Abstract
Summary: The assembly of bacterial ribosomes is viewed with increasing interest as a potential target for new antibiotics. The in vivo synthesis and assembly of ribosomes are briefly reviewed here, highlighting the many ways in which assembly can be perturbed. The process is compared with the model in vitro process from which much of our knowledge is derived. The coordinate synthesis of the ribosomal components is essential for their ordered and efficient assembly; antibiotics interfere with this coordination and therefore affect assembly. It has also been claimed that the binding of antibiotics to nascent ribosomes prevents their assembly. These two contrasting models of antibiotic action are compared and evaluated. Finally, the suitability and tractability of assembly as a drug target are assessed.
INTRODUCTION
The bacterial 70S ribosome is a complex 2.4-MDa ribonucleoprotein comprising two subunits of unequal size. These are designated the small, or 30S, subunit and the large, or 50S, subunit according to their sedimentation coefficients. In Escherichia coli, the small subunit contains one 16S rRNA and 21 different ribosomal proteins named S1 through S21 (S here is for small subunit), while the large subunit contains a 5S rRNA, a 23S rRNA, and 34 different proteins named L1 through L36 (L is for large subunit). Each component is present as a single copy, with the exception of two copies of the L12 protein and two copies of its N-terminally-acetylated derivative, L7 (67).
Despite early progress, our understanding of the assembly of these components has lagged behind other aspects of ribosome biochemistry. After a period of relative quiescence, the field has been enjoying a resurgence of interest, spurred in part by X-ray crystallographic structures of the subunits and by new techniques that are gaining traction on old questions. Further interest is provided by a growing number of reports of specific effects of antibiotics on ribosome assembly and the assertion that this is an equally important target to translation. In the prevailing climate of increasing antibiotic resistance, the discovery of new targets of antibiotic action is naturally of great interest. Recent reviews are available on more general aspects of the assembly of both subunits (88, 123) or the more studied 30S subunit (44, 182).
ASSEMBLY IN VIVO
The process of assembly in E. coli begins with transcription of the rRNA from one of the seven rRNA (rrn) operons. Each transcript encodes a copy of all three ribosomal RNAs in the order 16S, 23S, and 5S. The 16S gene is preceded by a 5′ “leader” sequence and followed by a “spacer” sequence that includes one or two tRNA genes. Some operons end with a second 5S gene and/or one or two more tRNA genes (162). The discarded leader and spacer sequences appear to play an early role in the processing of the rRNAs, as mutations in them can impair processing (7, 96, 161).
Rapid nucleolytic cleavages release the RNAs as oversized precursor forms called p16S, p23S, p5S, and ptRNA (p is for precursor). This difference in size is readily detectable for the p16S rRNA (but not the p23S rRNA) by sedimentation (at 17S). A succession of nucleases progressively trim the rRNAs to produce mature termini (94). The 16S and 23S rRNAs and several of the ribosomal proteins are modified during the assembly process, mainly by methylation (88). The processes of transcription, processing, modification, and assembly of the rRNA are intertwined and interdependent.
Observing Assembly In Vivo
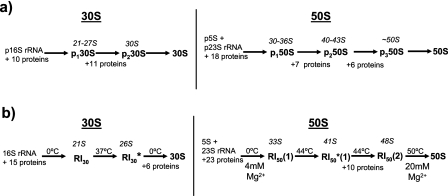
Only about 2 to 5% of rRNA is normally contained in assembling (immature) ribosomes in growing cells (97). Groups of ribosomal proteins add to the rRNA to form discrete precursor particles. The time taken for a pulse of [3H]uracil added to a growing culture to appear as newly formed rRNA in 70S ribosomes is a roughly constant fraction of the generation time at different growth rates and about 2 to 3 min during rapid growth (117). Early studies defined two sequential precursors to the 30S subunit (the p130S and p230S) and three sequential precursors to the 50S subunit (the p150S, p250S, and p350S) (97, 98). The process is shown schematically in Fig. 1a. Approximate sedimentation values are given for the precursors, but the exact values vary with the experimental conditions used, particularly the magnesium ion concentration (69).
