Abstract
Summary: The T-box mechanism is a common regulatory strategy used for modulating the expression of genes of amino acid metabolism-related operons in gram-positive bacteria, especially members of the Firmicutes. T-box regulation is usually based on a transcription attenuation mechanism in which an interaction between a specific uncharged tRNA and the 5′ region of the transcript stabilizes an antiterminator structure in preference to a terminator structure, thereby preventing transcription termination. Although single T-box regulatory elements are common, double or triple T-box arrangements are also observed, expanding the regulatory range of these elements. In the present study, we predict the functional implications of T-box regulation in genes encoding aminoacyl-tRNA synthetases, proteins of amino acid biosynthetic pathways, transporters, and regulatory proteins. We also consider the global impact of the use of this regulatory mechanism on cell physiology. Novel biochemical relationships between regulated genes and their corresponding metabolic pathways were revealed. Some of the genes identified, such as the quorum-sensing gene luxS, in members of the Lactobacillaceae were not previously predicted to be regulated by the T-box mechanism. Our analyses also predict an imbalance in tRNA sensing during the regulation of operons containing multiple aminoacyl-tRNA synthetase genes or biosynthetic genes involved in pathways common to more than one amino acid. Based on the distribution of T-box regulatory elements, we propose that this regulatory mechanism originated in a common ancestor of members of the Firmicutes, Chloroflexi, Deinococcus-Thermus group, and Actinobacteria and was transferred into the Deltaproteobacteria by horizontal gene transfer.
INTRODUCTION
The regulation of bacterial gene expression is often based on RNA recognition of an appropriate signal. The term “riboswitch” has been used to describe cis-acting RNA regulatory elements that undergo significant structural shifts in response to a specific regulatory signal. This recognition occurs in the absence of the action of an RNA binding protein or a translating ribosome. The shift in riboswitch structure regulates the expression of RNA sequences located downstream on that RNA. Riboswitch RNAs regulate the expression of genes encoding proteins with a broad range of functions in a variety of bacterial species. These regulated genes include those specifying enzymes concerned with the charging of amino acids onto tRNAs, the synthesis or transport of amino acids, and the synthesis of cofactors, nucleotides, and metal ions (55).
The T-box family of riboswitches commonly modulates the expression of many genes concerned with amino acid metabolism in gram-positive bacteria, especially members of the family Firmicutes. The T-box mechanism utilizes uncharged tRNA as the signal molecule. Genes in this family exhibit a conservation of a set of sequence and structural features in the untranslated “leader region” of the RNA upstream of the regulated coding sequences (43, 58). For most operons in the T-box family, segments of these upstream leader RNAs can fold to form either of two alternative hairpin structures, an intrinsic transcription terminator or a competing transcription antiterminator. The formation of the terminator hairpin results in a premature termination of transcription, reducing the transcription of the downstream coding region(s). In each T-box-regulated operon, the proper pairing of an appropriate uncharged tRNA with the leader RNA promotes the stabilization of the alternate antiterminator structure. This pairing prevents the formation of the terminator, which allows continued transcription into the downstream gene or genes of the operon (43, 56). The specific recognition of the cognate uncharged tRNA and the tRNA-directed formation of a transcription antiterminator can occur in the absence of any other cellular factor(s) (50). Like other riboswitches in which a leader RNA transcript senses a signal in regulating downstream gene expression, T-box RNAs can also control translation initiation. In transcripts with this capability, the terminator helix is replaced by a helix that can sequester the Shine-Dalgarno (SD) sequence of a downstream coding region, thereby inhibiting the initiation of translation rather than prematurely terminating transcription. T-box RNAs that regulate transcription termination are most commonly observed in low-G+C gram-positive bacteria, while T-box RNAs that control translation initiation predominate in high-G+C gram-positive bacteria and in gram-negative bacteria (organisms in which T-box-mediated regulation is less common) (47, 107).
The common feature of the T-box mechanism and other riboswitch mechanisms is the ability of a leader RNA to directly sense a signal molecule (an uncharged tRNA or some other specific molecule), resulting in a rearrangement of the leader RNA that determines whether the downstream coding sequences will be expressed. The T-box mechanism is particularly well suited for regulating the expression of genes encoding proteins involved in the aminoacylation of tRNA and amino acid biosynthesis and in amino acid transport. Aminoacylated tRNA is also sensed in some bacterial species using different RNA regulatory strategies. For example, in Escherichia coli, the trp transcription attenuation mechanism is used; in this mechanism, the translation of a leader peptide coding region modulates the formation of alternative leader RNA structures, determining whether or not transcription termination will occur in the leader region of the trp operon (117, 118). The major difference between the E. coli trp transcription attenuation mechanism and the T-box riboswitch mechanism is that tRNA charging is sensed indirectly by a translating ribosome in the former mechanism, compared to the direct binding of the tRNA to the leader RNA in the T-box mechanism.
The high level of conservation of the sequences and structural features of T-box leader RNAs permits the identification of many genes that are likely to be regulated by this mechanism. This information, in conjunction with the identification of the products of the downstream coding sequences, permits prediction of the specificity of their regulatory responses. Recently, Vitreschak et al. (107) reported the identification of 805 T-box leader sequences in 96 partial and completely sequenced bacterial genomes. In their paper, they provided an overview of the use of this regulatory strategy and discussed the evolutionary relationships of T-box leader sequences with regard to the origin of this highly conserved regulatory mechanism. Here, we report genomic analyses with a larger set of genomes using a different set of parameters. We identified 1,111 T-box leader sequences in 87 completely sequenced bacterial genomes; 472 of the genomes examined did not contain an identifiable T-box-controlled gene. This analysis of T-box elements allowed us to identify arrangements of this regulatory element that differ from its standard mode. We will discuss the predicted physiological roles of the identified T-box-regulated genes and the functional implications of T-box-mediated regulation for the functioning of the corresponding metabolic pathways. We will also discuss the distribution of T-box versus other regulatory mechanisms for genes concerned with different classes of amino acids. In addition to citing examples supporting the previously described role of T-box elements in regulating the expression of individual genes or operons, we predict that T-box-mediated regulation controls the synthesis of regulatory proteins that in turn regulate additional sets of genes. These features increase the overall impact of the T-box mechanism in modulating various cellular activities.
GENERAL FEATURES OF T-BOX RIBOSWITCH RNA
A T-box RNA consists of a segment of leader RNA with conserved features that allow recognition of, and pairing with, a specific uncharged tRNA (Fig. 1). These features include the capacity of the leader RNA to form alternative secondary structures, one of which can serve as an intrinsic transcription terminator or as an anti-SD (ASD) helix that pairs with an SD sequence, blocking translation initiation. The major structures formed within the T-box RNA, in addition to the segments that can form the terminator/antiterminator (or ASD/anti-ASD) elements, are designated stem I, stem II, the stem IIA/stem IIB pseudoknot, and stem III (47, 88) (Fig. 1A). The sequestration of sequences that form the 5′ strand of the terminator (or the ASD helix) into a competing antiterminator structure (or anti-ASD helix) allows the transcription or translation of the downstream coding sequence. In each T-box RNA, the terminator (or ASD helix) is predicted to be more stable than the competing antiterminator structure; therefore, binding of uncharged tRNA is required to stabilize the competing antiterminator (or anti-ASD helix) structure.
FIG. 1.
The T-box RNA regulatory system. (A) Structural model of the B. subtilis tyrS T-box leader RNA. The T-box element present in the B. subtilis tyrS leader region was originally described by Grundy and Henkin (see reference 43). The standard T-box leader RNA arrangement consists of three major elements, stem I, stem II, and stem III plus the stem IIA/stem IIB pseudoknot, and the competing terminator and antiterminator structures. The specifier loop, an internal bulge in stem I, contains the specifier sequence (boxed UAC residues complementary to the anticodon sequence of tRNATyr); the conserved purine (an adenine) following the specifier sequence is inside a green circle. The T-box sequence is unpaired in the terminator form and is paired in the antiterminator form (the antiterminator is shown to the right of the terminator). The sequence highlighted in blue shows the nucleotides involved in the antiterminator structure. The antiterminator structure has a bulge that interacts with the unpaired residues at the acceptor end of an uncharged tRNA. Nucleotide conservation in all 722 T-box sequences analyzed was evaluated using a multiple sequence alignment obtained from the Rfam database (42), and residues are color coded accordingly. (B) Model of the regulatory alternatives for the T-box mechanism. During the transcription of a leader region by RNA polymerase (red ovals), the nascent RNA folds into a structure competent for binding of the cognate tRNA at two sites. The binding of uncharged tRNA (top) to both the specifier sequence and the antiterminator bulge stabilizes the antiterminator (green RNA segment), preventing the formation of the terminator. This allows transcription to proceed into the downstream-regulated coding sequence (blue box). Charged tRNA (represented by Tyr attached to the 3′ end of the tRNA) can interact with the specifier sequence but cannot interact with the antiterminator; a failure to stabilize the antiterminator allows the formation of the terminator helix (red RNA segment), and transcription is terminated before the downstream coding region can be transcribed. Conserved elements of T-box RNAs are stem I (black), stem II (orange), the stem IIA/stem IIB pseudoknot (light blue), and stem III (purple).
Each T-box RNA is presumably the result of evolutionary selection, preparing it to respond to a specific uncharged tRNA. Specific tRNA binding requires the pairing of the tRNA anticodon with a single codon sequence designated the “specifier sequence” in the T-box RNA (Fig. 1A). The specifier sequence is positioned at a discrete location in the specifier loop within stem I (Fig. 1A). Due to the redundancy of the genetic code, each of the amino acids, with the exception of Met and Trp, is charged onto more than one tRNA species. In theory, any of these tRNA species could be sensed by the T-box mechanism. However, initial studies of T-box leader sequences (121), in agreement with results described previously by Vitreschak et al. (107) and our analyses of T-box elements in fully sequenced genomes, exhibited a strong bias toward the presence of a C in the third position of the specifier sequence of most T-box RNAs. This preference is not influenced by the codon usage or the tRNA abundance in a specific organism. For example, glycine (Gly)-specific T-box RNAs exhibit a strong preference for the GGC codon. This is the least used Gly codon in Bacillus halodurans but the most often used Gly codon in Bacillus clausii. Mutational studies of the Bacillus subtilis tyrS gene demonstrated that the replacement of the UAC Tyr codon with a UAU Tyr codon resulted in a dramatic decrease in levels of expression in vivo. Moreover, initial studies with the B. subtilis glyQS leader region suggested that the bias for C in the third position of the specifier sequence is due to structural constraints for codon and/or codon-anticodon pairing that differ from those involved in translation (E. Caserta, F. Grundy, and T. Henkin, unpublished results). The universally conserved U34 residue 5′ to the anticodon in tRNA was also predicted to pair with a conserved purine residue 3′ to the specifier sequence (83; F. Grundy and T. Henkin, unpublished data). This pairing is supported by the structural analysis of the B. subtilis glyQS leader RNA/tRNAGly complex (121).
The 14 most highly conserved residues of the entire T-box RNA represent the “T-box sequence” (AGGGUGGNACCGCG) (Fig. 1A); the recognition of this sequence in the leader regions of several aminoacyl-tRNA synthetase (aaRS) genes led to the initial prediction of a conserved regulatory mechanism (44). This sequence was shown to form the 5′ side of the antiterminator structure, including the 7-nucleotide bulge (43). Discrimination between uncharged and charged tRNAs is mediated by the pairing of the four unpaired residues at the 3′ end of the tRNA (5′-NCCA-3′) with the first four residues of the antiterminator bulge (5′-UGGN-3′); the N residue in the antiterminator bulge covaries with the corresponding position of the tRNA, a position that often plays an important role in tRNA identity for recognition by the cognate aaRS. The presence of the amino acid at the 3′ end of a charged tRNA prevents the interaction of its 3′ end with the antiterminator RNA; hence, the antiterminator structure does not form. Both charged and uncharged tRNAs can interact with the leader RNA at the specifier sequence, but only uncharged tRNA has been shown to stabilize the antiterminator sequence. Thus, importantly, each T-box sequence monitors the ratio between the charged and uncharged forms of a specific tRNA rather than the absolute amount of the uncharged tRNA (51). T-box leader RNAs are further characterized by the existence of a set of conserved primary sequence elements at specific locations relative to the structural features (43). Mutational studies have demonstrated that many of the sequence and structural features conserved in T-box leader RNAs are important for function (49, 88, 112), but overall primary sequence conservation is low. Interestingly, the two main features of tRNAs (the anticodon and the base immediately preceding the CCA at its 3′ end) that are recognized by the T-box RNAs are also usually central elements for recognition by the cognate aaRS (14). aaRSs have been grouped into two nonhomologous sets of enzymes, class I (LeuRS, IleRS, MetRS, TyrRS, GluRS, ValRS, ArgRS, LysRS1, and TrpRS) and class II (PheRS, SerRS, ThrRS, ProRS, AlaRS, HisRS, AspRS, AsnRS, LysRS2, and GlyRS) (113). Class I and class II aaRSs recognize different tRNA structural features. Whether these or other tRNA structural features are also recognized by the T-box system remains to be determined.
In addition to the classical T-box RNA arrangement described by Grundy and Henkin (47), a reduced version of the T-box RNA, which is predicted to regulate the initiation of translation in the Actinobacteria, has also been described (107). This type of T-box RNA contains a smaller variant of the stem I structure, where the specifier sequence is placed in the terminal loop. Unusual examples of partially duplicated T-box RNAs in which a single stem I is followed by double or triple copies of the antiterminator/terminator were also reported (107), but the function(s) of these extra copies is not yet understood.
Using previously described position-specific matrices associated with leader RNAs that have T-box features (1, 2), we performed genome-scale searches using the MAST (6) and covariance models of Rfam with the program cmsearch (27, 42). Although other genomic studies used only primary sequence conservation (111), the use of covariance models for RNA secondary-structure prediction improves the accuracy of riboswitch identification. In the specific case of the T boxes, covariance analyses facilitated the identification of the specifier sequence. In the present study, 559 fully sequenced bacterial genomes were examined and 1,111 T-box leader sequences were identified in 87 organisms. The criteria for concluding that adjacent genes are within the same operon were described previously by Janga et al. and Salgado et al. (65, 89). The genomic contexts of the significant matches identified were further analyzed using our Web GeConT server (19, 72). Our findings are organized according to the type of gene or genes that were identified downstream of a T-box element, as described in the following sections. The complete list of T-box genes with a known or predicted function is shown in Fig. 3 to 8 and 10. Specific examples are discussed below to emphasize particular points of interest.
FIG. 3.
