious parallels. Wzy proteins and initiating hexose-1-P transferases both contain multiple membrane-spanning regions. Notably, the PES domains of the OPX proteins, the periplasmic loops of Wzy, and the initiating transferases all have α-helical content, which may provide the common element in binding partners. If predictions of a scaffold role are indeed correct, the structures of isolated PCP proteins will provide a limited view. Experience with the tripartite drug efflux systems indicates that structures of complexes will be essential to resolve the functions and interactions. If PCP proteins are scaffolds for diverse proteins, their influence on polymerization may be indirect, and in a sense, the “polysaccharide copolymerase” designation is potentially something of a misnomer.
THE ENIGMA OF TYROSINE PHOSPHORYLATION OF PCP-2a PROTEINS
PCP-2a proteins, such as Wzc, and PCP-2b proteins have an additional feature not seen in either PCP-1 or PCP-3 proteins. These proteins possess a cytoplasmic C-terminal tyrosine kinase belonging to the BY kinase family (reviewed in reference 35). This domain is autophosphorylated at several tyrosine residues, and its modification is essential for high-molecular-weight CPS assembly (83, 117). In vitro phosphorylation has been proposed to occur in a two-step process (36). The first step involves phosphorylation of a conserved “regulatory” tyrosine located downstream of the Walker box motifs by an intramolecular mechanism. This is required to fully activate the kinase for the second step, involving intermolecular phosphorylation of a cluster of tyrosines at the extreme C terminus of the protein (117). The observation that Wzc exists as a tetramer (19) provides a structural context for intermolecular phosphorylation but does not explain the details, since the contact within the isolated tetramer is confined to the periplasmic domains whereas the cytosolic kinase domains are well separated (19) (Fig. 8a). The structure of the kinase domain of Etk, a closely related homolog of Wzc, was recently reported (59). The closest sequence homolog (outside PCP proteins) is MinD (60), an ATPase involved in bacterial cell division. The structure of Etk suggests that the unusual kinase mechanism of Wzc may be related to NTPases. Interestingly, purified Wzc protein also has ATPase activity (107). The site of the initial regulatory phosphorylation in Etk is Tyr574 (corresponding to Tyr569 in Wzc). The side chain of Tyr574 sterically hinders access to the catalytic site of the kinase, and Tyr574∼P would clash with the bound nucleotide. In the proposed working model, the initial intramolecular phosphorylation at Tyr574 creates a conformational change in which the charged Tyr574∼P moves toward adjacent positively charged residues in the structure. Tyr574 is not essential for function in the subsequent intermolecular phosphorylation of the C-terminal tyrosines. Whether this process is conserved in its entirety in other closely related PCP proteins is unclear, because a Tyr569Y → F mutation in Wzc from the K30 system does not abrogate either phosphorylation or function in CPS synthesis. Paradoxically, CPS biosynthesis also requires dephosphorylation of Wzc by its cognate phosphatase (117), Wzb. Current working models propose that Wzc must cycle between its phosphorylated and nonphosphorylated forms to sustain CPS biosynthesis (80, 83). The terminal domain of Wzc from E. coli K30 contains seven Tyr residues that can be phosphorylated, and purified Wzc has a mixture of phosphorylation states (20, 59). Mutagenesis studies suggest that a phosphorylation “load” rather than a specific residue (or residues) is important in overall function and that the extent of phosphorylation influences polymer size (83). In the proposed model (59), the charged state of the C-terminal domain modulates its propensity to interact with the positively charged arginine and lysine-containing “RK domain.” Such interactions, regardless of their origin are implied by the observed intermolecular transphosphorylation reactions.
A recent bioinformatics study described the properties of the BY kinase family and identified several PCP-2a proteins (43). However, the search parameters dictate that the PCP-2a proteins must all contain a canonical BY kinase domain. This does not address a basic question: how conserved is the overall phosphorylation process in PCP-2a proteins? Put another way, are all PCP-2a proteins BY kinases? While no functional data currently exist for many of the homologs, phylogenetic and sequence analyses provide fascinating insight into variations in PCP-2a proteins. Many have Walker box sequences that deviate from the canonical version (Table 1), and their influence on kinase activity is unknown. Some representatives (e.g., B. cenocepacia PCP proteins associated with group A OPX proteins) lack an identifiable C-terminal tyrosine residue or residues, despite being part of the group including the Wzc prototype. Many others lack an obvious candidate for a residue corresponding to Y574 in Etk. It is conceivable that these resemble site-directed Etk mutants that show constitutive activation, i.e., where the first intramolecular phosphorylation is unnecessary. Finally, there are PCP-2a proteins such as GumC in X. campestris and PCP-1 proteins such as OtnB in V. cholerae in which there is no kinase domain at all, yet these proteins clearly participate in the biosynthesis of CPS/EPS. One intriguing question is why PCP-2a proteins have a kinase domain when PCP-1 proteins do not, yet they both interact with essentially identical Wzy-dependent biosynthesis systems. One possibility is that phosphorylation is required to overcome some unknown barrier in the biosynthesis of very high-molecular-weight polymers, but such speculation is not well founded. For example, in X. campestris, xanthan gum is made in large amounts and with a size range of 106 to 107 Da, yet the process involves the “kinase-less” PCP-2a representative, GumC.