FIG. 1.
(a) In vivo assembly scheme for both subunits. Precursor names are given in boldface type, with their approximate sedimentation values above them in italic type. (b) In vitro assembly scheme with ionic and temperature changes during the process given.
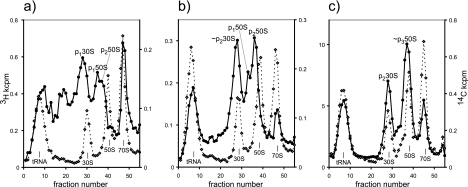
Figure 2 captures the assembly process by sucrose gradient centrifugation of pulse-labeled cell extracts and recapitulates data from earlier studies. Prior labeling with [14C]uracil for two generations provides a backdrop of completed 30S and 50S subunits followed by 70S ribosomes, with tRNA at the top of the gradients, while the precursors are revealed by pulse-labeling with [3H]uracil. The p130S precursor is indicated in Fig. 2a and increases in sedimentation until, as the p230S precursor, it coincides with the 30S reference peak (Fig. 2c). A mixture of material sedimenting between p130S and the tRNA at the first time point is incomplete 16S and 23S rRNA to which some ribosomal proteins have bound (43, 109). The p150S and p250S precursors are also indicated in Fig. 2a, with a discrete conversion of the former to the latter with time (Fig. 2b). The p250S then gradually increases in sedimentation (Fig. 2c) to form the p350S that superimposes on the 50S reference peak after 20 min (not shown). (Cosedimentation time scales are extended, as there is no chase, but a more detailed analysis of label entry to the 70S peak indicates an assembly time of 3 min.) Approximate sedimentation values are given in the legend; precursors that sediment at about 28S, 40S, and 45S here (in 10 mM Mg2+) correspond to the 21S, 32S, and 43S precursors reported in studies using lower magnesium concentrations (0.1 mM Mg) (69, 110). The higher magnesium concentration used here has the advantage that precursors to the 50S are well resolved from the peak of 30S subunits (and the p230S precursor that sediments with it).
FIG. 2.
Observation of assembly in vivo by pulse-labeling. The long-term 14C label is shown by a dashed line with open diamonds, and the pulse 3H label is shown by solid lines and circles. Counts per minute (103) (kcpm) detected by liquid scintillation counting are given on the y axes. E. coli strain A19 was grown, long-term labeled with [14C]uracil, and then pulse-labeled with [3H]uracil as described previously (40). Samples were harvested at intervals during pulse-labeling, lysed, and centrifuged, and radioactivity was detected in fractions as described previously (40). The identity of reference peaks is given on the plot at the base of each peak. Times of labeling in minutes (and generation time) were 20 s (0.01 generations) (a), 1 min (0.03 generations) (b), and 10 min (0.28 generations) (c). Approximate sedimentation values for the indicated precursor peaks were 28S, 40S, and 43S (a); 29S, 39S, and 45S (b); and 30S and 47S (c). (Panel b is reproduced from reference 40 with permission of the publisher.)
The final (p230S and p350S) precursors were estimated to be the longest-lived precursors (97). They contain all the proteins and cosediment with mature subunits, but their rRNAs retain additional terminal sequences that must be trimmed off to form the mature product. The precise details are not fully known, but the final rRNA processing events are thought to occur in polysomes (98, 111, 154, 160). As a result of this coupling, the maturation of the 17S rRNA to 16S is thought to depend on association of the p230S with 50S subunits. This blurs the distinction between 16S rRNA maturation, 30S subunit assembly, and subunit association so that defects in subunit association, or a shortage of 50S subunits with which to associate, can manifest as a delayed maturation of the 17S rRNA (and the 30S rRNA as a whole) (37, 112). Alternatively, stalled precursors to the 50S rRNA might conceivably sequester auxiliary factors required for the maturation of both subunits.