Aminoacyl-tRNA synthetase genes regulated by the T-box mechanism. From the set of T-box-regulated genes identified in our study, operons containing aaRS genes were grouped according to the amino acid class of the aaRS. Operons containing more than one different aaRS gene are shown under the amino acid category matching the predicted specifier sequence. In the exceptional case of leuS in C. hydrogenoformans, P. thermopropionicum, and S. wolfei, the T-box sequence, drawn in green, contains a tRNA gene. Organism nomenclature is as follows for members of the Firmicutes: A. met, “Alkaliphilus metalliredigens”; B. amy, Bacillus amyloliquefaciens; B. ant, Bacillus anthracis; B. cla, Bacillus clausii; B. cer, Bacillus cereus; B. hal, Bacillus halodurans; B. lic, Bacillus lichenformis; B. pum, Bacillus pumilus; B. sub, Bacillus subtilis; B. ste, Bacillus stearothermophilus; B. thu, Bacillus thuringiensis; C. ace, Clostridium acetobutylicum; C. bei, Clostridium beijerinckii; C. bot, Clostridium botulinum; C. dif, Clostridium difficile; C. hyd, Clostridium hydrogenoformans; C. klu, Clostridium kluyveri; C. nov, Clostridium novyi; C. per, Clostridium perfringens; C. tet, Clostridium tetani; C. the, Clostridium thermocellum; D. haf, Desulfitobacterium hafniense; D. red, Desulfotomaculus reducens; E. fae, Enterococcus faecalis; G. kau, Geobacillus kaustophilus; G. the, Geobacillus thermodenitrificans; L. aci, Lactobacillus acidophilus; L. bre, Lactobacillus brevis; L. cas, Lactobacillus casei; L. del, both Lactobacillus delbrueckii subsp. bulgaricus strains; L. del 11842, Lactobacillus delbrueckii subsp. bulgaricus strain ATCC 11842; L. del BAA365, Lactobacillus delbrueckii subsp. bulgaricus strain ATCC BAA365; L. gas, Lactobacillus gasseri; L. inn, Listeria innocua; L. joh, Lactobacillus johnsonii; L. lac, Lactococcus lactis; L. mes, Leuconostoc mesenteroides; L. mon, Listeria monocytogenes; L. pla, Lactobacillus plantarum; L. reu, Lactobacillus reuteri; L. sak, Lactobacillus sakei; L. sal, Lactobacillus salivarius; L. wel, Listeria welshimeri; M. cap, Mycoplasma capricolum; M. flo, Mesoplasma florum; M. myc, Mycoplasma mycoides; M. the, Moorella thermoacetica; O. ihe, Oceanobacillus iheyensis; O. oen, Oenococcus oeni; P. pen, Pediococcus pentosaceus; P. the, Pelotomaculum thermopropionicum; S. aga, Streptococcus agalactiae; S. aur, Staphylococcus aureus; S. epi, Staphylococcus epidermidis; S. gor, Streptococcus gordonii; S. hae, Staphylococcus haemolyticus; S. mut, Streptococcus mutans; S. pne, Streptococcus pneumoniae; S. pyo, Streptococcus pyogenes; S. san, Streptococcus sanguinis; S. sap, Staphylococcus saprophyticus; S. sui, Streptococcus suis; S. the, Streptococcus thermophilus; S. wol, Syntrophomonas wolfei; T. ten, Thermoanaerobacter tengcongensis. Organism nomenclature is as follows for members of the Actinobacteria: B. ado, Bifidobacterium adolescentis; B. lon, Bifidobacterium longum; C. eff, Corynebacterium efficiens; C. dip, Corynebacterium diphtheriae; C. glu, Corynebacterium glutamicum; C. jei, Corynebacterium jeikeium; M. avi, Mycobacterium avium; M. bov, Mycobacterium bovis; M. lep, Mycobacterium leprae; M. sme, Mycobacterium smegmatis; M. tub, Mycobacterium tuberculosis; M. ulc, Mycobacterium ulcerans; N. far, Nocardia farcinica; P. can, Propionibacterium acnes; R. xyl, Rubrobacter xylanophilus; S. ave, Streptomyces avermitilis; S. coe, Streptomyces coelicolor; T fus, Thermobifida fusca. Organism nomenclature is as follows for members of the Fusobacteria: F. nuc, Fusobacterium nucleatum. Organism nomenclature is as follows for members of the Deinococcus-Thermus group: D. geo, Deinococcus geothermalis; D. rad, Deinococcus radiodurans; T. the, Thermus thermophilus. Organism nomenclature is as follows for members of the Chlorobi: C. tep, Chlorobium tepidum; C. aur, Chloroflexus aurantiacus; C. hut, Cytophaga hutchinsonii. Organism nomenclature is as follows for members of the Chloroflexi: D. BAV1, “Dehalococcoides” sp. strain BAV1; D. CBDB1, Dehalococcoides sp. strain CBDB1; D. eth, “Dehalococcoides ethenogenes”; R. SM1, Roseiflexus sp. strain SM1; R. cas, Roseiflexus castenholzii. Organism nomenclature is as follows for members of the Proteobacteria: G. sul, Geobacter sulfurreducens; G. met, Geobacter metallireducens; G. ura, Geobacter uraniumreducens; P. car, Pelobacter carbinolicus; P. pro, Pelobacter propionicus. Operon predictions and the color code used for the different types of regulated genes are described in the legend of Fig. 2.
FIG. 8.
Genes involved in amino acid transport that are regulated by the T-box mechanism. The common designation for each class of transporter gene is shown inside each arrow. Genes that have not been annotated were labeled based on their corresponding COG numbers (i.e., a gene that belongs to COG4166 is drawn as an arrow with the number “4166”). Note that genes are named in accordance with the GenBank annotation and might not represent the real specificity of the transporter as revealed by the identification of the specifier codon in our T-box analysis. Organism abbreviations and gene color codes are described in the legends of Fig. 2 and 3.
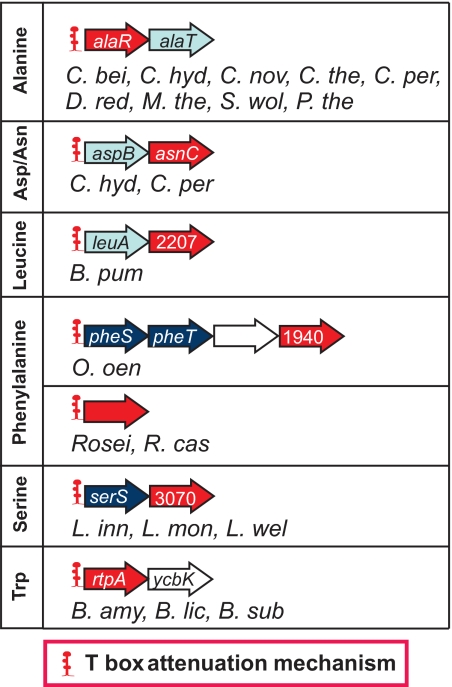
FIG. 10.
Regulatory genes controlled by the T-box mechanism. Organism abbreviations and color codes are described in the legends of Fig. 2 and 3. COG1940 is annotated as a “negative regulator of the xylose operon”; this annotation does not correspond to the function deduced from its Phe T-box specificity. COG3070 corresponds to a “regulator of competence-specific genes”; its true function is unknown, but it is predicted to be related to its Ser specifier sequence and the fact that is cotranscribed with serS. COG2207 is the family of AraC transcriptional regulators. The Roseiflexus (Rosei) sp. regulatory genes do not belong to any COG family but are annotated in GenBank as “putative transcriptional regulators, MerR family.”
AMINOACYL-tRNA SYNTHETASE GENES
The first group of genes to be identified that are regulated by the T-box mechanism encode aaRSs (43, 57). The accumulation of uncharged tRNATyr was shown to be the intracellular signal that leads to the activation of transcription of the B. subtilis tyrS gene, encoding TyrRS. Regulation was achieved by modulating the readthrough of the leader RNA transcription terminator. Similar findings were reported for other B. subtilis aaRS genes, including thrS, thrZ, leuS, and valS (20, 69, 82, 106), and T-box leader regions were identified upstream of additional aaRS genes, including pheS, tyrZ, and trpS, in Bacillus sp. (43). Subsequent analyses uncovered serS, ileS, glyQS, alaS, and hisS-aspS as additional T-box-regulated aaRS genes in the B. subtilis genome (17).
Our genomic analyses of the aaRS gene family revealed that the aaRS responsible for charging each of the 20 amino acids is regulated by the T-box mechanism in at least one firmicute species (Fig. 2 and 3). This mechanism appears to be the most commonly used regulatory mechanism for this family of genes in this group of organisms. T-box sequences were identified for members of both class I and class II aaRS enzymes. As previously mentioned, members of the two classes of aaRS enzymes are not homologous and are grouped based on the topology of the ATP binding domain. Class I proteins contain a Rossman fold, while class II enzymes possess an unrelated β-sheet arrangement (113, 114). Class I and class II aaRS enzymes are present in all organisms, and each type of tRNA is aminoacylated exclusively by a member of one of the two classes of aaRS, with the exception of tRNALys, which is aminoacylated by LysRS enzymes of both classes (113). A class II LysRS is present in all members of the Firmicutes and is encoded by a gene residing within a supraoperon containing genes involved in folate biosynthesis. This supraoperon does not contain a T-box sequence, and in B. subtilis, it is transcriptionally regulated in a growth phase-dependent manner (25). In addition to this class II LysRS, a gene encoding a class I LysRS (which is preceded by a T-box regulatory sequence) is present in Bacillus cereus, Bacillus thuringiensis, and Clostridium beijerinckii (Fig. 3).
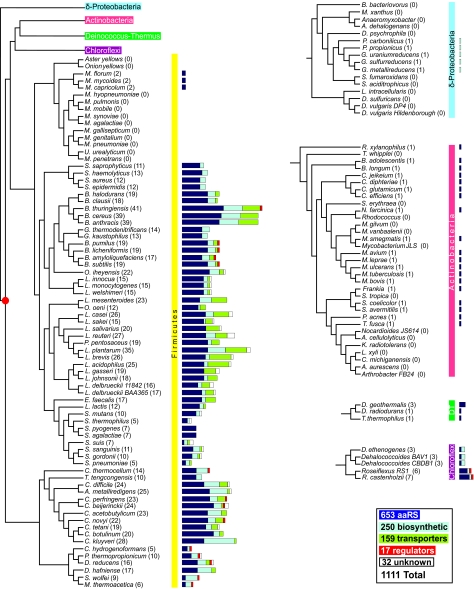
FIG. 2.
Distribution of T-box regions in different phylogenetic taxa. The phylogenetic tree for organisms relevant to our study was constructed based on the phylogenetic distances of aligned sequences from the concatenation of 31 proteins in 191 species, as previously described (18). Alignments were generated using the program MUSCLE (28), and phylogenetic reconstruction was performed using the PROTDIST program of the PHYLIP phylogeny inference package program (version 3.57c; J. Felsenstein, University of Washington, Seattle). Operons were predicted based on an analysis of intergenic distances, as described previously (76). Horizontal bar lengths are drawn to scale, reflecting the number of operons regulated by a T-box sequence; these are classified into one of the following groups: aminoacyl-tRNA synthetases (dark blue), amino acid biosynthetic genes (light blue), genes coding for regulatory proteins (red), transporter genes (green), and genes of unknown function (white). The most parsimonious scenario would place the initially evolved T-box regulatory sequence in a common ancestor of the Firmicutes, the Actinobacteria, the Chloroflexi, and the Deinococcus-Thermus (DT) group. The postulated origin is represented by a red dot in the tree. Names of Firmicutes are as follows: M. hyopneumoniae, Mycoplasma hyopneumoniae; M. pulmonis, Mycoplasma pulmonis; M. mobile, Mycoplasma mobile; M. synoviae, Mycoplasma synoviae; M. agalactiae, Mycoplasma agalactiae; M. gallisepticum, Mycoplasma gallisepticum; M. genitalium, Mycoplasma genitalium; M. pneumoniae, Mycoplasma pneumoniae; U. urealyticum, Ureaplasma urealyticum; M. penetrans, Mycoplasma penetrans. Names of Deltaproteobacteria are as follows: B. bacteriovorus, Bdellovibrio bacteriovorus; M. xanthus, Myxococcus xanthus; A. dehalogenans, Anaeromyxobacter dehalogenans; D. psychrophila, Desulfotalea psychrophila; S. fumaroxidans, Syntrophobacter fumaroxidans; S. aciditrophicus, Syntrophus aciditrophicus; L. intracellularis, Lawsonia intracellularis; D. sulfuricans, Desulfovibrio desulfuricans; D. vulgaris, Desulfovibrio vulgaris. Names of Actinobacteria are as follows: T. whipplei, Tropheryma whipplei; M. gilvum, Mycobacterium gilvum; M. vanbaalenii, Mycobacterium vanbaalenii; S. tropica, Salinispora tropica; A. cellulolyticus, Acidothermus cellulolyticus; K. radiotolerans, Kineococcus radiotolerans; L. xyli, Leifsonia xyli; C. michiganensis, Clavibacter michiganensis; A. aurescens, Arthrobacter aurescens. Additional names are listed in the legend to Fig. 3.
Generally, T-box-regulated aaRS genes are located in monocistronic transcriptional units, with three major exceptions: (i) when they encode different polypeptides in a heterodimeric enzyme, like glyQS; (ii) when they are associated with biosynthetic genes, such as cysS, which is located within the cysteine biosynthetic operon in some members of the Firmicutes; and (iii) when two aaRS enzymes for different amino acids are encoded in the same operon, such as the hisS-aspS operon, encoding HisRS and AspRS (Fig. 3). The coexpression of biosynthesis genes with aaRS genes of the same amino acid class represents an efficient use of the T-box regulatory mechanism, as both sets of genes respond in concert with the same tRNA. In contrast, the cotranscription of aaRS genes of different amino acid classes represents a potential regulatory problem, as the expression of each gene would be expected to respond individually to its cognate uncharged tRNA. A possible solution is apparent in some hisS-aspS aaRS operons in which the specifier loop of the T-box element contains the sequence GACAC, which has Asp (GAC) and His (CAC) codons that overlap by one nucleotide. The predicted RNA secondary structure places GAC in the most appropriate position for interaction with the tRNA anticodon, suggesting that the transcription of both hisS and aspS would depend on the accumulation of uncharged tRNAAsp. This could cause a deficiency in the sensing of tRNAHis, resulting in a failure to increase the synthesis of HisRS when increased charging of tRNAHis is required. However, in B. halodurans, Bacillus licheniformis, and Clostridium thermocellum, the stem I region could adopt an alternative secondary structure in which the His (CAC) codon is accessible for interactions with tRNAHis, allowing the transcription of the operon to respond to each of these uncharged tRNA species. This hypothesis requires experimental validation.