There are several other possible roles for phosphorylation of PCP-2a proteins in the biosynthesis of CPS/EPS. One attractive possibility is that the operation of the Wza-Wzc complex is dependent on the phosphorylation state of Wzc. Wza does form a complex with kinase-defective Wzc (20), but potential effects of phosphorylation on the conformation of the Wza-Wzc complex have not been investigated yet. For example, phosphorylation could regulate opening of the OPX channel. However, the conservation of phosphorylation-dephosphorylation in gram-positive assembly systems (where clearly no OPX is involved), as well as the variant gram-negative systems where no phosphorylation is likely, tends to argue against a role for phosphorylation confined to regulating the export pathway.
An alternative possibility is that the kinase activity of PCP-2a (and PCP-2b) proteins is involved primarily in regulating the early steps of biosynthesis that are conserved in all of the relevant assembly systems. The activity of the kinase is certainly dependent on the transmembrane part of the protein (and the intervening periplasmic domain) (106, 117). The transmembrane component has recently been referred to as the transactivator domain (43). However, this activity does not rule out the transmembrane and periplasmic regions (the conserved feature of all PCP proteins) having an additional role(s) that is independent of the active kinase. In simple terms, there are currently no data to conclude that the two domains necessarily play identical roles in EPS/CPS biosynthesis. Multiple functions would remain undetected due to the CPS/EPS-null phenotypes arising from most PCP deletions. It is now known that PCP-2a and PCP-2b kinases can phosphorylate heterologous substrates, including enzymes involved in the biosynthesis of sugar-nucleotide precursors used in polysaccharide biosynthesis. One example in E. coli K-12 colanic acid biosynthesis is Ugd (uridine-5-diphospho-glucose dehydrogenase), which is required for the common precursor UDP-glucuronic acid (37, 55). The corresponding BY kinases from gram-positive CPS biosynthesis systems have additional targets that may include the enzymes that transfer the first hexose-1-phosphate to und-P to initiate biosynthesis of the und-PP-linked intermediate (74). It is unclear if this is sufficient to account for the phenotype of kinase defects in CPS/EPS biosynthesis in gram-negative bacteria. For example, these modifications might slow the assembly of und-PP-linked repeat units and alter the flow through the polymerization system, giving rise to the massive reduction in chain length seen in wzc mutants. Evidence from mutagenic dissection of the C-terminal tyrosine cluster certainly suggests that Wzc derivatives with different potential phosphorylation loads do alter the size distribution of the polymeric product in E. coli group 1 K30 CPS (83). A similar phenomenon has been reported for the phosphorylation state/potential in E. coli K-12 colanic acid production (80). In this scenario, the role of the kinase domain is analogous to two-component regulatory systems, in which changes in the periplasm are communicated to a response regulator in the cytoplasm via a histidine kinase containing two transmembrane regions (109). In the CPS/EPS situation, the phosphorylated biosynthesis proteins would be analogous to the response regulator. However, there is a growing body of evidence implicating the BY kinases in diverse physiological responses (35, 56), and these may not necessarily all involve the kinase activity per se. Thus, one has to entertain the possibility that the role of autokinase activity in CPS assembly is indirect and that it acts through an (as-yet-unidentified) partner that cooperates with the CPS-specific machinery in the overall process. Of note, PCP-3 proteins lack the kinase domain, and in group 2 CPS assembly systems, there is no feedback control between the export and biosynthesis steps; E. coli kpsD and kpsE mutants still synthesize polysaccharide that accumulates in the periplasm (12, 85, 102). There is clearly much still to learn concerning the role of the autokinase.