Auxiliary Factors
Aside from enzymes that chemically modify the rRNA and proteins, a growing number of proteins are thought to act as auxiliary factors to facilitate assembly in vivo (3, 15, 183). Many of these factors act late in assembly and have overlapping functions. Mutations in several factors (RimM, RimN, and RbfA) and GTPases (Era, CgtAE, RsgA, and EngA) affect 17S rRNA maturation and the ability of the 30S subunits to associate with the 50S (47, 76, 82, 89, 100, 142, 143). A deficiency of some of them (RimM, RbfA, and Era) is complemented by the overexpression of another (RbfA, Era, and KsgA, respectively).
For the 50S, depletion of any of the proteins CgtAE, EngA, and RrmJ results in immature 50S subunits that unfold at low magnesium concentrations (66, 86). Two putative ATP-dependent RNA helicases, SrmB and CsdA, were isolated as suppressors of cold-sensitive mutants and may facilitate structural transitions by melting rRNA structures at low temperatures; the deficiency of either one results in the accumulation of a large-subunit precursor (36, 37). The heat shock chaperones GroEL and DnaK influence assembly during growth above 30°C (2, 58, 59). A deficiency in various factors (RrmJ, DnaK, CgtA, SrmB, and CsdA) results in precursors to the 50S that are commonly deficient in at least four proteins (L16, L25, L28, and L33) (36, 37, 58, 66, 86). In the completed subunit, these four proteins are located near tRNA binding sites, the maturation of which is likely an important step in late assembly.
Assembly In Vitro
Despite the many additional factors and processes that contribute to assembly in vivo, ribosomal subunits can be assembled in vitro simply by incubating the mature components together under appropriate conditions (124, 172). The precursors from the in vitro process and conditions required for their formation are depicted in Fig. 1b. Comparison of the in vivo and in vitro schemes at once suggests similar features, with an interconversion of a series of precursors of similar sedimentation values in both schemes, by the addition of discrete groups of proteins. However, the process is slow and inefficient compared to that in vivo. The first precursors, the RI30 and RI50(1), form at 0°C by the binding of a subset of proteins to the 16S or 5S and 23S rRNAs, respectively. Both precursors then require incubation at a higher temperature to effect a conformational change to form the more compact (and faster-sedimenting) RI30* and RI50*(1) particles. This change allows the binding of further proteins to complete the 30S subunit, while the 50S subunit requires continued incubation with proteins, followed by a second heating step at 55°C and higher magnesium ion concentrations for completion. Conditions for the formation of the 50S rRNA are thus rather unphysiological. For the 30S subunit, a distinct RI30 precursor is generated only at low temperatures (0°C to 15°C) (173); kinetic experiments at higher temperatures showed an uninterrupted incorporation of protein, suggesting that the generation of this in vitro precursor at a low temperature is somewhat artificial (168). Furthermore, the final precursors to both subunits in vivo have no counterpart in vitro, as the latter are assembled from mature rRNA.
The 16S rRNA folds into a secondary structure that can be neatly divided into three major domains. Each domain assembles with its associated proteins to form a discrete morphological feature of the subunit (see, e.g., 107), and these can be assembled independently in vitro (1, 141, 181). Such autonomous assembly prevails in vitro for the whole E. coli subunit, where the individual domains have been observed (by electron microscopy) to assemble independently and then coalesce together to form the mature subunit (108). For the 50S subunit, the different domains of 23S rRNA are interwoven so that such modular assembly is not possible (5, 174). Modular assembly is one of the many ways in which the assembly of the smaller 30S subunit is simpler than that of the 50S.
Assembly of Individual Proteins
Comparison of the protein catalogs of the in vivo and in vitro precursors (55, 71, 80) and kinetic studies of the order of their assembly (133) in vivo and in vitro (79, 135, 168) reveal a general consensus for the first eight or so proteins to assemble for both subunits but divergence thereafter, suggesting alternative late-assembly pathways in vivo and in vitro.
The detailed roles of individual proteins during assembly in vitro were deduced by omitting them from assembly reactions. The assembly of other proteins is often reduced as a result, revealing a web of protein interdependencies for each subunit known as the assembly map (44, 70, 73, 138). Assembly in vitro is hierarchical: “primary” binding proteins bind independently to the RNA, followed by secondary binders (that require the former to bind first) and then tertiary binders that require one or two primary or secondary binders to be bound first. In vivo, deletion of the gene encoding certain proteins can also result in accumulation of precursors deficient in other proteins, but the relationships revealed can be different from those in vitro.