A second example in which aaRS genes for two different amino acids are regulated by one T-box RNA sequence is found in several Clostridium species (Clostridium acetobutylicum, C. beijerinckii, C. difficile, C. perfringens, and C. tetani). In these organisms, cysS and proS are in the same transcriptional unit, which appears to be regulated by a T-box sequence responding only to tRNAPro; this is predicted to create an imbalance favoring the sensing of tRNAPro (Fig. 3). In C. tetani, the Cys biosynthetic genes are also cotranscribed in a cysS-proS operon with a Pro specifier sequence (Fig. 3), making this potential imbalance even more significant, as both Cys biosynthesis and tRNACys charging would therefore be predicted to be regulated in response to the availability of Pro rather than Cys. It is unclear whether there is an additional regulatory strategy (e.g., at the level of transcription initiation or translation) that overcomes this potential problem. Another example of regulation of an operon by an unexpected tRNA is found in the yurG-serAS operon in Pelotomaculum thermopropionicum, which includes genes involved in the biosynthesis of Ser as well as the SerRS gene yet is regulated by a Gly (GGC) T box. This is likely to be explained by the efficient interconversion of glycine and serine in bacterial cells (see below).
Although, in general, there is only one copy of each aaRS gene per genome, there are a few organisms in which multiple copies of an aaRS gene are present. These include tyrS/tyrZ and thrS/thrZ in B. subtilis, each of which is regulated by the T-box mechanism. Bacillus anthracis, B. cereus, and B. thuringiensis have two genes encoding AspRS, one of which is monocistronic, whereas the other is present in a hisS-aspS operon. These two aaRS genes are both predicted to be regulated by the T-box mechanism using a leader RNA with an Asp specifier sequence. These organisms (among others) also encode a third AspRS-related aaRS that mischarges tRNAAsn with Asp to generate Asp-tRNAAsn, which is subsequently converted to Asn-tRNAAsn by an amidotransferase enzyme complex. This mischarging AspRS gene is preceded by a T-box leader that is predicted to respond to uncharged tRNAAsn, consistent with its function (Fig. 3).
aaRS genes are generally regulated by a single T-box element, although examples with tandem T-box arrangements have been identified (Fig. 3). An extreme example is represented by the B. subtilis thrZ gene, encoding an isozyme of ThrRS (80). The leader region of the thrZ operon contains three T-box elements as direct repeats. This arrangement results in tighter regulation, since the binding of three molecules of uncharged tRNAThr is required for transcription to proceed through all three leader region terminators (38, 82). The thrS gene, which encodes the major ThrRS isoenzyme, is regulated by a single T-box element and therefore can be expressed when uncharged tRNAThr first begins to accumulate; ThrRS activity increases the pool of charged tRNAThr, and as a consequence, the transcription of thrZ is rarely induced under normal growth conditions. A similar arrangement is found in B. clausii and Bacillus pumilus, where the thrZ gene is regulated by tandem T-box sequences. This T-box distribution is also found in the leader regions of some operons containing biosynthetic genes (see below).
The coupling of transcription of each aaRS gene with the charging of its cognate tRNA appears to be metabolically beneficial. Reduced levels of an aaRS relative to its uncharged tRNA substrates decrease the extent of charging of the corresponding tRNAs, which promotes the stalling of the translational machinery and eventually triggers mRNA degradation and the stringent response. The increased level of expression of each aaRS when charging of the cognate tRNA is low allows the maintenance of protein synthesis. However, the synthesis of excess levels of an individual aaRS relative to its uncharged tRNA substrate increases the risk of mischarging of a noncognate tRNA. The ability of the T-box mechanism to monitor the ratio between its substrate (uncharged tRNA) and its product (charged tRNA) allows the synthesis of each aaRS to precisely match the physiological requirements of the cell.
The phylogenetic distribution of T-box sequences revealed by our analysis agrees with data from the recent study by Vitreschak et al. (107) showing that the number of aaRS genes regulated by a T box in gram-positive bacteria is highly variable, ranging from high among the Bacillaceae, where most aaRS genes are regulated by a T-box sequence, to low in the Actinomycetes, where the T-box regulation of aaRS genes is uncommon (Fig. 2). This variability may reflect differences in the evolutionary history of each organism as well as differences in their environmental niches.
T-BOX REGULATION OF AMINO ACID BIOSYNTHETIC GENES
The biosynthesis of amino acids is energetically costly; accordingly, the expression of amino acid biosynthetic genes is generally highly regulated. The mechanisms used to regulate the expression of these genes have evolved to respond to changes in the intracellular levels of their free amino acids and/or the relative levels of their corresponding nonaminoacylated and aminoacylated tRNAs (118). In gram-negative bacteria, this regulation often utilizes regulatory proteins that control transcription initiation in response to amino acid availability as well as leader regions that mediate transcription attenuation via leader peptide coding region translation, which is sensitive to the availability of specific charged tRNAs (116, 117). In gram-positive bacteria, regulatory proteins are also used to sense amino acids (29, 97, 98). For example, in B. subtilis and its close relatives, the biosynthetic genes of the trp operon are regulated by tryptophan (Trp) and the TRAP RNA binding protein, which regulates transcription termination (5, 40). In contrast, in many other members of the Firmicutes, the transcriptional regulation of the trp operon is mediated by a T-box RNA that responds to uncharged tRNATrp. The regulation of biosynthetic operons by the T-box mechanism might have evolved in response to metabolic demands, as revealed by our initial genomic analysis of the trp biosynthetic operons of gram-positive bacteria (52) and extended in the present study to genes concerned with other amino acid biosynthetic pathways.
Regulation of Serine and Glycine Biosynthetic Genes by the T-Box and gcvT Riboswitches
The serA gene encodes the enzyme that catalyzes the first reaction in the serine (Ser) biosynthetic pathway. The mechanism of regulation of serA in most members of the Firmicutes is unknown, although in B. clausii, B. halodurans, C. acetobutylicum, and C. tetani, a Ser T-box sequence is located in the serA regulatory region (Fig. 4). In B. clausii and B. halodurans, a Ser (AGC) codon is present in the specifier loop of the serA T-box RNA, while in the other two organisms, the Ser (UCC) codon is present. In C. acetobutylicum and C. tetani, serA is in an operon that also encodes a probable serine-pyruvate/aspartate aminotransferase. This operon also contains the serS gene, encoding SerRS, in C. acetobutylicum. In B. subtilis, the serA paralog yoaD is regulated by the S-box riboswitch, which responds to S-adenosylmethionine (SAM) rather than a tRNA (45).
FIG. 4.
Variety of mechanisms used in regulating methionine and serine biosynthetic genes of B. subtilis and other bacteria. The regulatory mechanisms and operon arrangements found in B. subtilis (left column) are compared with those of T-box-regulated operons in other members of the Firmicutes (right column). Where genes of other organisms share the same regulatory mechanism as B. subtilis, the names of these organisms are indicated in the B. subtilis column. The graphic representation of each type of regulatory element is indicated in the red box in each subfigure. No attempt was made to identify pathways exhibiting feedback inhibition of enzyme activity; only those reported in the literature for B. subtilis are indicated. The regulatory proteins shown represent their corresponding binding sites in the operon. Genes that have not been annotated were labeled based on their corresponding COG numbers (i.e., a gene that belongs to COG1878 is drawn as an arrow containing the number “1878”). Organism abbreviations and color codes are described in the legends of Fig. 2 and 3.
The serC and serB genes encode the enzymes that catalyze the second and third reactions in this pathway, respectively. No information on their regulation is currently available, but our analyses suggest that in C. thermocellum and Desulfitobacterium hafniense, the serC gene is regulated by a Ser (UCC) T-box RNA. In Lactococcus lactis and Streptococcus mutans, serB is located in the his operon and appears to be regulated by a T-box sequence that responds to tRNAHis. Ser can also be synthesized from pyruvate by a one-step enzymatic reaction; the gene encoding this enzyme (designated yurG in B. subtilis) is cotranscribed with serA in C. acetobutylicum, Clostridium botulinum, Clostridium kluyveri, Clostridium novyi, C. tetani, Desulfotomaculum reducens, and Syntrophomonas wolfei. This gene is regulated by a single Ser (UCC) T box in all of the clostridia mentioned above except C. kluyveri, where regulation is controlled by tandem Ser (UCC) T boxes (Fig. 4).
Ser can also be synthesized from glycine by GlyA (46). The glyA gene is commonly regulated by the Gly-responsive gcvT riboswitch, which results in increased expression when glycine is abundant (1, 7, 70). Interconversion of Gly and Ser by GlyA is the primary pathway used for Gly synthesis in bacteria (46, 99). This pathway also produces 5,10-methylenetetrahydrofolate, a major contributor of the one-carbon unit in the formation of methionine (Met), purines, and thymine (75).
The only Gly T boxes identified so far that regulate biosynthetic operons are found in Moorella thermoacetica (yurG-serA) and in P. thermopropionicum (yurG-serAS) (Fig. 5). Although YurG and SerA are directly involved in Ser biosynthesis, the biological relevance of the regulation of these operons in response to uncharged tRNAGly accumulation might be that an increase in the pool of Ser (which can be converted to Gly) would also increase the pool of glycine.
FIG. 5.
Variety of mechanisms used in regulating leucine, isoleucine, valine, and histidine biosynthetic genes of B. subtilis and other bacteria. The color code used for the different types of regulated genes and abbreviations of organisms are described in the legend of Fig. 2. The graphic representation of each type of regulatory element is described in the legend of Fig. 4.
Pathways for Synthesis of the Sulfur-Containing Amino Acids Methionine and Cysteine Are Regulated by S-Box and T-Box Riboswitches
The biosynthesis of Met and cysteine (Cys) can utilize a number of alternate pathways and is regulated by a wide variety of mechanisms, including the T-box and S-box mechanisms. The SAM-responsive S-box riboswitch also regulates the expression of the metK gene, encoding SAM synthetase, reflecting the importance of Met not only as an amino acid but also as a precursor of SAM. A third type of riboswitch, the SMK box, regulates metK expression in lactic acid bacteria including Enterococcus, Streptococcus, and Lactococcus spp. in response to SAM (33). In contrast, DNA binding transcription factors such as MtaR, MetR, and CmbR are utilized in streptococci (67, 86), in common with the regulatory pattern found in E. coli.
Homoserine is a key intermediate in the biosynthesis of Met and Cys, and it also participates in the biosynthesis of Gly and threonine (Thr) (46). Homoserine is derived from aspartate (Asp) by the action of homoserine dehydrogenase, which is encoded by the hom gene. Since the product of the hom gene participates in multiple biosynthetic pathways, it is not surprising to find that this gene is regulated by several different mechanisms. For example, in B. clausii, B. halodurans, D. hafniense, and Thermoanaerobacter tengcongensis, the expression of the hom gene is regulated by an S-box riboswitch in response to SAM, while in C. difficile, the hom gene is regulated by tandem Thr-responsive T boxes (Fig. 5) (46). In B. anthracis, B. cereus, and B. thuringiensis, there are two paralogous copies of the hom gene. One of these hom genes is contranscribed with the metY and metA genes and is regulated by an S-box riboswitch, while the other is cotranscribed with the thrB and thrC genes and is regulated by tandem T boxes, each with a Thr specifier sequence (Fig. 5).
In B. subtilis, homoserine is acetylated by MetA followed by a reaction with Cys to form cystathionine. The metA gene is regulated by the S-box mechanism in a number of organisms, including C. difficile and Staphylococcus sp., while the regulatory mechanism in B. subtilis remains unknown. In contrast, in Lactobacillus plantarum, the metA gene is cotranscribed with a gene encoding O-acetylhomoserine (thiol)-lyase in an operon regulated by the T-box mechanism using a Met specifier sequence. Other organisms (including E. coli) utilize MetA to convert homoserine to O-succinylhomoserine (41, 46). Further steps are catalyzed by cystathionine γ-synthase (YjcI/MetI) and cystathionine β-lyase (YjcJ/MetJ). The genes encoding these two enzymes are cotranscribed in B. subtilis and are regulated by a SAM-responsive S-box riboswitch (45, 46), while in L. plantarum, Leuconostoc mesenteroides, Oenococcus oeni, and Staphylococcus sp., yjcI and yjcJ are regulated by a T box with a Met specifier sequence (Fig. 4). Previously, the identification of Met T boxes in biosynthetic genes had been restricted to the Lactobacillaceae (37, 107), but we can now extend this regulation to the staphylococci.
The final step in the Met biosynthetic pathway is catalyzed by methionine synthase. There are two classes of this enzyme, the B12-dependent class, encoded by metH, and the B12-independent class, encoded by metE. The metE gene is regulated by a T box with a Met specifier sequence in Lactobacillus casei and L. plantarum (Fig. 4), while it is regulated by a SAM-responsive S box in B. subtilis, C. acetobutylicum (45, 46, 56), B. clausii, Listeria innocua, and Listeria monocytogenes. The metH gene is present in a few members of the Firmicutes, and it is regulated by an S-box riboswitch in C. acetobutylicum and Oceanobacillus iheyensis as well as in Deinococcus geothermalis, a member of the Deinococcus-Thermus group. The orthologous yxjG and yxjH genes, which are related to metE, are regulated by an S box in B. subtilis (45). Orthologs of yxjG/yxjH are regulated by T-box elements with a Met specifier sequence in Enterococcus faecalis (45) and many members of the Lactobacillales. The conversion of homocysteine to Met also requires methylenetetrahydrofolate reductase, encoded by the S-box-regulated yitJ gene in B. subtilis. yitJ is cotranscribed with metE in L. innocua and L. monocytogenes and is regulated by the S-box mechanism, but in L. mesenteroides, yitJ is monocistronic and is regulated by a Met-responsive T-box element (Fig. 4). Met can also be synthesized by the recycling of methylthioadenosine, which is generated as a by-product of polyamine biosynthesis, in a number of organisms including B. subtilis (46, 77, 95). Genes involved in this pathway in Bacillus sp. are regulated by SAM via the S-box riboswitch (45).
Homocysteine can also be recycled via the activated methyl cycle (22). This pathway employs the LuxS protein, which recycles the toxic intermediate S-adenosylhomocysteine to yield homocysteine as well as 4,5-dihydroxy-2,3-pentanedione (22). 4,5-Dihydroxy-2,3-pentanedione is then spontaneously rearranged into autoinducer-2, which is a key molecule in quorum sensing (92). In some members of the Lactobacillaceae, luxS is regulated by a Met-specific T-box RNA (Fig. 4). This is the first reported example of a gene involved in quorum sensing regulated by a T-box riboswitch.