CONCLUDING REMARKS
Over the last few years, considerable progress has been made in understanding the structural biology of OPX and PCP proteins. Despite this, there are still significant open questions concerning the interactions between PCP proteins and their OPX partners. These will be solved only by high-resolution structures of complexes, and the tools for this are in place. Since PCP-3 and PCP-2a proteins interface with different CPS/EPS biosynthesis processes, it seems unlikely that a single system will allow conclusions about all. The analyses presented here suggest that there may be extensive variation in the details of OPX-PCP interactions and in the contribution(s) of the PCP proteins to the overall assembly process. Phylogenetic analyses identify those systems that merit more investigation.
It is important to recognize that not all EPS biosynthesis systems involve PCP and OPX proteins. Exceptions include poly-N-acetylglucosamine (42), cellulose (95), and alginate (92). The proteins involved in their biosynthesis have been identified, but the nature of the acceptor for chain formation, mechanism(s) of polymerization, and details of export are largely unknown. It is conceivable that they share a common mode of assembly and that this will differ from the systems described here.
ADDENDUM IN PROOF
While this paper was in proof, an interesting review focusing on PCP proteins was published (R. Morona, L. Purins, A. Tocilj, A. Matte, and M. Cygler, Trends Biochem. 34:78-84, 2008). Morona et al. reported findings consistent with our own concerning the predicted secondary structures of PCP proteins in the context of the PCP-1 crystal structure.
Acknowledgments
Work in our laboratories is supported by the Canadian Institutes of Health Research (C.W.), the Wellcome Trust (J.H.N. and C.W.), and the Biotechnology and Biological Sciences Research Council (J.H.N.). C.W. holds a Canada Research Chair.
REFERENCES
- 1.Akama, H., T. Matsuura, S. Kashiwagi, H. Yoneyama, S. Narita, T. Tsukihara, A. Nakagawa, and T. Nakae. 2004. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 27925939-25942. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrecubieta, C., T. C. Hammarton, B. Barrett, S. Chareonsudjai, N. Hodson, D. Rainey, and I. S. Roberts. 2001. The transport of group 2 capsular polysaccharides across the periplasmic space in Escherichia coli. Roles for the KpsE and KpsD proteins. J. Biol. Chem. 2764245-4250. [DOI] [PubMed] [Google Scholar]
- 4.Barr, K., J. Klena, and P. D. Rick. 1999. The modality of enterobacterial common antigen polysaccharide chain lengths is regulated by o349 of the wec gene cluster of Escherichia coli K-12. J. Bacteriol. 1816564-6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastin, D. A., G. Stevenson, P. K. Brown, A. Haase, and P. R. Reeves. 1993. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol. Microbiol. 7725-734. [DOI] [PubMed] [Google Scholar]
- 6.Bavro, V. N., Z. Pietras, N. Furnham, L. Perez-Cano, J. Fernandez-Recio, X. Y. Pei, R. Misra, and B. Luisi. 2008. Assembly and channel opening in a bacterial drug efflux machine. Mol. Cell 30114-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayer, M. E. 1991. Zones of membrane adhesion in the cryofixed envelope of Escherichia coli. J. Struct. Biol. 107268-280. [DOI] [PubMed] [Google Scholar]
- 8.Bayer, M. E., and H. Thurow. 1977. Polysaccharide capsule of Escherichia coli: microscope study of its size, structure, and sites of synthesis. J. Bacteriol. 130911-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker, A., and A. Pühler. 1998. Specific amino acid substitutions in the proline-rich motif of the Rhizobium meliloti ExoP protein result in enhanced production of low-molecular-weight succinoglycan at the expense of high-molecular-weight succinoglycan. J. Bacteriol. 180395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beis, K., R. F. Collins, R. C. Ford, A. B. Kamis, C. Whitfield, and J. H. Naismith. 2004. Three-dimensional structure of Wza, the protein required for translocation of group 1 capsular polysaccharide across the outer membrane of Escherichia coli. J. Biol. Chem. 27928227-28732. [DOI] [PubMed] [Google Scholar]
- 11.Bik, E. M., A. E. Bunschoten, R. J. L. Willems, A. C. Y. Chang, and F. R. Mooi. 1996. Genetic organization and functional analysis of the otn DNA essential for cell-wall polysaccharide synthesis in Vibrio cholerae O139. Mol. Microbiol. 20799-811. [DOI] [PubMed] [Google Scholar]