For instance, the effects of the absence of either protein L27 or protein L28 in vivo were very different from those predicted by the in vitro work. The deletion of the gene for protein L27 results in the accumulation of a 40S precursor lacking not only L27 but also L16, L20, and L21 (184). The L21 and L27 proteins share an operon (85), but the synthesis of L21 appears to be normal, and resupply of L27 from a plasmid restores normal assembly. In vitro studies position L27 late in assembly without influence on other proteins (73). How the early assembling L20 is affected is thus unclear. Furthermore, 23S rRNA was also detected sedimenting at about 30S, suggesting an even earlier assembly defect. Conversely, protein L30 depends on both L20 and L21 for assembly in vitro (73) yet is present in the particle despite their absence.
Several mutants were available for the operon that encodes proteins L28 and L33 (20, 21, 40, 104). Extensive studies of ribosome assembly in these mutants together show that a shortage or absence of L28, or a C-terminal frameshifted version, causes an accumulation of an abnormal 47S precursor to the 50S subunit that is deficient not only in L28 but also in L16, L25, L27, and L33 (19, 104, 105, 113). Protein L33 shows a slight dependence on L28 for assembly in vitro, but the others do not. However, they all cluster around protein L15 in the in vitro assembly map, and therefore, the assembly of this group may be more cooperative in vivo than in vitro. Although neither L28 nor L33 was ascribed an important role in vitro, if both are absent, no 50S subunits are made in vivo (106).
As both L27 and L28 are thought to be late-assembling proteins, multiple pathways in late assembly may help account for the differences described here. Given the consensus around early binding proteins, it is more surprising that half of the in vitro primary binding proteins of the 30S subunit (S15, S17, and S20) were found to be individually dispensable in vivo by gene deletion despite their crucial role in vitro (16, 93). Thus, although in vitro assembly shows general features that are reflected in vivo, the two processes differ in their details, particularly with regard to the importance of individual proteins. This point is returned to below in considering in vitro assays for assembly inhibition.
CONTROL OF SYNTHESIS OF RIBOSOME COMPONENTS
The coordinate synthesis of ribosomal components is essential for their efficient assembly. Ribosome production consumes up to 40% of the cell's energy in rapidly growing bacteria and is therefore tightly regulated on several levels. To achieve optimal growth, the cell must balance production of the translation machinery with production of its substrates so that the supply of the ATP and amino acids required for tRNA charging and the GTP required for translation factor function are matched to demand. The synthesis of ribosomes increases with the square of the growth rate, a phenomenon called growth rate control (12). It must also respond to nutritional changes such as outgrowth or stationary phase. Ribosome synthesis and its control are driven primarily by the synthesis of the rRNA. This is subject to several overlapping regulatory mechanisms that have been difficult to disentangle and are still not fully understood.
Each rrn operon has tandem promoters called P1 and P2. Activities of both promoters are subject to change (120), but the upstream P1 is the more responsive, and during fast growth, it is two- to fivefold more active than P2. The lifetime of the RNA polymerase open complex is unusually short on these promoters (146), conferring sensitivity to the concentration of the initiating nucleotide, which is ATP, bar one case of GTP (62). Nus-based antitermination operates to protect these long untranslated transcripts from premature termination (127, 151, 159).
During early-log-phase growth, or a nutritional shift-up, the synthesis of rRNA is greatly stimulated. This is partly due to the FIS (factor for inversion stimulation) protein (125), which binds to an “upstream activating sequence” preceding the P1 promoter. As stationary phase is approached, HN-S and another nucleoid structuring protein, LRP (leucine-responsive regulatory protein), act synergistically on this region to repress initiation (136).