As noted above, the Cys biosynthetic gene cysE and the cysteinyl-tRNA synthetase gene cysS are cotranscribed and regulated by a T-box sequence with a Cys (UGC) codon in many members of the Firmicutes (35) (Fig. 5). Cys can also be synthesized by the reduction of sulfate compounds such as thiosulfate, which can be converted to S-sulfocysteine (46). This reaction is carried out by cysteine synthase, the product of cysK, which also participates in the conversion of Ser to Cys. The cysK gene is regulated in response to tRNACys by the T-box mechanism in C. acetobutylicum, C. beijerinckii, C. botulinum, C. kluyveri, C. perfringens, L. plantarum, and Staphylococcus epidermidis (Fig. 5). The transcription of cysK is regulated by the CymR DNA binding protein in B. subtilis (29).
Some bacterial species, including B. subtilis, can also synthesize Cys by the reverse trans-sulfuration pathway using Met as a precursor (71, 86). The two genes involved in these reactions are yrhA (encoding cystathionine β-synthase) and yrhB (encoding cystathionine γ-lyase), which are commonly found in the same transcriptional unit. In C. acetobutylicum (46), this operon is regulated by the T-box mechanism using a Cys specifier sequence, consistent with its role in Cys biosynthesis (Fig. 5). It is interesting that yrhB is closely related to the yjcI and yjcJ genes, which encode enzymes that catalyze the conversion of Cys to homocysteine and which respond to Met (or SAM) accumulation rather than to an increase in the Cys level.
The Branched-Chain Amino Acids Isoleucine, Leucine, and Valine and Their Relationship to the Pantothenate Pathway
Pyruvate is the common precursor of the branched-chain amino acids (BCAAs) isoleucine (Ile), leucine (Leu), and valine (Val). The biosynthetic pathways for these amino acids share the first four enzymes, encoded by ilvC, ilvD, ilvE, and ilvB-ilvN, the products of which form a heterodimeric enzyme complex. In the Firmicutes, these genes are rarely organized in a single transcription unit: ilvB, ilvN, and ilvC are commonly found within the ilvBNC-leuABCD operon, while ilvD and ilvE are usually monocistronic. In addition, B. anthracis, B. cereus, and B. thuringiensis contain paralogous copies of the ilvB, ilvN, and ilvC genes that are cotranscribed within the ilvEBNCDA operon. These operons are regulated by tandem T-box riboswitches, both of which contain an Ile (AUC) specifier sequence (Fig. 6). Therefore, their expression would require a more substantial decrease in tRNAIle charging since two molecules of uncharged tRNAIle are necessary to promote the readthrough of both terminators. This operon could be considered as a backup when tRNAIle charging is critically low, by analogy with the regulation of the B. subtilis ThrRS genes (38, 82).
FIG. 6.
Variety of mechanisms used in regulating alanine, arginine, asparagine, aspartate, cysteine, glycine, and threonine biosynthetic genes of B. subtilis and other bacteria. The color code used for the different types of regulated genes and abbreviations of organisms are described in the legend of Fig. 2. The graphic representation of each type of regulatory element is described in the legend of Fig. 4.
In B. subtilis and its closest relatives, the ilv and leu genes are organized in a single operon, ilvBNC-leuABCD, which is preceded by a T-box leader that responds to uncharged tRNALeu (Fig. 6). This could result in an imbalance during growth under Leu-rich conditions if Ile and/or Val were limiting. A second level of regulation uses the CodY DNA binding protein to repress ilvB promoter activity in response to the availability of Ile and Val. This ensures that the intracellular levels of all three BCAAs have an impact on the regulation of this operon (96). The ilvBNC-leuABCD operon in B. subtilis is also regulated by TnrA, which responds to nitrogen limitation (32), and CcpA, which is active in glucose-grown cells and prevents repression by CodY (34, 36, 109).
C. beijerinckii has two ilv transcription units belonging to the T-box regulon, ilvH-leuACDB (Ile specifier sequence) and ilvBH (Val specifier sequence). In C. thermocellum, leuA is regulated by tandem Leu T boxes (Fig. 6). The genome of this organism also contains a Leu T-box-regulated transcriptional unit annotated as specifying an uncharacterized cyclic AMP-dependent synthase and ligase; we suggest that either this gene has an unknown role in BCAA metabolism or it is incorrectly annotated. Finally, C. kluyveri has a large number of operons predicted to be regulated by T boxes that respond to BCAAs; most of these genes correspond to canonical BCAA biosynthetic genes, but others represent unexpected examples of T-box-regulated genes and will be discussed below.
The regulation of BCAA biosynthesis by the T-box mechanism is not restricted to the Firmicutes. The leuA genes of Geobacter and Pelobacter spp. are regulated by a Leu T box (Fig. 6). In contrast to the conclusions reported by Vitreschak et al. (107), we suggest that the DNA region responsible for this regulatory mechanism could have been acquired by horizontal gene transfer (HGT) since this would be the more parsimonious scenario considering the great phylogenetic distances between the Firmicutes and the Deltaproteobacteria (see below) (Fig. 2).
We also identified genes in L. plantarum, L. reuteri, and L. salivarius that participate in pantothenate biosynthesis and are predicted to be regulated by BCAA T boxes (Fig. 6). This finding is particularly interesting considering that the biosyntheses of Val, Leu, and pantothenate share α-ketoisovalerate as a common precursor (110). One possible explanation for the use of this type of regulation for the pantothenate (pan) biosynthetic genes would include the sensing of a metabolic stage requiring an increased synthesis of the BCAAs and their precursors.
Histidine Biosynthesis: Possible Consequences of a Weak tRNA-T-Box Interaction
In almost all bacteria for which sequence information is available, including all members of the Firmicutes, the genes for histidine (His) biosynthesis are clustered in a single operon (4). The first reaction in the pathway, the condensation of ATP with 5-phosphoribosyl-1-pyrophosphate to form N′-5′-phosphoribosyl-ATP, is catalyzed by the enzyme N′-5′-phosphoribosyl-ATP transferase (4). This reaction is performed by an octameric enzyme complex composed of polypeptides encoded by the hisG and hisZ genes, which often are the first two genes in the operon (11, 23, 24). The majority of these his genes are arranged in the order hisZGDBHAFIE. The his operon is only rarely regulated by the T-box mechanism; most examples are found in members of the Lactobacillales, including L. lactis (24) and S. mutans, both of which show the unusual hisCZGC-serB-hisB-ymdC-hisHAFIK gene order (Fig. 6). In contrast, the his operons of L. casei, L. plantarum, and L. mesenteroides are regulated by T-box RNAs with a His (CAC) specifier sequence despite the presence of the hisZGDBHAFIEC gene organization typical of his operons that are not regulated by the T-box mechanism. The rarity of T-box regulation for the genes of this pathway, as well as for HisRS genes, may be due to the fact that the acceptor arm of tRNAHis has only three unpaired nucleotides (CCA) at its 3′ end; tRNAHis therefore lacks one of the unpaired nucleotides involved in the interaction of tRNAs with the antiterminator bulge. The absence of the fourth position of pairing could potentially result in a lower efficiency of tRNA-dependent antitermination. In gram-negative bacteria, the his biosynthetic genes are also clustered within a his operon, and the transcription of this operon is regulated by transcription attenuation in response to the accumulation of uncharged tRNAHis, which is sensed by the translation of sequential his codons in a leader peptide coding region (115). The activity of the HisG enzyme is also highly regulated by feedback inhibition (4).
Aromatic Amino Acid Biosynthesis: Prediction of Tight Regulation by Tandem T Boxes
Chorismate is the common precursor of all three aromatic amino acids. It can yield anthranilate, which is used to synthesize Trp, or prephenate, which is the common precursor of phenylalanine (Phe) and tyrosine (Tyr). Several of the proteins participating in chorismate biosynthesis are encoded by genes that are located in the aro-trp supraoperon in B. subtilis and its closest relatives (40, 59, 74). In contrast, in the other members of the Firmicutes, these genes are generally not clustered with the biosynthetic trp genes, and they exhibit a very diverse chromosomal organization (Fig. 7) (52, 53).
FIG. 7.
Variety of mechanisms used in regulating phenylalanine, proline, tryptophan, and tyrosine biosynthetic genes of B. subtilis and other bacteria. The color code used for the different types of regulated gene and abbreviations of organisms are described in the legend of Fig. 2. The graphic representation of each type of regulatory element is described in the legend of Fig. 4. TRAP, in addition to transcriptionally regulating the trp operon in B. subtilis and its closest relatives, can also regulate trpE translation by binding to the trpE leader RNA and promoting the formation of a secondary structure that sequesters the SD sequence, inhibiting translation initiation (73).
aroA encodes 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase, which catalyzes the first step in chorismate formation. This gene is regulated by the T-box mechanism (sensing tRNATyr, tRNAPhe, or tRNATrp) or by another mechanism that is yet to be determined. Examples of aroA genes that are regulated by a T-box sequence are found in B. anthracis, B. cereus, and B. thuringiensis (Tyr specifier sequence); C. thermocellum and M. thermoacetica (Phe specifier sequence); and T. tengcongensis and Dehalococcoides sp. (Trp specifier sequence) (Fig. 7). Alternatively, examples of aroA genes that are regulated by Trp-activated TRAP RNA binding can be found in S. wolfei and Carboxydothermus hydrogenoformans. Since chorismate is the precursor of all three aromatic amino acids, a regulatory mechanism that senses only a single amino acid could result in imbalanced regulation if only one of the aromatic amino acids is available. To deal with this imbalance, some organisms have paralogous copies of aroA genes that may respond to other aromatic amino acids. This expectation is supported by the genomic context of the aroA paralogs, which are clustered with phe biosynthetic genes in S. wolfei and C. hydrogenoformans and with tyr biosynthetic genes in C. hydrogenoformans, C. thermocellum, and M. thermoacetica. The number of aroA genes per organism varies, from one in Bacillus sp. to five in S. wolfei. In S. wolfei, one copy of aroA is cotranscribed with pheA and is regulated by a Phe (UUC) T box (Fig. 7), while a second copy of aroA is cotranscribed with the trp biosynthetic genes and is regulated by Trp-activated TRAP.
AroF catalyzes the last common step in the aromatic pathway, the reaction that results in the synthesis of chorismate. In B. anthracis, B. cereus, and B. thuringiensis, the gene that encodes this enzyme is part of the aroF-hisC-tyrA-aroE operon, which, like aroA, is regulated by a T-box sequence that responds to tRNATyr. As noted above, this could result in a deficiency in the sensing of Trp and Phe. The regulation of expression of the aroF gene has not yet been described for the remaining members of the Firmicutes, with the exception of B. subtilis, where aroF is in the aro-trp supraoperon.
Chorsimate can be converted to prephenate by the aromatic aminotransferase AroH. Prephenate can be converted to tyrosine and phenylalanine by the enzymatic reactions carried out by the PheA/TyrA, HisC, and HisH enzymes. The transcription of the genes encoding these enzymes is regulated by different schemes among the Firmicutes. For example, in B. anthracis, B. cereus, and B. thuringiensis, the transcription of tyrA is regulated by a Tyr (UAC) T-box RNA, whereas in D. hafniense and S. wolfei, pheA is regulated by a Phe (UUC) T-box RNA, and in L. casei, L. lactis, L. plantarum, and S. mutans, hisC and hisH (which encode the enzymes that catalyze some of the last steps of the His, Tyr, and Phe pathways) are located within the his operon and are regulated by a His (CAC) T-box RNA (Fig. 6 and 7).
In B. anthracis, B. cereus, and B. thuringiensis, there is an additional route for the synthesis of Tyr from Phe via the participation of the biopterin compound. This reaction is performed by enzymes encoded in the pah-dcoH operon, which is regulated by tandem Tyr (UAC) T-box RNAs that are predicted to respond to tRNATyr (Fig. 7).
After the common aromatic pathway diverges, seven enzymatic reactions lead to Trp biosynthesis. The regulation of this pathway is particularly important due to the high energetic cost of Trp synthesis. The trp operon of the gram-negative bacterium E. coli has been thoroughly analyzed, and it has been established that the regulatory mechanisms used sense both tRNATrp charging and the intracellular level of free Trp (reviewed in reference 118). These two signals are also sensed by the gram-positive bacterium B. subtilis but by regulatory mechanisms different from those used by E. coli (74, 118). In B. subtilis, free Trp inhibits trp operon expression by activating the TRAP protein, which binds to trp leader RNA and promotes transcription termination (39, 40). The level of charged tRNATrp is sensed in B. subtilis by the rtpA-ycbK operon (15, 16, 104, 105). The first gene of this operon encodes an anti-TRAP (AT) protein, which can inhibit Trp-activated TRAP and prevent it from terminating transcription in the trp operon (105). The expression of the AT operon is transcriptionally regulated by uncharged tRNATrp via the T-box mechanism (104, 105). In B. subtilis, AT synthesis is also regulated translationally by a leader peptide-coding region containing three Trp codons (15, 16). In this case, whenever the charged tRNATrp level is insufficient to allow the rapid translation of the three leader peptide Trp codons, the ribosome synthesizing the leader peptide stalls, exposing the rtpA SD region for an efficient initiation of translation of the rtpA coding region. When there is sufficient Trp to allow the ribosome synthesizing the leader peptide to reach its stop codon, this ribosome blocks the adjacent rtpA SD sequence, inhibiting AT synthesis (16). Although AT has been found only in B. subtilis, Bacillus amyloliquefaciens, and B. licheniformis, the distribution of TRAP is more widespread (reviewed in reference 53). It is not clear whether organisms that use TRAP but not AT have an alternate mechanism for sensing the level of charged tRNATrp.
In contrast to the regulatory mechanisms used by B. subtilis and its close relatives, the trp biosynthetic genes of the vast majority of the Firmicutes are generally concerned solely with Trp biosynthesis and are organized as a single operon, which is regulated by a T-box element in response to the accumulation of uncharged tRNATrp (52, 53) (Fig. 7). It is not known if these organisms can sense free Trp as a regulatory signal. The trp biosynthetic genes of C. thermocellum are organized into two different operons. This constitutes an interesting example of the coordinate regulation of the Trp biosynthetic pathway genes by two different regulatory mechanisms: TRAP, sensing l-Trp, regulates the trpEGDCF operon, and a T box, sensing tRNATrp, regulates the trpBA operon (Fig. 7). The regulatory elements of these two operons were incorrectly assigned in our previous genome analysis of the evolution and regulation of the trp biosynthetic genes (74).