- 12.Bronner, D., V. Sieberth, C. Pazzani, I. S. Roberts, G. J. Boulnois, B. Jann, and K. Jann. 1993. Expression of the capsular K5 polysaccharide of Escherichia coli: biochemical and electron microscopic analyses of mutants with defects in region 1 of the K5 gene cluster. J. Bacteriol. 1755984-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryson, K., L. J. McGuffin, R. L. Marsden, J. J. Ward, J. S. Sodhi, and D. T. Jones. 2005. Protein structure prediction servers at University College London. Nucleic Acids Res. 33W36-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campisano, A., C. Schroeder, M. Schemionek, J. Overhage, and B. H. Rehm. 2006. PslD is a secreted protein required for biofilm formation by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 723066-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatzidaki-Livanis, M., M. J. Coyne, H. Roche-Hakansson, and L. E. Comstock. 2008. Expression of a uniquely regulated extracellular polysaccharide confers a large-capsule phenotype to Bacteroides fragilis. J. Bacteriol. 1901020-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng, J., A. Z. Randall, M. J. Sweredoski, and P. Baldi. 2005. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 33W72-W76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke, B. R., R. Pearce, and I. S. Roberts. 1999. Genetic organization of the Escherichia coli K10 capsule gene cluster: identification and characterization of two conserved regions in group III capsule gene clusters encoding polysaccharide transport functions. J. Bacteriol. 1812279-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole, C., J. D. Barber, and G. J. Barton. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36W197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins, R. F., K. Beis, B. R. Clarke, R. C. Ford, M. Hulley, J. H. Naismith, and C. Whitfield. 2006. Periplasmic protein-protein contacts in the inner membrane protein Wzc form a tetrameric complex required for the assembly of Escherichia coli group 1 capsules. J. Biol. Chem. 2812144-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins, R. F., K. Beis, C. Dong, C. H. Botting, C. McDonnell, R. C. Ford, B. R. Clarke, C. Whitfield, and J. H. Naismith. 2007. The 3D structure of a periplasm-spanning platform required for assembly of group 1 capsular polysaccharides in Escherichia coli. Proc. Natl. Acad. Sci. USA 1042390-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox, A. D., J.-R. Brisson, V. Varma, and M. B. Perry. 1996. Structural analysis of the lipopolysaccharide from Vibrio cholerae O139. Carbohydr, Res. 56143-58. [DOI] [PubMed] [Google Scholar]
- 22.Coyne, M. J., K. G. Weinacht, C. M. Krinos, and L. E. Comstock. 2003. Mpi recombinase globally modulates the surface architecture of a human commensal bacterium. Proc. Natl. Acad. Sci. USA 10010446-10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dams-Kozlowska, H., and D. L. Kaplan. 2007. Protein engineering of Wzc to generate new emulsan analogs. Appl. Environ. Microbiol. 734020-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dams-Kozlowska, H., N. Sainath, and D. L. Kaplan. 2008. Construction of a chimeric gene cluster for the biosynthesis of apoemulsan with altered molecular weight. Appl. Microbiol. Biotechnol. 78677-683. [DOI] [PubMed] [Google Scholar]
- 25.Das, D., Q. S. Xu, J. Y. Lee, I. Ankoudinova, C. Huang, Y. Lou, A. DeGiovanni, R. Kim, and S. H. Kim. 2007. Crystal structure of the multidrug efflux transporter AcrB at 3.1Å resolution reveals the N-terminal region with conserved amino acids. J. Struct. Biol. 158494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong, C., K. Beis, J. Nesper, A. L. Brunkan-Lamontagne, B. R. Clarke, C. Whitfield, and J. H. Naismith. 2006. Wza, the translocon for E. coli capsular polysaccharides, defines a new class of membrane protein. Nature 444226-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummelsmith, J., and C. Whitfield. 1999. Gene products required for surface expression of the capsular form of the group 1 K antigen in Escherichia coli (O9a:K30). Mol. Microbiol. 311321-1332. [DOI] [PubMed] [Google Scholar]
- 28.Drummelsmith, J., and C. Whitfield. 2000. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli (O9a:K30) requires a multimeric complex in the outer membrane. EMBO J. 1957-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eswaran, J., E. Koronakis, M. K. Higgins, C. Hughes, and V. Koronakis. 2004. Three's company: component structures bring a closer view of tripartite drug efflux pumps. Curr. Opin. Struct. Biol. 14741-747. [DOI] [PubMed] [Google Scholar]
- 30.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5164-166. [Google Scholar]
- 31.Ferrières, L., S. N. Aslam, R. M. Cooper, and D. J. Clarke. 2007. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology 1531070-1080. [DOI] [PubMed] [Google Scholar]
- 32.Reference deleted.