Stringent Control
Another form of control, called stringent control, occurs in response to the abrupt withdrawal of a required amino acid. This causes a shortage of the corresponding aminoacyl-tRNA. The binding of the deacylated form of the tRNA to the ribosomal A site prompts the synthesis of the alarmone guanosine tetraphosphate (or pentaphosphate) from GDP (or GTP) by a ribosome-associated protein called RelA (68). The pentaphosphate is rapidly converted to tetraphosphate by guanosine pentaphosphatase (Gpp). Guanosine tetra- or pentaphosphate [(p)ppGpp] interacts with RNA polymerase to affect its activity on different promoters so that different groups of genes are up- or downregulated (24). Initiation at the P1 rRNA promoter is dramatically reduced.
During an aminoacyl-tRNA shortage, the stringent system thus prevents the wasteful synthesis of rRNA and ribosomes. Mutants defective in this control (“relaxed” mutants) allow RNA synthesis to continue unabated during amino acid starvation (see, e.g., reference 45). Continued rRNA transcription during translation inhibition disrupts ribosomal assembly, since rRNA transcripts cannot assemble into mature subunits without ribosomal proteins. Consequently, abnormal ribosome precursors accumulate during amino acid starvation of relaxed mutants (165, 166). Resupply of the missing amino acid results in the conversion of the precursors to mature 30S and 50S subunits (69, 103).
Growth Rate Control and Feedback Control
The primary explanation for growth rate control is that it is also mediated by ppGpp but in this instance made by a second synthetase called SpoT in response to more general nutritional limitations (24) such as carbon, energy, or groups of amino acids (rather than starvation for a single amino acid as with RelA). Steady-state levels of ppGpp decrease as the growth rate increases, stimulating rRNA synthesis from the P1 promoter. SpoT has both ppGpp synthetase and hydrolase activities, but how these activities are balanced under different conditions is not yet clear. Interestingly, the synthetase activity is highly unstable and therefore requires continual protein synthesis (121), while the hydrolase activity does not. P1 promoter activity is sensitive to levels of eight amino acids in the cell (188), and it was suggested that this reflects reduced SpoT synthetase activity, perhaps caused by some effect of the amino acids on the translation of its mRNA that alters the balance of its synthetase and hydrolase activities.
In addition, the increased availability of RNA polymerase during growth in rich medium may passively favor initiation from rrn promoters relative to other promoters (95). Levels of the FIS protein are controlled by ppGpp (126) and increase with the growth rate, also contributing to control. The steady-state growth rate control of rrn expression is robust and can compensate when one of its elements is disabled (139). Reports of whether ppGpp alone can account for growth rate control are conflicting (63, 72). Auxiliary mechanisms certainly exist, but the extents of their contributions have yet to be fully established.
The most important of these auxiliary mechanisms is known as “feedback control,” in which rRNA synthesis responds to the translational capacity of the cell by somehow sensing ribosomes engaged in protein synthesis. This is based on the observation that if rrn gene dosage is artificially increased, rRNA synthesis rates per gene are reduced to compensate, provided that the extra genes encode functional rRNA products (87). Conversely, deletion of rrn genes increases expression from those remaining (42). Furthermore, reduced initiation on mRNAs, caused either by mutation of the anti-Shine-Dalgarno sequence of the 16S rRNA or by limiting initiation factor IF-2, derepresses the synthesis of rRNA, suggesting that ribosomes must be capable of initiation in order to cause feedback repression (39, 185). Ribosomes with impaired rates of elongation also cause an upregulation of ribosome synthesis (118), and the inhibition of ribosome assembly by limiting levels of specific ribosomal proteins reduces translational activity and derepresses rRNA synthesis (167). Since translation consumes GTP and ATP, translational activity may also be monitored by sensing nucleotide availability.
The discussion described above emphasizes the multilayered complexity of controls of rRNA synthesis and highlights many connections with amino acid supply and translational activity. Several reviews are available for a more detailed discussion of attempts to fit the many experimental observations into a coherent model (52, 84, 163, 179).
Synthesis of Ribosomal Proteins
The synthesis of ribosomal proteins is linked to that of rRNA by a form of negative feedback called “translational autoregulation.” In this mechanism, one of the ribosomal protein products of an operon has a dual RNA binding capability. It binds to rRNA as part of the normal assembly process but, if present in excess of rRNA with which to assemble, can bind instead to the mRNA that encodes it (usually within the leader sequence) to prevent not only its own translation but also that of other proteins encoded by the operon (99); details differ for each operon, with many variants of this general scheme (187). Translation “coupling” is known to operate in most ribosomal protein operons, at least for some of the genes (187), so their translation is contingent on the translation of the upstream genes. In addition, failure to be translated leaves an mRNA more exposed to nucleases and shortens its half-life (156).