Tandem T boxes are found in the trp operons of several members of the Firmicutes (52, 53). The presence of tandem T boxes implies that these organisms require the accumulation of a higher relative level of uncharged tRNATrp to allow an appreciable expression of the trp operon, since the binding of multiple tRNATrp molecules is needed for transcriptional readthrough (52). Interestingly, the trp operon of these species is a discrete Trp biosynthetic unit containing only the seven Trp pathway genes, unlike the aro supraoperon, which contains only six of these genes in combination with other aromatic amino acid biosynthetic genes. This gene organization is consistent with a tight, specific response to tRNATrp.
Several organisms appear to lack crucial portions of the Trp biosynthetic pathway. The facultative pathogen C. novyi has only the trpB gene, which encodes a polypeptide that catalyzes the last reaction in Trp biosynthesis. Although in this organism, the trp pathway is incomplete, this gene is in an operon regulated by a Trp T box (Fig. 7). It seems likely that this organism acquires indole from its host and converts it to Trp by the action of TrpB. The lactic acid bacterium L. casei lacks the trpE and trpG genes, which is consistent with its habitat in the human gut and mouth, where it presumably can find an adequate supply of Trp precursors. The trpDCFBA operon of this organism is regulated by a Trp T box (Fig. 7).
T-box elements were also found upstream of the trp genes in members of the group Chloroflexus. In Dehalococoides sp., T-box regulation was observed in the trp operon as well as in a trpB paralog, which encodes an alternative tryptophan synthetase beta subunit (53). On the other hand, in Roseiflexus sp., only the trpB paralog gene is regulated by a Trp T box, and the mechanism used for the regulation of the trp operon is unknown. Several members of the Firmicutes including C. kluyveri, C. novyi, D. hafniense, Streptococcus gordonii, and Streptococcus sanguinis also have a trpB paralog that is regulated by a Trp T box (Fig. 7).
Biosynthetic Genes for Aspartate and Asparagine, Key Precursors of Many Other Amino Acids
The Krebs cycle intermediate oxaloacetate is the common precursor of Asp and asparagine (Asn). Oxaloacetate is converted to Asp by an aspartate aminotransferase, the product of the aspB gene. In B. subtilis, the expression of aspB does not respond to the presence of Asp and appears to be constitutive (8, 63). All low-G+C gram-positive bacteria have multiple genes encoding proteins similar to AspB (8), although most of these genes have not been characterized biochemically. The aspB genes and the majority of their paralogs are not regulated by a T-box mechanism, although in C. hydrogenoformans and C. perfringens, aspB is cotranscribed with asnC (which encodes a putative transcriptional regulator) in an operon preceded by an Asp T-box sequence (Fig. 5). Since Asp is a key metabolite in the synthesis of many other amino acids, such as Asn, lysine (Lys), Thr, Met, and Ile, it is not surprising that the genes responsible for its synthesis are often expressed constitutively.
Asp can also be synthesized by the action of asparaginase, encoded by the genes of the ansAB operon (8, 101). This operon is expressed during vegetative growth in B. subtilis but does not contribute significantly to Asp synthesis (8). Its regulation does not depend on the T-box mechanism, but instead, it is subject to strong repression by the Asn-responsive transcriptional regulator AsnR during the early stages of sporulation (102).
In the Firmicutes, Asn can be synthesized from Asp, either in its free form or after it is charged onto tRNAAsn (by AspRS), or from glutamine (Gln). In the first strategy, Asn is synthesized from Asp by an asparagine synthetase, AsnA (8). In Lactobacillus delbrueckii subsp. bulgaricus (66), L. plantarum, and L. reuteri, asnA is cotranscribed with asnS (encoding AsnRS) and is regulated by an Asn T box, whereas in B. anthracis, B. cereus, B. thuringiensis, C. perfringens, C. tetani, Lactobacillus acidophilus, Lactobacillus brevis, Lactobacillus gasseri, Lactobacillus johnsonii, and Pediococcus pentosaceus, asnA is monocistronic and regulated by an Asn T-box element (Fig. 5).
The second route to Asn synthesis is by the transamidation of Asp. Using this mechanism, a nondiscriminating AspRS charges Asp not only onto tRNAAsp but also onto tRNAAsn. Subsequently, a tRNA-dependent Asp-tRNAAsn amidotransferase converts Asp to Asn to form Asn-tRNAAsn (61, 62). This heterotrimeric enzyme, encoded by the gatCAB genes, also carries out the transamidation of Glu-tRNAGln to Gln-tRNAGln (61). In C. acetobutylicum, the gatCAB genes are cotranscribed with the gene encoding the nondiscriminatory AspRS in a transcription unit regulated by an Asn T box, a regulatory pattern that is consistent with the role of this operon in the generation of Asn-tRNAAsn (Fig. 5).
Asn can also be synthesized from Gln by a glutamine amidotransferase. Three enzymes of this class, AsnB, AsnH, and AsnO, have been found in B. subtilis (8, 120), and none of these is regulated by the T-box mechanism; in contrast, asnB in C. kluyveri is regulated by an Asn T box (Fig. 5). In B. subtilis, asnB and asnH are expressed almost constitutively, although the level of asnH expression decreases somewhat in response to the accumulation of excess Asn. asnO expression is restricted to stationary phase and is dependent on the σE sporulation sigma factor (8, 120). In other members of the Firmicutes, the mechanism of regulation of these genes has not yet been described.
Alanine Biosynthesis Involves T-Box Regulation of Operons Containing Biosynthetic and Regulatory Genes
Alanine (Ala) is synthesized by the transamination of pyruvate. In several members of the Clostridium group, the aminotransferase gene alaT is cotranscribed with alaR, a putative transcriptional regulator of the Lrp/AsnC family (8) (Fig. 5) (see below). The alaRT operon in these organisms contains a T-box sequence with an unusual Ala specifier sequence (GCA or GCC), in contrast to most other tRNAAla-regulated T-box genes that contain a GCU specifier sequence. The common use of GCU is an exception to the preference for codons ending in C for most tRNA classes. We note that in several of these organisms, alaT is incorrectly annotated as aspB (alaT and aspB are 38% identical). The identification of the specifier sequence of the regulatory sequence, and recognition of the similarity to B. subtilis alaR-alaT (8), permits us to assign these genes to the alanine biosynthetic pathway (in agreement with data described previously by Vitreschak et al.) (107).
Threonine Biosynthesis
In all bacteria, the biosynthesis of Thr from Asp involves five steps that are catalyzed by enzymes encoded by the thrD, asd, hom, thrB, and thrC genes (8, 78, 79). As previously mentioned, the hom gene also participates in the biosynthesis of Met (46), and this gene is regulated either by the S-box riboswitch mechanism in response to SAM or by a T-box mechanism that responds to tRNAThr or tRNAMet. S-box-regulated hom genes are found in B. clausii, B. halodurans, D. hafniense, and T. tengcongensis, while hom genes with Thr T-box regulation are in C. difficile (46), C. kluyveri, B. anthracis, B. cereus, and B. thuringiensis. In the last three organisms, hom is cotranscribed with thrB and thrC, and the operon is preceded by two tandem T boxes, each responding to tRNAThr (Fig. 5). In addition, Hom activity can be repressed by feedback inhibition in response to the presence of methionine, isoleucine, and possibly threonine, as has been shown for B. subtilis (119).
Proline Biosynthesis
In B. subtilis, proline (Pro) is synthesized from Gln in three enzymatic steps (8). The first reaction is carried out by ProB (and its paralog, ProJ), which catalyzes the conversion of Gln to γ-glutamyl phosphate. The second reaction involves the synthesis of Δ1-pyrroline 5-carboxylate by the action of ProA. The two genes encoding these proteins are cotranscribed, and their transcription is regulated by the T-box mechanism using a Pro (CCC or CCU) specifier sequence in B. clausii, B. halodurans, B. licheniformis, B. subtilis (17), B. anthracis, B. cereus, B. thuringiensis, and D. hafniense (Fig. 7). The third reaction in Pro synthesis is catalyzed by Δ1-pyrroline 5-carboxylate reductase, encoded by proC. In addition to ProC, three other proteins (ProI, ProG, and ProH) carry out a ProC-like function. The activity of any of these enzymes is sufficient for Pro biosynthesis (8). In B. clausii, B. halodurans, and D. hafniense, proA and proB are organized with proI in a single operon, whereas in B. anthracis, B. cereus, and B. thuringiensis, the proBA operon is transcribed divergently from proC (3, 9). proI is monocistronic in some members of the Firmicutes and is regulated by a Pro (CCU) T box in B. licheniformis, B. pumilus, and B. subtilis (Fig. 7). The proHJ operon is transcribed under osmotic stress conditions from a σA-type promoter in B. subtilis. Its transcription guarantees the synthesis of a higher intracellular level of Pro, presumably for use as an osmoprotectant (12). The comER gene, a homolog of proG, is expressed during vegetative growth (54) and also during sporulation from a σE-type promoter (30). The complex regulation of the pro genes in the Firmicutes contrasts with their constitutive expression in E. coli (8) and may relate to the dual role of Pro in protein synthesis and osmoprotection in the Firmicutes.
Regulation of Arginine Biosynthesis in the Firmicutes Is Mediated Predominantly by a DNA Binding Transcriptional Repressor Protein
The B. subtilis genes that are responsible for synthesizing arginine (Arg) from glutamate (Glu) are organized into two operons, argCJBD-carAB-argF and argGH. Both operons are commonly regulated by the ArgR DNA binding transcriptional repressor protein, which uses Arg as its corepressor (8). argR/ahrC-like genes have been detected in the genomes of all low-G+C gram-positive bacteria except C. difficile (8). In this organism, Arg (AGA) T-box riboswitches were identified in the regulatory regions of the argCJBMF, argG, and argH transcriptional units (Fig. 5). It is unclear why C. difficile is unique in its use of the T-box mechanism for the regulation of Arg biosynthesis. Analysis of additional genomes may reveal other organisms in this group.
Amino Acid Biosynthetic Pathway Genes That Are Not Regulated by the T-Box Mechanism
Analysis of 559 fully sequenced bacterial genomes failed to detect T-box regulation for operons involved in the biosynthetic pathways for Lys, Gln, and Glu. In these pathways, gene regulation is performed by a broad repertoire of regulatory mechanisms that include (i) DNA binding transcriptional regulatory proteins, e.g., the GltC repressor used for regulating operons involved in Glu biosynthesis (10); (ii) metabolite binding riboswitches, e.g., the Lys-responsive L box used for regulating operons of Lys biosynthesis (48, 85); and (iii) posttranslational regulation by feedback inhibition, as occurs in the regulation of synthesis of glutamine synthetase, a key enzyme in the Gln biosynthetic pathway (26, 93). It is possible that examples of T-box-regulated genes for these pathways will be identified as additional genome sequences become available.
REGULATION OF AMINO ACID TRANSPORTER GENES
Many genes encoding proteins involved in amino acid transport have been identified in operons regulated by the T-box mechanism. As these transport proteins increase the intracellular pool of their respective amino acids, regulation in response to tRNA charging provides an attractive mechanism for coupling the expression of a transporter gene to intracellular pools of its substrate. In the present study, we identified 34 different families of orthologous genes (COGs) encoding amino acid transporters that are regulated by a T-box sequence. T-box regulation of transporter genes appears to occur exclusively in the Firmicutes (Fig. 2 and 8).
Most of the transporters whose synthesis is regulated by the T-box mechanism are annotated as BCAA transporters or as members of the ABC transporters. The majority of the transporter gene operons possess only one T-box sequence, although there are a few examples with tandem T-box sequences (Fig. 8). These transporter genes are found in either monocistronic or polycistronic operons, where the genes presumably encode different subunits of the same transmembrane protein complex. Transporter genes are occasionally cotranscribed with biosynthetic genes. This has been observed for genes of the Met biosynthetic pathway in some members of the Lactobacillales. For example, the metQ transporter gene is cotranscribed with metA, which encodes a homoserine trans-succinylase, in L. delbrueckii. This operon also contains a gene that encodes the LuxS S-adenosyl homocysteine recycling protein (Fig. 5).
Analysis of the amino acid sequence of a transporter protein does not always provide sufficient information to allow the prediction of its substrate. The identification of a T-box sequence (and its corresponding specifier sequence) upstream of a transporter gene therefore provides valuable information not only on which uncharged tRNA regulates the expression of the gene but also for the prediction of the amino acid likely to be transported. For example, the yvbW gene of B. subtilis, annotated as a “hypothetical protein,” is related to the “gamma-aminobutyrate permease” group of genes. This gene has a T-box leader sequence with a Leu (CUC) specifier sequence. Consistent with the prediction that yvbW encodes a transporter of Leu or a related compound, yvbW expression was shown to be induced upon Leu limitation (87). Some recent reports have annotated the specificity of a large group of amino acid transporters that had previously been poorly characterized and were classified only as “hypothetical proteins” or “BCAA permeases” (107, 111). We further characterized the T-box regulation of these groups of amino acid transporters and observed that ∼70% of these genes are regulated by tRNAIle, while ∼15% respond to tRNAThr; the remainder are regulated by tRNALeu, tRNAVal, and tRNAPhe. This suggests that while most of these genes are likely to encode BCAA permeases, other members of this group may transport other substrates.
The predictive power of a T-box leader sequence can be used to identify incorrect gene annotations. For example, an orthologous operon of uncharacterized proteins present in members of the Lactobacillales contains genes related to ABC-type transporters. These operons are regulated by a T-box element with a His (CAC) specifier sequence, suggesting that the product is a His transporter. In L. plantarum, the genes of this operon are annotated in the GenBank database as Gln transporters (glnPQH), which is likely to be incorrect. We also identified a number of other transporter genes misannotated as glnPQH homologs; we predict from their T-box leader specifier sequences that they are involved in the transport of Asp, Asn, Phe, Cys, Met, and Ser (Fig. 8). Correcting errors of this type is important, as further annotation is likely to lead to the repeated misannotation of related genes in other genomes.