- 33.Ganapathiraju, M., C. J. Jursa, H. A. Karimi, and J. Klein-Seetharaman. 2007. TMpro web server and web service: transmembrane helix prediction through amino acid property analysis. Bioinformatics 232795-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez, J. E., C. E. Semino, L. X. Wang, L. E. Castellano-Torres, and G. C. Walker. 1998. Biosynthetic control of molecular weight in the polymerization of the octasaccharide subunits of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 9513477-13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grangeasse, C., A. J. Cozzone, J. Deutscher, and I. Mijakovic. 2007. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem. Sci. 3286-94. [DOI] [PubMed] [Google Scholar]
- 36.Grangeasse, C., P. Doublet, and A. J. Cozzone. 2002. Tyrosine phosphorylation of protein kinase Wzc from Escherichia coli K12 occurs through a two-step process. J. Biol. Chem. 2777127-7135. [DOI] [PubMed] [Google Scholar]
- 37.Grangeasse, C., B. Obadia, I. Mijakovic, J. Deutscher, A. J. Cozzone, and P. Doublet. 2003. Autophosphorylation of the Escherichia coli protein kinase Wzc regulates tyrosine phosphorylation of Ugd, a UDP-glucose dehydrogenase. J. Biol. Chem. 27839323-39329. [DOI] [PubMed] [Google Scholar]
- 38.Higgins, M. K., E. Bokma, E. Koronakis, C. Hughes, and V. Koronakis. 2004. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA 1019994-9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofman, K., and W. Stoffel. 1993. TMBASE—a database of membrane-spanning protein segments. Biol. Chem. Hoppe-Seyler 374166. [Google Scholar]
- 40.Huang, X. Q., R. C. Hardison, and W. Miller. 1990. A space-efficient algorithm for local similarities. Comput. Appl. Biosci. 6373-381. [DOI] [PubMed] [Google Scholar]
- 41.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23254-267. [DOI] [PubMed] [Google Scholar]
- 42.Itoh, Y., J. D. Rice, C. Goller, A. Pannuri, J. Taylor, J. Meisner, T. J. Beveridge, J. F. Preston III, and T. Romeo. 2008. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-β-1,6-N-acetyl-d-glucosamine. J. Bacteriol. 1903670-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jadeau, F., E. Bechet, A. J. Cozzone, G. Deleage, C. Grangeasse, and C. Combet. 2008. Identification of the idiosyncratic bacterial protein tyrosine kinase (BY-kinase) family signature. Bioinformatics 242427-2430. [DOI] [PubMed] [Google Scholar]
- 44.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292195-202. [DOI] [PubMed] [Google Scholar]
- 45.Kajimura, J., A. Rahman, and P. D. Rick. 2005. Assembly of cyclic enterobacterial common antigen in Escherichia coli K-12. J. Bacteriol. 1876917-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlyshev, A. V., D. Linton, N. A. Gregson, A. J. Lastovica, and B. W. Wren. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35529-541. [DOI] [PubMed] [Google Scholar]
- 47.Keenleyside, W. J., D. Bronner, K. Jann, B. Jann, and C. Whitfield. 1993. Coexpression of colanic acid and serotype-specific capsular polysaccharides in Escherichia coli strains with group II K antigens. J. Bacteriol. 1756725-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein, G., C. Dartigalongue, and S. Raina. 2003. Phosphorylation-mediated regulation of heat shock response in Escherichia coli. Mol. Microbiol. 48269-285. [DOI] [PubMed] [Google Scholar]
- 49.Knirel, Y. A., L. Paredes, P. E. Jansson, A. Weintraub, G. Widmalm, and M. J. Albert. 1995. Structure of the capsular polysaccharide of Vibrio cholerae O139 synonym Bengal containing d-galactose 4,6-cyclophosphate. Eur. J. Biochem. 232391-396. [DOI] [PubMed] [Google Scholar]
- 50.Knirel, Y. A., G. Widmalm, S. N. Senchenkova, P. E. Jansson, and A. Weintraub. 1997. Structural studies on the short-chain lipopolysaccharide of Vibrio cholerae O139 Bengal. Eur. J. Biochem. 247402-410. [DOI] [PubMed] [Google Scholar]
- 51.Koronakis, V., J. Eswaran, and C. Hughes. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73467-489. [DOI] [PubMed] [Google Scholar]
- 52.Kröncke, K.-D., J. R. Golecki, and K. Jann. 1990. Further electron microscopic studies on the expression of Escherichia coli group II capsules. J. Bacteriol. 1723469-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuhn, H.-M., U. Meier-Dieter, and H. Mayer. 1988. ECA, the enterobacterial common antigen. FEMS Microbiol. Rev. 54195-222. [DOI] [PubMed] [Google Scholar]