Many ribosomal protein operons encode proteins from both subunits. This has the important consequence that the autoregulatory protein, which belongs to one ribosomal subunit, can also regulate the production of proteins that belong to the other subunit. Thus, protein L4 regulates the production of six proteins of its own subunit and four of the small subunit, and protein S8 regulates two other S proteins and five proteins of the large subunit, while protein S4 regulates two S proteins and one L protein. Consequently, when protein S4 was overexpressed (with S11), this repressed the synthesis of both L17 and S13, severely affecting the assembly of both subunits (167). Similarly, if the assembly of a regulatory protein is prevented for any reason, excess free protein might reduce the synthesis of its coregulated proteins. Since genes for proteins of both subunits are intermingled, and the translation of downstream genes is coupled to those upstream, nonsense mutations cause polarity that can affect the synthesis of proteins belonging to both subunits (22). Thus, a mutation in S14 that affected the assembly of both subunits was explained by reduced levels of proteins encoded downstream, including L6, L18, and L30 of the large subunit (50). A mutant protein L22 that confers erythromycin resistance also affected the assembly of both subunits (132). It is not known if this is a direct effect, but possible alternatives are effects on the expression of the downstream gene for the important assembly protein S17 (75) or on the assembly of the important regulator L4, whose assembly is enhanced by L22 in vitro (73, 140). As a result of such interconnections, it has sometimes been difficult to draw hard conclusions, and measurements of the synthesis rates of each protein are important to understand the exact cause of assembly defects.
Overexpression of either 16S or 23S rRNA from a plasmid derepressed synthesis from those ribosomal protein operons whose regulator binds the overexpressed rRNA species. This caused noncoordinate synthesis of proteins from each subunit, with an apparent effect on the assembly of both subunits (186). In addition, mutants can cause an upregulation of rRNA synthesis via the feedback regulation model. This is thought to explain the effects of a mutant S5 protein that affected the assembly of both subunits (65, 122). Protein S5 is not a regulatory protein, but the mutation was found to affect translation initiation, and it was suggested that this derepresses feedback control of rRNA synthesis, leading to an excess of rRNA over ribosomal protein that impairs assembly of both subunits (54, 186). It would be of interest to test whether other mutants affected in initiation show the same effects. At least one mutant, affected in 50S assembly, oversynthesizes both 16S and 23S rRNA yet assembles the 30S subunit fairly normally (112); further studies may provide a consensus.
However, in most cases, the defective assembly of one subunit has little effect on that of the other. For instance, temperature-sensitive mutants of S17 or S4 make no 30S at all at the nonpermissive temperature but still make 50S subunits. Despite the wholesale degradation of 30S components in the S4 mutant, 50S assembly shows only a slowed rate that could be expected during slower growth (75, 117, 130, 131).
Antitermination of rRNA transcripts is also tied to levels of free ribosomal proteins. At least two ribosomal proteins are part of the nus antitermination system acting on rrn transcription. These are proteins S10 (NusE) (127) and S4 (171). A shortage of these proteins would thus impair antitermination and reduce the synthesis of rRNA. Conversely, protein S1 is reported to antagonize the binding of nus factors to their RNA binding sites (119).
EFFECTS OF ANTIBIOTICS ON RIBOSOME SYNTHESIS
Antibiotics acting on translation are predicted to have several effects pertinent to rRNA synthesis. If they target the ribosome or its associated translation factors (and not aminoacyl-tRNA synthetases), levels of tRNA charging will not decline but will increase by reduced consumption. The RelA synthetase would not be induced, and the SpoT synthetase levels cannot be sustained by continual protein synthesis. Levels of ppGpp would then decline due to continued SpoT hydrolase activity. This expected reduction in ppGpp has been observed with most ribosomal inhibitors tested (6, 101). Increases in ATP and GTP levels by reduced consumption in translation were also predicted and have been demonstrated at least for spectinomycin and chloramphenicol (145).