Shared Regulatory Mechanisms for Biosynthetic and Transporter Genes
Comparisons of the mechanisms employed in regulating gene expression have revealed that in many cases, a common regulatory mechanism is used (e.g., involving a T-box, L-box, or SAM riboswitch or the TRAP RNA binding protein) by an organism to regulate both the corresponding biosynthetic genes and amino acid transporter genes (Fig. 9). For example, in B. subtilis and closely related species, both the trp operon and the trpP tryptophan transporter gene are regulated by the TRAP RNA binding protein (90). However, in C. acetobutylicum, C. beijerinckii, C. kluyveri, C. novyi, and S. gordonii, the trp biosynthetic operon and trp transporter gene are regulated by the T-box mechanism. A second example can be found in C. difficile, which regulates both Arg biosynthetic and transporter genes by the T-box mechanism. In contrast, in other organisms, the T-box mechanism is used only for the ArgRS gene, and the biosynthetic and transporter genes are regulated by the ArgR transcriptional repressor. Similarly, in a large number of members of the Firmicutes, both the Lys biosynthetic operon and the Lys transport operon are regulated by a lysine-responsive L-box riboswitch (1, 48, 85), and Met biosynthetic and transport operons are regulated by a SAM-responsive S-box riboswitch (37, 58, 86) (Fig. 9).
FIG. 9.
Common strategy in regulating amino acid transporter genes and biosynthetic genes. Both amino acid transporters and biosynthetic enzymes can fulfill the need of an organism for certain amino acids. This results in a tendency to coordinate the expression of the genes for these classes of proteins by using a shared regulatory mechanism. As shown for three amino acid-related genes, this tendency is specific for each phylogenetic clade. The regulatory elements were identified using our Riboswitch Web server (RibEx) (1) and previously reported data (2). [1], the SMK riboswitch regulates metK genes in lactic acid bacteria, including Enterococcus, Streptococcus, and Lactococcus spp. (see reference 33). [2], in streptococci, unlike other members of the Firmicutes, methionine biosynthesis and transport are controlled by protein transcription factors, in this case, MtaR, MetR, and CmbR (see reference 67). Organism names and gene color codes are described in the legends of Fig. 2 and 3.
Due to the prevalence of the T box as a regulatory element in the Firmicutes, the most common use of shared regulation by biosynthetic and transporter genes involves the T-box mechanism. All of these examples may reflect the need of the organism to acquire the missing amino acid by coordinating biosynthesis with transport. It is also possible that T-box riboswitches that regulate different classes of genes in the same organism in response to the same effector tRNA may be differentially sensitive to the level of that uncharged tRNA and thereby allow differential regulation. Differential responses to SAM pools (which correlate with variability in the affinity of the riboswitch RNA for SAM) were observed for S-box-regulated genes in B. subtilis; this results in the expression of genes encoding a Met transporter when SAM pools are relatively high, whereas the expression of genes involved in Met biosynthesis remain repressed until SAM pools drop to a lower level (103). A similar variability in affinity for the cognate uncharged tRNA could allow an increased synthesis of the corresponding transporter prior to the induction of the genes for the full biosynthetic pathway. The possibility of differential regulation using the T-box mechanism requires further experimental study.
REGULATION OF SYNTHESIS OF REGULATORY PROTEINS
The expression of several operons encoding regulatory proteins is regulated by the T-box regulatory mechanism. In these examples, T-box-mediated control has a broader effect, as it influences the expression of all of the genes responding to each regulatory protein. Almost all regulatory genes predicted to be controlled by a T-box sequence are cotranscribed with an aaRS gene or with genes involved in amino acid synthesis or transport (Fig. 10). Thus, in L. innocua, L. monocytogenes, and Listeria welshimeri, a gene annotated as a “regulator of competence-specific genes” is cotranscribed with serS under the control of a Ser T-box sequence. The function of this putative regulatory protein is unknown, but it is likely that it plays a role in the regulation of serine metabolism. Similarly, in O. oeni, a gene annotated as a putative regulator is cotranscribed with pheST (encoding PheRS) and is regulated by a Phe T box. Examples of regulatory proteins that are cotranscribed with biosynthetic genes are found in the Ala T-box-regulated alaRT operon in members of the Clostridium group. The alaR gene encodes a transcriptional regulator of the Lrp/AsnC family, members of which are involved in modulating a variety of metabolic functions including the catabolism and anabolism of certain amino acids (13). The second gene of the operon, alaT, encodes an alanine transaminase, which can synthesize Ala from pyruvate (these genes are often misannotated as asnC aspB) (see above). In B. pumilus, an AraC family transcriptional regulator is cotranscribed with the biosynthetic gene leuA and is regulated by a Leu (CUC) T-box region (Fig. 10). Members of the AraC family regulate genes involved predominately in carbon source catabolism, the stress response, and virulence (60). An AraC family protein was reported to affect the regulation of a Val biosynthetic operon in Pseudomonas aeruginosa (100), which is consistent with our finding of an AraC-like regulator involved in the biosynthesis of Leu. In Roseiflexus sp., a monocistronic gene annotated as a “putative transcriptional regulator of the MerR family” is regulated by a Phe (UUC) T box. In this case, the presence of the T-box regulatory sequence provides the only clue relating this gene to Phe metabolism.
As noted above, the T-box mechanism plays an important secondary role in the regulatory events that modulate the synthesis of Trp in B. subtilis. A Trp T-box sequence regulates the transcription of rtpA, the gene encoding the AT regulatory protein, which modulates the activity of the major Trp-activated regulatory protein, TRAP, in response to the accumulation of uncharged tRNATrp (Fig. 10) (105). AT synthesis is also influenced by the translation of the three Trp codons in the coding region for a 10-residue regulatory leader peptide located in the at operon (16). Hence, a T-box regulatory region that senses the level of uncharged tRNATrp helps to define the overall fate of the regulatory cascade that modulates trp operon expression. The rtpA gene, encoding the AT regulatory protein, is cotranscribed with the ycbK gene, which encodes a presumed tryptophan transport protein, providing another example of the coordinate regulation of amino acid biosynthesis and transport (91).
OTHER IMPORTANT FEATURES OF THE T-BOX MECHANISM
ileS Is the Gene Most Widely Regulated by the T-Box Mechanism
In our computer analysis of 559 genome sequences, ileS, encoding IleRS, was the gene most often predicted to be regulated by the T-box mechanism. Our search identified T-box-regulated ileS genes in 57 members of the Firmicutes, 19 members of the Actinobacteria, 5 members of the Chloroflexi, and 2 members of the Deinococcus-Thermus group. The presence of a T-box region preceding ileS is most evident in some groups of the pathogenic Firmicutes (organisms that often lack genes for amino acid biosynthesis and rarely use the T-box regulatory mechanism) and in members of the Actinobacteria, which have predominantly only one T-box-regulated gene per genome. In each of these groups, the few T-box-regulated genes include ileS (Fig. 2 and 3).
Over- and Underrepresentation of T-Box Regions in Genomes
Among the organisms for which complete genomes are currently available, B. thuringiensis appears to employ the T-box mechanism most widely (Fig. 2). We identified 40 putative T-box-regulated operons in its genome; these represent more than 1% of its transcriptional units. The closely related species B. anthracis and B. cereus also appear to use the T box frequently, with 38 putative T boxes in their genomes. A likely explanation for the abundance of T-box-regulated genes in these organisms is their expanded capacity for amino acid and peptide uptake. B. thuringiensis and its closest relatives appear to encode a large number of ABC-type peptide binding proteins and BCAA transporters. This property may reflect the adaptation of these organisms to a protein-rich environment, as exists in decaying animal matter (84).
Single versus Tandem T-Box Elements
A T-box regulatory region is generally present as a single unit. However, as noted above, there are several examples where tandem T-box sequences are evident, most commonly upstream of biosynthetic operons. Within this group, the operons encoding the enzymes of the trp pathway illustrate this bias, since of 42 organisms with T-box-regulated trp operons, 12 have tandem T boxes (Fig. 7). This preference is also observed in operons used for BCAA biosynthesis although to a lesser extent. As mentioned previously, the thrZ gene of B. subtilis, encoding a paralog of ThrRS, is the only known example of a leader sequence with three tandem T-box elements; the presumed tight repression of expression of this gene ensures that the thrS-encoded major ThrRS is active under normal conditions, with thrZ expression occurring only when the levels of Thr-tRNAThr drop very low (38, 82).
T-Box Sequences Containing a tRNA Gene
Our genome searches for T-box sequences revealed that in certain members of the Clostridia (S. wolfei, P. thermopropionicum, and C. hydrogenoformans), leuS is preceded by a Leu T-box region, which contains a tRNAAla gene embedded within the T-box RNA sequence. This tRNA-encoding gene appears to be integrated within stem III, preceding the antiterminator structure. The tRNA presumably specified by this region is predicted to be functional since it contains all of the conserved features of tRNAAla, including those nucleotides of the tRNA anticodon and the G-U pair in the acceptor stem that are required for tRNAAla charging by AlaRS. We propose that this tRNA could be excised from the RNA by posttranscriptional processing after the terminator-antiterminator decision is made. This processing event would therefore have no effect on the transcription termination mechanism, although it could affect the stability of the leader transcript and/or the readthrough transcript. Alternatively, the excision of the tRNA before RNA polymerase reaches the termination site could prevent the appropriate interaction with the regulatory tRNA, resulting in termination. Future experimental studies will be required to determine if this type of tRNA sequence insertion in a T-box region has any effect on the T-box regulatory response.
EVOLUTIONARY ORIGIN OF T-BOX ELEMENTS
The analysis of the phylogenetic distribution of T boxes shown in Fig. 2 suggests that the T-box mechanism could have arisen in a common ancestor of the Firmicutes, the Chloroflexi, Deinococcus-Thermus group, and the Actinobacteria. However, a plausible explanation for the acquisition of the T-box mechanism by organisms in the Deltaproteobacteria is by HGT, given that very few (and closely related) members of the Deltaproteobacteria have a T box in their genomes, and there are many phylogenetic clades between the Deltaproteobacteria and the aforementioned phyla in which the T-box mechanism is absent.
Geobacter sp. and its relatives represent one group of the Deltaproteobacteria in which T-box sequences have been identified. The leuA genes of Geobacter sp. and Pelobacter sp. are monocistronic and exhibit 50% amino acid sequence similarity with the monocistronic and T-box-regulated leuA of the firmicute C. acetobutylicum. Since these organisms share the same ecological niche, it is likely that a common ancestor of these Geobacter/Pelobacter spp. could have acquired the leuA gene and its associated T-box element by HGT. It is noteworthy that C. acetobutylicum and G. sulfurreducens are syntrophic organisms, where C. acetobutylicum provides acetate, a carbon compound required for growth by G. sulfurreducens (21). The close contact of these organisms in their natural habitat increases the probability of HGT.
Examples of likely HGT are not restricted to organisms of different phyla but can also be traced within the Firmicutes. In Streptococcus thermophilus and S. mutans, small open reading frames encoding proteins of ∼100 amino acids in length are located upstream of the trpEGDCFBA operons. These putative coding sequences exhibit statistically significant similarity to the T-box-regulated pheA chorismate mutase gene of D. hafniense and S. wolfei but show no significant similarity to the non-T-box-regulated pheA genes of their respective genomes. We speculate that this small open reading frame along with its T-box regulatory region could have been horizontally transferred and inserted upstream of the trpEGDCFBA operons of S. thermophilus and S. mutans.
EXPECTED INSIGHTS ON T-BOX REGULATION FROM ANALYSES OF NEW GENOME SEQUENCES
Despite the large number of sequenced genomes currently available, we predict that new findings regarding the distribution of T-box regulatory regions will emerge as additional genome sequences become available for analysis. To support this prediction, we highlight deductions that are based on analyses of the recently sequenced genome of C. kluyveri.
C. kluyveri appears to have exceptional metabolic capabilities, including extremely active sulfur metabolism and the ability to grow anaerobically on ethanol and acetate as sole carbon and energy sources (94). We identified 12 transcription units regulated by T-box sequences predicted to respond to BCAAs. Two of these correspond to aaRS genes (leuS and ileS), and one encodes a Leu transporter. Of the remaining nine transcriptional units, six contain known BCAA biosynthetic genes (ilvE, ilvCB, ilvC, ilvI, and two copies of leuA). One copy of leuA and the ilvI gene are regulated by tandem T boxes, which is indicative of tighter regulation (Fig. 6). We also identified an Ile T-box-regulated operon annotated as the porCDAB operon, which encodes proteins similar to the subunits of pyruvate:ferredoxin oxidoreductase (POR) (Fig. 6). This enzyme catalyzes the thiamine pyrophosphate-dependent oxidative decarboxylation of pyruvate to form acetyl-CoA and CO2 (64), a reaction not apparently related to BCAA biosynthesis. Val and Ile are synthesized via the same five-step pathway except for the first step, which, for Ile biosynthesis, is a thiamine pyrophosphate-dependent reaction (normally carried out by IlvA) that resembles those carried out by the proteins encoded in the por operon (108). Based on the identification of the Ile T-box sequence upstream of the porCDAB operon in C. kluyveri, we propose that the enzymes encoded by this operon are involved in the first step of Ile biosynthesis. S. wolfei represents a similar situation, in which a porCDAB operon regulated by an Ile T box could be involved in Ile biosynthesis. We note that S. wolfei has another copy of the porCDAB operon that is regulated by a Leu T box; the possible relationship to Leu biosynthesis is unclear, since the proposed porCDAB-encoded step is apparently specific to Ile biosynthesis (Fig. 6).
C. kluyveri also has two copies of the etfBA operon, which encodes the subunits of an electron transfer flavoprotein that participates in a cycle for the reduction of 5-crotonyl-CoA to 5-butyryl-CoA (94). In this cycle, seven NADH molecules are oxidized to NAD+. One of these etf operons is regulated by an Ile T-box sequence (Fig. 6). We do not completely understand the physiological significance of the regulation of this electron transfer module in response to Ile availability. We note that Ile is one of the major regulators of basic metabolic processes in B. subtilis via its interaction with the CodY regulatory protein. One possibility is that reduced levels of Ile may signal the activation of electron flow toward amino acid biosynthesis. Another possibility is that the above-mentioned POR complex has a high requirement for electron donors, especially in organisms that have limited redox potential, and the etfBA-encoded electron transfer flavoprotein could serve as the electron donor.
In addition to the above-mentioned T-box-regulated operons, C. kluyveri also has a BCAA-responsive T-box operon that encodes the product of fhsA (formate-tetrahydrofolate [THF] ligase), fchA (methenyl-THF cyclohydrolase), and folD (THF dehydrogenase), a participant in the biosynthesis of THF. This operon is regulated by a Val T box (Fig. 6), consistent with the observation that valS is cotranscribed with folC in the majority of the Firmicutes (Fig. 3). As mentioned above, THF is an important cofactor that donates a one-carbon unit during the synthesis of Met, purines, thymine, and pantothenic acid. Pantothenic acid biosynthesis branches off from the Val pathway at keto-valine. The first enzyme of this pathway is a hydroxymethyltransferase, which uses methylene THF as a cofactor. THF may be synthesized when the Val pools are low to ensure the availability of THF for the biosynthesis of pantothenic acid and Val.