- 54.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 10114919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lacour, S., E. Bechet, A. J. Cozzone, I. Mijakovic, and C. Grangeasse. 2008. Tyrosine phosphorylation of the UDP-glucose dehydrogenase of Escherichia coli is at the crossroads of colanic acid synthesis and polymyxin resistance. PLoS One 3e3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lacour, S., P. Doublet, B. Obadia, A. J. Cozzone, and C. Grangeasse. 2006. A novel role for protein-tyrosine kinase Etk from Escherichia coli K-12 related to polymyxin resistance. Res. Microbiol. 157637-641. [DOI] [PubMed] [Google Scholar]
- 57.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 232947-2948. [DOI] [PubMed] [Google Scholar]
- 58.Larue, K., M. S. Kimber, R. C. Ford, and C. Whitfield. Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J. Biol. Chem., in press. [DOI] [PMC free article] [PubMed]
- 59.Lee, D. C., J. Zheng, Y. M. She, and Z. Jia. 2008. Structure of Escherichia coli tyrosine kinase Etk reveals a novel activation mechanism. EMBO J. 271758-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 31741-72. [DOI] [PubMed] [Google Scholar]
- 61.Liu, C. H., S. M. Lee, J. M. Vanlare, D. L. Kasper, and S. K. Mazmanian. 2008. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc. Natl. Acad. Sci. USA 1053951-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lobedanz, S., E. Bokma, M. F. Symmons, E. Koronakis, C. Hughes, and V. Koronakis. 2007. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc. Natl. Acad. Sci. USA 1044612-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacLachlan, P. R., W. J. Keenleyside, C. Dodgson, and C. Whitfield. 1993. Formation of the K30 (group I) capsule in Escherichia coli O9:K30 does not require attachment to lipopolysaccharide lipid A-core. J. Bacteriol. 1757515-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mao, Y., M. P. Doyle, and J. Chen. 2006. Role of colanic acid exopolysaccharide in the survival of enterohaemorrhagic Escherichia coli O157:H7 in simulated gastrointestinal fluids. Lett. Appl. Microbiol. 42642-647. [DOI] [PubMed] [Google Scholar]
- 65.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33D192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marlovits, T. C., T. Kubori, A. Sukhan, D. R. Thomas, J. E. Galan, and V. M. Unger. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 3061040-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marolda, C. L., E. R. Haggerty, M. Lung, and M. A. Valvano. 2008. Functional analysis of predicted coiled-coil regions in the Escherichia coli K-12 O-antigen polysaccharide chain length determinant Wzz. J. Bacteriol. 1902128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marolda, C. L., L. D. Tatar, C. Alaimo, M. Aebi, and M. A. Valvano. 2006. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide O antigen. J. Bacteriol. 1885124-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marolda, C. L., J. Vicarioli, and M. A. Valvano. 2004. Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 1504095-4105. [DOI] [PubMed] [Google Scholar]
- 70.Matias, V. R., A. Al-Amoudi, J. Dubochet, and T. J. Beveridge. 2003. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 1856112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mazmanian, S. K., J. L. Round, and D. L. Kasper. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453620-625. [DOI] [PubMed] [Google Scholar]
- 72.McNulty, C., J. Thompson, B. Barrett, L. Lord, C. Andersen, and I. S. Roberts. 2006. The cell surface expression of group 2 capsular polysaccharides in Escherichia coli: the role of KpsD, RhsA and a multi-protein complex at the pole of the cell. Mol. Microbiol. 59907-922. [DOI] [PubMed] [Google Scholar]
- 73.Mikolosko, J., K. Bobyk, H. I. Zgurskaya, and P. Ghosh. 2006. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 14577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Minic, Z., C. Marie, C. Delorme, J. M. Faurie, G. Mercier, D. Ehrlich, and P. Renault. 2007. Control of EpsE, the phosphoglycosyltransferase initiating exopolysaccharide synthesis in Streptococcus thermophilus, by EpsD tyrosine kinase. J. Bacteriol. 1891351-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morona, R., L. Van Den Bosch, and C. Daniels. 2000. Evaluation of Wzz/MPA1/MPA2 proteins based on the presence of coiled-coil regions. Microbiology 1461-4. [DOI] [PubMed] [Google Scholar]
- 76.Morona, R., L. Van Den Bosch, and P. A. Manning. 1995. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J. Bacteriol. 1771059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419587-593. [DOI] [PubMed] [Google Scholar]