CONCLUSIONS
The discovery of the T-box regulatory mechanism (43, 44, 56, 57) and our prediction of its widespread use in regulating genes involved in amino acid charging, biosynthesis, and transport in gram-positive bacteria demonstrate the apparent importance of sensing the extent of charging of specific tRNAs in regulating the expression of genes involved in amino acid metabolism. Early bacterial evolution was undoubtedly influenced by the fact that amino acids are expensive to synthesize and that they can be used for a variety of essential processes in addition to protein synthesis. Therefore, in regulating the expression of genes concerned with amino acid biosynthesis, charging, and transport, it was also essential to sense the availability of individual aminoacylated tRNAs, as the level of a charged or uncharged tRNA is a more accurate measure of whether an additional amino acid must be provided to maintain protein synthesis. Aminoacyl-tRNA synthetases usually recognize two features of their tRNA substrates, the anticodon sequence and the acceptor end sequence; it was therefore evolutionarily efficient for each T-box regulatory sequence to be able to detect these same features. The T-box mechanism illustrates the logical elegance of genome changes and developments that must have occurred during evolution to allow a single type of molecule (e.g., tRNA) to serve multiple functions.
In the present study, comparative genomics was used to identify all of the bacterial genes of amino acid metabolism that are predicted to be regulated by the T-box mechanism. From our findings, we conclude that the T-box mechanism is the most prominent RNA-based regulatory mechanism known to be employed by members of the Firmicutes. This mechanism is used to a lesser extent by members of the Chloroflexi, the Deinococcus-Thermus group, and the Actinobacteria. Based on the distribution of T-box sequences in bacterial genomes, we hypothesize that this regulatory system originated in a common ancestor of members of these phyla and that its use expanded in the Firmicutes, followed by HGT to a very few members of the Deltaproteobacteria.
Sequence analysis of T-box regions allows the prediction of probable recent events in T-box evolution. A notable example is provided by the Leu T-box sequences upstream of leuS in some members of the Clostridia. These T-box sequences contain an apparently intact gene that specifies tRNAAla inserted into a portion of stem III that is highly variable and insensitive to mutation. The presence of this tRNA gene in only a small subclass of leuS T-box sequences in a related group of organisms suggests that its insertion is likely to have been a recent event that preserved both tRNA and leader RNA function because of a posttranscriptional processing event that releases the mature tRNA from either the terminated or readthrough transcript.
The list of genes known to be involved in amino acid biosynthesis that are regulated by the T-box mechanism was increased to include genes in the Ala and Gly pathways. In addition, the T-box regulation of genes involved in the biosynthesis of Ser, Pro, Arg, Met, Cys, Ile, Leu, Val, Tyr, Phe, Trp, His, Asn, Asp, and Thr was confirmed. We also predict that the luxS gene, which is involved in quorum sensing and Met biosynthesis, is regulated by the T-box mechanism in some members of the Lactobacillales. For most T-box-regulated genes, such as those involved in tRNA charging, amino acid transport, and amino acid biosynthesis, the relationship between the specificity of the T-box sequence and the corresponding regulated genes is obvious. In some instances, novel biochemical relationships between the regulated genes and their corresponding metabolic pathway were revealed; C. kluyveri provides an interesting example.
It is likely that the genes encoding aminoacyl-tRNA synthetases, amino acid biosynthetic proteins, amino acid transport proteins, and key regulatory proteins may require differential responses to modulations in tRNA aminoacylation. The arrangement of T-box leader sequences as single, double, or triple copies expands the regulatory range for this mechanism, as the presence of tandem copies requires the independent binding of additional uncharged tRNA molecules to promote transcription readthrough. Regulation at the translational level, as predicted for the Actinobacteria, may also result in variability in the sensitivity of the system. It is also likely that individual transcriptional units may utilize regulatory mechanisms in addition to the T-box mechanism to ensure tighter regulation or a response to multiple regulatory signals, as has been observed for the B. subtilis ilv-leu operon (97).
Our analyses revealed an imbalance in tRNA sensing during the regulation of expression of operons containing multiple aaRS genes or biosynthetic genes involved in pathways common to more than one amino acid. This potential regulatory imbalance may be the consequence of (i) the phylogenetic origin of the operon, as noted by Vitreschak et al. (107); (ii) an incomplete or transitory stage in the evolutionary process leading to operon organization or regulation; or (iii) the ecological niche of the organism. In these cases, other regulatory mechanisms may act in concert with the T-box mechanism to ensure an appropriate physiological response. Alternatively, the organism may contain additional copies of individual genes that are subject to a different regulatory response.
In contrast to metabolite binding riboswitches that generally recognize several unique features of a single ligand, the T-box system allows a specific response of each regulated transcriptional unit to a deficiency of a single charged tRNA. A change in the regulatory response can be achieved by minor changes in the specifier sequence (and antiterminator bulge) of each T-box sequence to allow the recognition of a new uncharged tRNA class. This high degree of flexibility, without a loss of specificity, is probably responsible for the abundant use of the T-box mechanism in regulating gene expression in the Firmicutes. Also notable in terms of the global impact of this mechanism on cell physiology is the use of the T-box system to regulate the synthesis of proteins involved in the regulation of other gene families, including an enzyme involved in quorum sensing.
Acknowledgments
We acknowledge the contributions of our students and postdoctoral research associates, past and present, to the work described in this review. We also thank Ricardo Ciria, Christian Eduardo Martínez, and Arturo Ocadiz for computer support and Shirley Ainsworth for bibliographical assistance. We thank Roy Jensen, Mariana Peimbert, Rosa María Gutiérrez-Ríos, Cei Abreu-Goodger, Aubin Arroyo, and Dmitry Rodionov for their critical comments.
This work was supported by CONACyT grants 60127-Q and SALUD-2007-C01-68992 to E.M., NIH grant R01GM47823 to T.M.H., and NSF grant MCB-0615390 to C.Y.
REFERENCES
- 1.Abreu-Goodger, C., and E. Merino. 2005. RibEx: a Web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 33W690-W692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abreu-Goodger, C., N. Ontiveros-Palacios, R. Ciria, and E. Merino. 2004. Conserved regulatory motifs in bacteria: riboswitches and beyond. Trends Genet. 20475-479. [DOI] [PubMed] [Google Scholar]
- 3.Ahn, K. S., and R. G. Wake. 1991. Variations and coding features of the sequence spanning the replication terminus of Bacillus subtilis 168 and W23 chromosomes. Gene 98107-112. [DOI] [PubMed] [Google Scholar]
- 4.Alifano, P., R. Fani, P. Lio, A. Lazcano, M. Bazzicalupo, M. S. Carlomagno, and C. B. Bruni. 1996. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol. Rev. 6044-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babitzke, P., and P. Gollnick. 2001. Posttranscription initiation control of tryptophan metabolism in Bacillus subtilis by the trp RNA-binding attenuation protein (TRAP), anti-TRAP, and RNA structure. J. Bacteriol. 1835795-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey, T. L., and M. Gribskov. 1998. Combining evidence using P-values: application to sequence homology searches. Bioinformatics 1448-54. [DOI] [PubMed] [Google Scholar]
- 7.Barrick, J. E., K. A. Corbino, W. C. Winkler, A. Nahvi, M. Mandal, J. Collins, M. Lee, A. Roth, N. Sudarsan, I. Jona, J. K. Wickiser, and R. R. Breaker. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. USA 1016421-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belitsky, B. R. 2002. Biosynthesis of amino acids of the glutamate and aspartate families, alanine, and polyamines, p. 203-231. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 9.Belitsky, B. R., J. Brill, E. Bremer, and A. L. Sonenshein. 2001. Multiple genes for the last step of proline biosynthesis in Bacillus subtilis. J. Bacteriol. 1834389-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belitsky, B. R., L. V. Wray, Jr., S. H. Fisher, D. E. Bohannon, and A. L. Sonenshein. 2000. Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression. J. Bacteriol. 1825939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bovee, M. L., K. S. Champagne, B. Demeler, and C. S. Francklyn. 2002. The quaternary structure of the HisZ-HisG N-1-(5′-phosphoribosyl)-ATP transferase from Lactococcus lactis. Biochemistry 4111838-11846. [DOI] [PubMed] [Google Scholar]
- 12.Bremer, E. 2002. Adaptation to changing osmolarity, p. 385-391. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 13.Brinkman, A. B., T. J. G. Ettema, W. M. de Vos, and J. van der Oost. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48287-294. [DOI] [PubMed] [Google Scholar]
- 14.Carter, C. W., Jr. 2008. Whence the genetic code? Thawing the ‘frozen accident.’ Heredity 100339-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, G., and C. Yanofsky. 2003. Tandem transcription and translation regulatory sensing of uncharged tryptophan tRNA. Science 301211-213. [DOI] [PubMed] [Google Scholar]
- 16.Chen, G., and C. Yanofsky. 2004. Features of a leader peptide coding region that regulate translation initiation for the anti-TRAP protein of B. subtilis. Mol. Cell 13703-711. [DOI] [PubMed] [Google Scholar]
- 17.Chopin, A., V. Biaudet, and S. D. Ehrlich. 1998. Analysis of the Bacillus subtilis genome sequence reveals nine new T-box leaders. Mol. Microbiol. 29662-664. [DOI] [PubMed] [Google Scholar]
- 18.Ciccarelli, F. D., T. Doerks, C. von Mering, C. J. Creevey, B. Snel, and P. Bork. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 3111283-1287. [DOI] [PubMed] [Google Scholar]
- 19.Ciria, R., C. Abreu-Goodger, E. Morett, and E. Merino. 2004. GeConT: gene context analysis. Bioinformatics 202307-2308. [DOI] [PubMed] [Google Scholar]
- 20.Condon, C., H. Putzer, and M. Grunberg-Manago. 1996. Processing of the leader mRNA plays a major role in the induction of thrS expression following threonine starvation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 936992-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cord-Ruwisch, R., D. R. Lovley, and B. Schink. 1998. Growth of Geobacter sulfurreducens with acetate in syntrophic cooperation with hydrogen-oxidizing anaerobic partners. Appl. Environ. Microbiol. 642232-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Keersmaecker, S. C., K. Sonck, and J. Vanderleyden. 2006. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 14114-119. [DOI] [PubMed] [Google Scholar]
- 23.Delorme, C., S. D. Ehrlich, and P. Renault. 1992. Histidine biosynthesis genes in Lactococcus lactis subsp. lactis. J. Bacteriol. 1746571-6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delorme, C., S. D. Ehrlich, and P. Renault. 1999. Regulation of expression of the Lactococcus lactis histidine operon. J. Bacteriol. 1812026-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Saizieu, A., P. Vankan, C. Vockler, and A. P. van Loon. 1997. The trp RNA-binding attenuation protein (TRAP) regulates the steady-state levels of transcripts of the Bacillus subtilis folate operon. Microbiology 143979-989. [DOI] [PubMed] [Google Scholar]
- 26.Deuel, T. F., and S. Prusiner. 1974. Regulation of glutamine synthetase from Bacillus subtilis by divalent cations, feedback inhibitors, and L-glutamine. J. Biol. Chem. 249257-264. [PubMed] [Google Scholar]
- 27.Eddy, S. R. 2002. A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinformatics 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Even, S., P. Burguiere, S. Auger, O. Soutourina, A. Danchin, and I. Martin-Verstraete. 2006. Global control of cysteine metabolism by CymR in Bacillus subtilis. J. Bacteriol. 1882184-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 978063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32.Fisher, S. H., and M. Debarbouille. 2002. Nitrogen source utilization and its regulation, p. 181-192. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 33.Fuchs, R. T., F. J. Grundy, and T. M. Henkin. 2006. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 13226-233. [DOI] [PubMed] [Google Scholar]
- 34.Fujita, Y., Y. Miwa, A. Galinier, and J. Deutscher. 1995. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol. Microbiol. 17953-960. [DOI] [PubMed] [Google Scholar]
- 35.Gagnon, Y., R. Breton, H. Putzer, M. Pelchat, M. Grunberg-Manago, and J. Lapointe. 1994. Clustering and co-transcription of the Bacillus subtilis genes encoding the aminoacyl-tRNA synthetases specific for glutamate and for cysteine and the first enzyme for cysteine biosynthesis. J. Biol. Chem. 2697473-7482. [PubMed] [Google Scholar]
- 36.Galinier, A., J. Haiech, M. C. Kilhoffer, M. Jaquinod, J. Stulke, J. Deutscher, and I. Martin-Verstraete. 1997. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc. Natl. Acad. Sci. USA 948439-8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gelfand, M. S. 2006. Evolution of transcriptional regulatory networks in microbial genomes. Curr. Opin. Struct. Biol. 16420-429. [DOI] [PubMed] [Google Scholar]
- 38.Gendron, N., H. Putzer, and M. Grunberg-Manago. 1994. Expression of both Bacillus subtilis threonyl-tRNA synthetase genes is autogenously regulated. J. Bacteriol. 176486-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gollnick, P., P. Babitzke, A. Antson, and C. Yanofsky. 2005. Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis. Annu. Rev. Genet. 3947-68. [DOI] [PubMed] [Google Scholar]
- 40.Gollnick, P., P. Babitzke, E. Merino, and C. Yanofsky. 2002. Aromatic amino acid metabolism in Bacillus subtilis, p. 233-244. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 41.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 42.Griffiths-Jones, S., A. Bateman, M. Marshall, A. Khanna, and S. R. Eddy. 2003. Rfam: an RNA family database. Nucleic Acids Res. 31439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grundy, F. J., and T. M. Henkin. 1993. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74475-482. [DOI] [PubMed] [Google Scholar]
- 44.Grundy, F. J., and T. M. Henkin. 1994. Conservation of a transcription antitermination mechanism in aminoacyl-tRNA synthetase and amino acid biosynthesis genes in gram-positive bacteria. J. Mol. Biol. 235798-804. [DOI] [PubMed] [Google Scholar]
- 45.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30737-749. [DOI] [PubMed] [Google Scholar]
- 46.Grundy, F. J., and T. M. Henkin. 2002. Synthesis of serine, glycine, cysteine, and methionine, p. 245-254. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 47.Grundy, F. J., and T. M. Henkin. 2003. The T box and S box transcription termination control systems. Front. Biosci. 8D20-D31. [DOI] [PubMed] [Google Scholar]
- 48.Grundy, F. J., S. C. Lehman, and T. M. Henkin. 2003. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl. Acad. Sci. USA 10012057-12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grundy, F. J., T. R. Moir, M. T. Haldeman, and T. M. Henkin. 2002. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: disparity between conservation and functional requirements. Nucleic Acids Res. 301646-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grundy, F. J., W. C. Winkler, and T. M. Henkin. 2002. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc. Natl. Acad. Sci. USA 9911121-11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grundy, F. J., M. R. Yousef, and T. M. Henkin. 2005. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J. Mol. Biol. 34673-81. [DOI] [PubMed] [Google Scholar]
- 52.Gutierrez-Preciado, A., R. A. Jensen, C. Yanofsky, and E. Merino. 2005. New insights into regulation of the tryptophan biosynthetic operon in gram-positive bacteria. Trends Genet. 21432-436. [DOI] [PubMed] [Google Scholar]
- 53.Gutierrez-Preciado, A., C. Yanofsky, and E. Merino. 2007. Comparison of tryptophan biosynthetic operon regulation in different gram-positive bacterial species. Trends Genet. 23422-427. [DOI] [PubMed] [Google Scholar]
- 54.Hahn, J., G. Inamine, Y. Kozlov, and D. Dubnau. 1993. Characterization of comE, a late competence operon of Bacillus subtilis required for the binding and uptake of transforming DNA. Mol. Microbiol. 1099-111. [DOI] [PubMed] [Google Scholar]
- 55.Henkin, T. M. 2008. Riboswitch RNAs: using RNA to sense cellular metabolism Genes Dev. 223383-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henkin, T. M. 1994. tRNA-directed transcription antitermination. Mol. Microbiol. 13381-387. [DOI] [PubMed] [Google Scholar]
- 57.Henkin, T. M., B. L. Glass, and F. J. Grundy. 1992. Analysis of the Bacillus subtilis tyrS gene: conservation of a regulatory sequence in multiple tRNA synthetase genes. J. Bacteriol. 1741299-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henkin, T. M., and F. J. Grundy. 2006. Sensing metabolic signals with nascent RNA transcripts: the T box and S box riboswitches as paradigms. Cold Spring Harb. Symp. Quant. Biol. 71231-237. [DOI] [PubMed] [Google Scholar]
- 59.Henner, D. J., and C. Yanofsky. 1993. Biosynthesis of aromatic amino acids, p. 269-280. In R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, DC.
- 60.Ibarra, J. A., E. Perez-Rueda, L. Segovia, and J. L. Puente. 2008. The DNA-binding domain as a functional indicator: the case of the AraC/XylS family of transcription factors. Genetica 13365-76. [DOI] [PubMed] [Google Scholar]
- 61.Ibba, M., H. D. Becker, C. Stathopoulos, D. L. Tumbula, and D. Soll. 2000. The adaptor hypothesis revisited. Trends Biochem. Sci. 25311-316. [DOI] [PubMed] [Google Scholar]
- 62.Ibba, M., and D. Soll. 2000. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69617-650. [DOI] [PubMed] [Google Scholar]
- 63.Iijima, T., M. D. Diesterhaft, and E. Freese. 1977. Sodium effect of growth on aspartate and genetic analysis of a Bacillus subtilis mutant with high aspartase activity. J. Bacteriol. 1291440-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ikeda, T., T. Ochiai, S. Morita, A. Nishiyama, E. Yamada, H. Arai, M. Ishii, and Y. Igarashi. 2006. Anabolic five subunit-type pyruvate:ferredoxin oxidoreductase from Hydrogenobacter thermophilus TK-6. Biochem. Biophys. Res. Commun. 34076-82. [DOI] [PubMed] [Google Scholar]
- 65.Janga, S. C., W. F. Lamboy, A. M. Huerta, and G. Moreno-Hagelsieb. 2006. The distinctive signatures of promoter regions and operon junctions across prokaryotes. Nucleic Acids Res. 343980-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim, S. I., J. E. Germond, D. Pridmore, and D. Soll. 1996. Lactobacillus bulgaricus asparagine synthetase and asparaginyl-tRNA synthetase: coregulation by transcription antitermination? J. Bacteriol. 1782459-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kovaleva, G. Y., and M. S. Gelfand. 2007. Transcriptional regulation of the methionine and cysteine transport and metabolism in streptococci. FEMS Microbiol. Lett. 276207-215. [DOI] [PubMed] [Google Scholar]
- 68.Reference deleted.
- 69.Luo, D., J. Leautey, M. Grunberg-Manago, and H. Putzer. 1997. Structure and regulation of expression of the Bacillus subtilis valyl-tRNA synthetase gene. J. Bacteriol. 1792472-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mandal, M., M. Lee, J. E. Barrick, Z. Weinberg, G. M. Emilsson, W. L. Ruzzo, and R. R. Breaker. 2004. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306275-279. [DOI] [PubMed] [Google Scholar]
- 71.Marcos, A. T., K. Kosalkova, R. E. Cardoza, F. Fierro, S. Gutierrez, and J. F. Martin. 2001. Characterization of the reverse transsulfuration gene mecB of Acremonium chrysogenum, which encodes a functional cystathionine-gamma-lyase. Mol. Gen. Genet. 264746-754. [DOI] [PubMed] [Google Scholar]
- 72.Martinez-Guerrero, C. E., R. Ciria, C. Abreu-Goodger, G. Moreno-Hagelsieb, and E. Merino. 2008. GeConT 2: gene context analysis for orthologous proteins, conserved domains and metabolic pathways. Nucleic Acids Res. 36W176-W180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Merino, E., P. Babitzke, and C. Yanofsky. 1995. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J. Bacteriol. 1776362-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Merino, E., R. A. Jensen, and C. Yanofsky. 2008. Evolution of bacterial trp operons and their regulation. Curr. Opin. Microbiol. 1178-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moat, A. G., J. W. Foster, and M. P. Spector. 2002. Biosynthesis and metabolism of amino acids, p. 503-544. In A. G. Moat, J. W. Foster, and M. P. Spector (ed.), Microbial physiology. Wiley-Liss, Inc., New York, NY.
- 76.Moreno-Hagelsieb, G., and J. Collado-Vides. 2002. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics 18(Suppl. 1)S329-S336. [DOI] [PubMed] [Google Scholar]
- 77.Murphy, B. A., F. J. Grundy, and T. M. Henkin. 2002. Prediction of gene function in methylthioadenosine recycling from regulatory signals. J. Bacteriol. 1842314-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parsot, C. 1986. Evolution of biosynthetic pathways: a common ancestor for threonine synthase, threonine dehydratase and D-serine dehydratase. EMBO J. 53013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parsot, C., and G. N. Cohen. 1988. Cloning and nucleotide sequence of the Bacillus subtilis hom gene coding for homoserine dehydrogenase. Structural and evolutionary relationships with Escherichia coli aspartokinases-homoserine dehydrogenases I and II. J. Biol. Chem. 26314654-14660. [PubMed] [Google Scholar]
- 80.Putzer, H., A. A. Brakhage, and M. Grunberg-Manago. 1990. Independent genes for two threonyl-tRNA synthetases in Bacillus subtilis. J. Bacteriol. 1724593-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Putzer, H., C. Condon, D. Brechemier-Baey, R. Brito, and M. Grunberg-Manago. 2002. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res. 303026-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Putzer, H., N. Gendron, and M. Grunberg-Manago. 1992. Co-ordinate expression of the two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J. 113117-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Putzer, H., S. Laalami, A. A. Brakhage, C. Condon, and M. Grunberg-Manago. 1995. Aminoacyl-tRNA synthetase gene regulation in Bacillus subtilis: induction, repression and growth-rate regulation. Mol. Microbiol. 16709-718. [DOI] [PubMed] [Google Scholar]
- 84.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. Deboy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. X. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 42381-86. [DOI] [PubMed] [Google Scholar]
- 85.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res. 316748-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2004. Comparative genomics of the methionine metabolism in gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 323340-3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rollins, S. M. 2002. The mRNA/tRNA interaction promoting T box transcriptional antitermination. Ph.D. thesis. The Ohio State University, Columbus.
- 88.Rollins, S. M., F. J. Grundy, and T. M. Henkin. 1997. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol. 25411-421. [DOI] [PubMed] [Google Scholar]
- 89.Salgado, H., G. Moreno-Hagelsieb, T. F. Smith, and J. Collado-Vides. 2000. Operons in Escherichia coli: genomic analyses and predictions. Proc. Natl. Acad. Sci. USA 976652-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sarsero, J. P., E. Merino, and C. Yanofsky. 2000. A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport. J. Bacteriol. 1822329-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sarsero, J. P., E. Merino, and C. Yanofsky. 2000. A Bacillus subtilis operon containing genes of unknown function senses tRNATrp charging and regulates expression of the genes of tryptophan biosynthesis. Proc. Natl. Acad. Sci. USA 972656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41463-476. [DOI] [PubMed] [Google Scholar]
- 93.Schreier, H. J. 1993. Biosynthesis of glutamine and glutamate and assimilation of ammonia, p. 281-298. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, DC.
- 94.Seedorf, H., W. F. Fricke, B. Veith, H. Bruggemann, H. Liesegang, A. Strittmatter, M. Miethke, W. Buckel, J. Hinderberger, F. Li, C. Hagemeier, R. K. Thauer, and G. Gottschalk. 2008. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl. Acad. Sci. USA 1052128-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sekowska, A., and A. Danchin. 1999. Identification of yrrU as the methylthioadenosine nucleosidase gene in Bacillus subtilis. DNA Res. 6255-264. [DOI] [PubMed] [Google Scholar]
- 96.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53599-611. [DOI] [PubMed] [Google Scholar]
- 97.Shivers, R. P., and A. L. Sonenshein. 2005. Bacillus subtilis ilvB operon: an intersection of global regulons. Mol. Microbiol. 561549-1559. [DOI] [PubMed] [Google Scholar]
- 98.Smith, M. C., A. Mountain, and S. Baumberg. 1986. Cloning in Escherichia coli of a Bacillus subtilis arginine repressor gene through its ability to confer structural stability on a fragment carrying genes of arginine biosynthesis. Mol. Gen. Genet. 205176-182. [DOI] [PubMed] [Google Scholar]
- 99.Stauffer, G. V. 1996. Biosynthesis of serine, glycine, and one-carbon units, p. 506-513. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 100.Steele, M. I., D. Lorenz, K. Hatter, A. Park, and J. R. Sokatch. 1992. Characterization of the mmsAB operon of Pseudomonas aeruginosa PAO encoding methylmalonate-semialdehyde dehydrogenase and 3-hydroxyisobutyrate dehydrogenase. J. Biol. Chem. 26713585-13592. [PubMed] [Google Scholar]
- 101.Sun, D., and P. Setlow. 1993. Cloning and nucleotide sequence of the Bacillus subtilis ansR gene, which encodes a repressor of the ans operon coding for l-asparaginase and l-aspartase. J. Bacteriol. 1752501-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun, D. X., and P. Setlow. 1991. Cloning, nucleotide sequence, and expression of the Bacillus subtilis ans operon, which codes for l-asparaginase and l-aspartase. J. Bacteriol. 1733831-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tomsic, J., B. A. McDaniel, F. J. Grundy, and T. M. Henkin. 2008. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in Bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. J. Bacteriol. 190823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Valbuzzi, A., P. Gollnick, P. Babitzke, and C. Yanofsky. 2002. The anti-trp RNA-binding attenuation protein (anti-TRAP), AT, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein. J. Biol. Chem. 27710608-10613. [DOI] [PubMed] [Google Scholar]
- 105.Valbuzzi, A., and C. Yanofsky. 2001. Inhibition of the B. subtilis regulatory protein TRAP by the TRAP-inhibitory protein, AT. Science 2932057-2059. [DOI] [PubMed] [Google Scholar]
- 106.Vander Horn, P. B., and S. A. Zahler. 1992. Cloning and nucleotide sequence of the leucyl-tRNA synthetase gene of Bacillus subtilis. J. Bacteriol. 1743928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vitreschak, A. G., A. A. Mironov, V. A. Lyubetsky, and M. S. Gelfand. 2008. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 14717-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Voet, D., and J. G. Voet. 1995. Biochemistry, 2nd ed., p. 764-776. John Wiley & Sons, Inc., Hoboken, NJ.
- 109.Voskuil, M. I., and G. H. Chambliss. 1996. Significance of HPr in catabolite repression of α-amylase. J. Bacteriol. 1787014-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Webb, M. E., A. G. Smith, and C. Abell. 2004. Biosynthesis of pantothenate. Nat. Prod. Rep. 21695-721. [DOI] [PubMed] [Google Scholar]
- 111.Wels, M., K. T. Groot, M. Kleerebezem, R. J. Siezen, and C. Francke. 2008. An in silico analysis of T-box regulated genes and T-box evolution in prokaryotes, with emphasis on prediction of substrate specificity of transporters. BMC Genomics 9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Winkler, W. C., F. J. Grundy, B. A. Murphy, and T. M. Henkin. 2001. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA 71165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Woese, C. R., G. J. Olsen, M. Ibba, and D. Soll. 2000. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 64202-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wolf, Y. I., L. Aravind, N. V. Grishin, and E. V. Koonin. 1999. Evolution of aminoacyl-tRNA synthetases—analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 9689-710. [PubMed] [Google Scholar]
- 115.Yanofsky, C. 1981. Attenuation in the control of expression of bacterial operons. Nature 289751-758. [DOI] [PubMed] [Google Scholar]
- 116.Yanofsky, C. 1988. Transcription attenuation. J. Biol. Chem. 263609-612. [PubMed] [Google Scholar]
- 117.Yanofsky, C. 2000. Transcription attenuation: once viewed as a novel regulatory strategy. J. Bacteriol. 1821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yanofsky, C. 2004. The different roles of tryptophan transfer RNA in regulating trp operon expression in E. coli versus B. subtilis. Trends Genet. 20367-374. [DOI] [PubMed] [Google Scholar]
- 119.Yeggy, J. P., and D. P. Stahly. 1980. Sporulation and regulation of homoserine dehydrogenase in Bacillus subtilis. Can. J. Microbiol. 261386-1391. [DOI] [PubMed] [Google Scholar]
- 120.Yoshida, K., Y. Fujita, and S. D. Ehrlich. 1999. Three asparagine synthetase genes of Bacillus subtilis. J. Bacteriol. 1816081-6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yousef, M. R., F. J. Grundy, and T. M. Henkin. 2005. Structural transitions induced by the interaction between tRNA(Gly) and the Bacillus subtilis glyQS T box leader RNA. J. Mol. Biol. 349273-287. [DOI] [PubMed] [Google Scholar]