- 78.Murray, G. L., S. R. Attridge, and R. Morona. 2006. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 1882735-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nesper, J., C. M. Hill, A. Paiment, G. Harauz, K. Beis, J. H. Naismith, and C. Whitfield. 2003. Translocation of group 1 capsular polysaccharide in Escherichia coli serotype K30. Structural and functional analysis of the outer membrane lipoprotein Wza. J. Biol. Chem. 27849763-49772. [DOI] [PubMed] [Google Scholar]
- 80.Obadia, B., S. Lacour, P. Doublet, H. Baubichon-Cortay, A. J. Cozzone, and C. Grangeasse. 2007. Influence of tyrosine-kinase Wzc activity on colanic acid production in Escherichia coli K12 cells. J. Mol. Biol. 36742-53. [DOI] [PubMed] [Google Scholar]
- 81.Olivares-Illana, V., P. Meyer, E. Bechet, V. Gueguen-Chaignon, D. Soulat, S. Lazereg-Riquier, I. Mijakovic, J. Deutscher, A. J. Cozzone, O. Laprevote, S. Morera, C. Grangeasse, and S. Nessler. 2008. Structural basis for the regulation mechanism of the tyrosine kinase CapB from Staphylococcus aureus. PLoS Biol. 6e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ouali, M., and R. D. King. 2000. Cascaded multiple classifiers for secondary structure prediction. Protein Sci. 91162-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paiment, A., J. Hocking, and C. Whitfield. 2002. Impact of phosphorylation of specific residues in the tyrosine autokinase, Wzc, on its activity in assembly of group 1 capsules in Escherichia coli. J. Bacteriol. 1846437-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paulsen, I. T., A. M. Beness, and M. H. J. Saier. 1997. Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology 1432685-2699. [DOI] [PubMed] [Google Scholar]
- 85.Pazzani, C., C. Rosenow, G. J. Boulnois, D. Bronner, K. Jann, and I. S. Roberts. 1993. Molecular analysis of region 1 of Escherichia coli K5 antigen gene cluster: a region encoding proteins involved in cell surface expression of capsular polysaccharide. J. Bacteriol. 1755978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peleg, A., Y. Shifrin, O. Ilan, C. Nadler-Yona, S. Nov, S. Koby, K. Baruch, S. Altuvia, M. Elgrably-Weiss, C. M. Abe, S. Knutton, M. A. Saper, and I. Rosenshine. 2005. Identification of an Escherichia coli operon required for formation of the O-antigen capsule. J. Bacteriol. 1875259-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Phoenix, D. A., K. Brandenburg, F. Harris, U. Seydel, T. Hammerton, and I. S. Roberts. 2001. An investigation into the membrane-interactive potential of the Escherichia coli KpsE C-terminus. Biochem. Biophys. Res. Commun. 285976-980. [DOI] [PubMed] [Google Scholar]
- 88.Purins, L., L. Van Den Bosch, V. Richardson, and R. Morona. 2008. Coiled-coil regions play a role in the function of the Shigella flexneri O-antigen chain length regulator WzzpHS2. Microbiology 1541104-1116. [DOI] [PubMed] [Google Scholar]
- 89.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rahn, A., J. Drummelsmith, and C. Whitfield. 1999. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 1812307-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reid, A. N., and C. Whitfield. 2005. Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J. Bacteriol. 1875470-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Remminghorst, U., and B. H. Rehm. 2006. Bacterial alginates: from biosynthesis to applications. Biotechnol. Lett. 281701-1712. [DOI] [PubMed] [Google Scholar]
- 93.Reuber, T. L., and G. C. Walker. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74269-280. [DOI] [PubMed] [Google Scholar]
- 94.Rigg, G. P., B. Barrett, and I. S. Roberts. 1998. The localization of KpsC, S and T and KfiA, C and D proteins involved in the biosynthesis of the Escherichia coli K5 capsular polysaccharide: evidence for a membrane-bound complex. Microbiol. 1442905-2914. [DOI] [PubMed] [Google Scholar]
- 95.Romling, U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153205-212. [DOI] [PubMed] [Google Scholar]
- 96.Ryder, C., M. Byrd, and D. J. Wozniak. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saldias, M. S., K. Patel, C. L. Marolda, M. Bittner, I. Contreras, and M. A. Valvano. 2008. Distinct functional domains of the Salmonella enterica WbaP transferase that is involved in the initiation reaction for synthesis of the O antigen subunit. Microbiology 154440-453. [DOI] [PubMed] [Google Scholar]
- 98.Seeger, M. A., A. Schiefner, T. Eicher, F. Verrey, K. Diederichs, and K. M. Pos. 2006. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 3131295-1298. [DOI] [PubMed] [Google Scholar]
- 99.Seeger, M. A., C. von Ballmoos, T. Eicher, L. Brandstatter, F. Verrey, K. Diederichs, and K. M. Pos. 2008. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat. Struct. Mol. Biol. 15199-205. [DOI] [PubMed] [Google Scholar]
- 100.Sennhauser, G., P. Amstutz, C. Briand, O. Storchenegger, and M. G. Grutter. 2007. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 5e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shifrin, Y., A. Peleg, O. Ilan, C. Nadler, S. Kobi, K. Baruch, G. Yerushalmi, T. Berdichevsky, S. Altuvia, M. Elgrably-Weiss, C. Abe, S. Knutton, C. Sasakawa, J. M. Ritchie, M. K. Waldor, and I. Rosenshine. 2008. Transient shielding of intimin and the type III secretion system of enterohemorrhagic and enteropathogenic Escherichia coli by a group 4 capsule. J. Bacteriol. 1905063-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Silver, R. P., W. Aaronson, and W. F. Vann. 1987. Translocation of capsular polysaccharides in pathogenic strains of Escherichia coli requires a 60-kilodalton periplasmic protein. J. Bacteriol. 1695489-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Silver, R. P., K. Prior, C. Nsahlai, and L. F. Wright. 2001. ABC transporters and the export of capsular polysaccharides from gram-negative bacteria. Res. Microbiol. 152357-364. [DOI] [PubMed] [Google Scholar]
- 104.Smith, C. S., A. Hinz, D. Bodenmiller, D. E. Larson, and Y. V. Brun. 2003. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J. Bacteriol. 1851432-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6175-182. [PubMed] [Google Scholar]
- 106.Soulat, D., J. M. Jault, B. Duclos, C. Geourjon, A. J. Cozzone, and C. Grangeasse. 2006. Staphylococcus aureus operates protein-tyrosine phosphorylation through a specific mechanism. J. Biol. Chem. 28114048-14056. [DOI] [PubMed] [Google Scholar]
- 107.Soulat, D., J. M. Jault, C. Geourjon, P. Gouet, A. J. Cozzone, and C. Grangeasse. 2007. Tyrosine-kinase Wzc from Escherichia coli possesses an ATPase activity regulated by autophosphorylation. FEMS Microbiol. Lett. 274252-259. [DOI] [PubMed] [Google Scholar]
- 108.Steenbergen, S. M., and E. R. Vimr. 2008. Biosynthesis of the Escherichia coli K1 group 2 polysialic acid capsule occurs within a protected cytoplasmic compartment. Mol. Microbiol. 681252-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Szurmant, H., R. A. White, and J. A. Hoch. 2007. Sensor complexes regulating two-component signal transduction. Curr. Opin. Struct. Biol. 17706-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tang, K. H., H. Guo, W. Yi, M. D. Tsai, and P. G. Wang. 2007. Investigation of the conformational states of Wzz and the Wzz:O-antigen complex under near-physiological conditions. Biochemistry 4611744-11752. [DOI] [PubMed] [Google Scholar]
- 111.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 176487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tocilj, A., C. Munger, A. Proteau, R. Morona, L. Purins, E. Ajamian, J. Wagner, M. Papadopoulos, L. Van Den Bosch, J. L. Rubinstein, J. Fethiere, A. Matte, and M. Cygler. 2008. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat. Struct. Mol. Biol. 15130-138. [DOI] [PubMed] [Google Scholar]
- 113.Törnroth-Horsefield, S., P. Gourdon, R. Horsefield, L. Brive, N. Yamamoto, H. Mori, A. Snijder, and R. Neutze. 2007. Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure 151663-1673. [DOI] [PubMed] [Google Scholar]
- 114.Vorholter, F. J., S. Schneiker, A. Goesmann, L. Krause, T. Bekel, O. Kaiser, B. Linke, T. Patschkowski, C. Ruckert, J. Schmid, V. K. Sidhu, V. Sieber, A. Tauch, S. A. Watt, B. Weisshaar, A. Becker, K. Niehaus, and A. Puhler. 2008. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 13433-45. [DOI] [PubMed] [Google Scholar]
- 115.Whitfield, C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 7539-68. [DOI] [PubMed] [Google Scholar]
- 116.Whitfield, C., E. R. Vimr, J. W. Costerton, and F. A. Troy. 1984. Protein synthesis is required for in vivo activation of polysialic acid capsule synthesis in Escherichia coli K1. J. Bacteriol. 159321-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wugeditsch, T., A. Paiment, J. Hocking, J. Drummelsmith, C. Forrester, and C. Whitfield. 2001. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J. Biol. Chem. 2762361-2371. [DOI] [PubMed] [Google Scholar]