The inhibition of protein synthesis by antibiotics that target the ribosome (or translation factors) thus allows bulk RNA synthesis to continue and tends to increase the proportion of rRNA synthesis, as demonstrated previously for streptomycin (57), erythromycin, chloramphenicol (13, 152), sparsomycin (157), fusidic acid (6), and puromycin (101). (In some cases, measurements of bulk RNA accumulation are used to approximate rRNA levels, since the majority of RNA in the cell is rRNA; tRNA is largely coregulated with it, and mRNA levels are relatively invariant [12].)
At low levels of translation inhibition, cells can compensate by upregulating the synthesis of ribosomal proteins (51), but as inhibition increases, this compensation fails to keep pace. Furthermore, the inhibition of translation can expose mRNA to degradation, resulting in polarity and the noncoordinate synthesis of ribosomal proteins from longer operons, exacerbating the shortage of ribosomal proteins further, as shown for chloramphenicol (51).
Such an imbalance causes the expected accumulation of ribosome precursors just as it does during the “relaxed” response to amino acid starvation. Previously reported examples include chloramphenicol (164), puromycin (81), chlortetracycline (77), and streptomycin (57). As expected, assembly of both subunits was affected.
Some Effects of Erythromycin on Assembly In Vivo
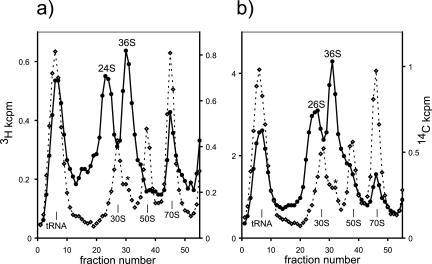
A pulse-labeling experiment similar to that shown in Fig. 2 but conducted 10 min after the addition of erythromycin to the growing culture is shown in Fig. 3 (B. A. Maguire and D. G. Wild, unpublished data). At this moderate concentration of erythromycin (50 μg/ml), growth and protein synthesis (as the incorporation of [3H]lysine per unit of growth) were reduced by 25%, and RNA synthesis (as [14C]uracil incorporation per unit of growth) was increased by 40%. The precursors to both subunits seen at both labeling times are clearly less advanced than in the earliest time point in the absence of antibiotic. The assembly of both subunits is inhibited, presumably as a result of imbalanced RNA and protein synthesis. The profiles show an accumulation of material akin to the p130S and p150S precursors, sedimenting at 24S and 36S, respectively (Fig. 3a and b). There is little earlier material visible between this and the tRNA peak, suggesting accumulation at a bottleneck. In addition to this change in kinetics, the accumulation of the precursor to the 50S is evident in the reference 14C-labeled profiles (marked with an asterisk), with a concomitant decrease in the 50S subunit peak. (Although a minor component, the 50S precursor is easily seen because most of the 14C-labeled subunits are in polysomes that have pelleted.) A simple explanation for precursor accumulation is that the assembly of both subunits is impaired by protein limitation, but because of its greater complexity, that of the 50S subunit is more affected. The conversion of RI50(1) to RI50*(1) precursors in vitro requires that a specific subset of proteins already be bound to the 23S rRNA in order to allow the conformational rearrangement. A similar situation may exist in vivo so that the p150S-like material cannot convert to p250S for a lack of the required proteins and therefore accumulates.
FIG. 3.
Effect of erythromycin on assembly in vivo. Methods are described in the legend of Fig. 2, but erythromycin was added to a concentration of 50 μg/ml 10 min before the addition of the 3H pulse-label. A precursor that accumulates sufficiently to be visible in both 14C-labeled and absorbance profiles is marked by an asterisk. Times of labeling in minutes (and generations) were 1 min 10 s (0.02 generations) (a) and 4 min (0.09 generations) (b). Approximate sedimentation values are given on the plots for material akin to the p130S and p150S precursors. kcpm, counts per minute (103).
Such a discrete conformational change is not seen for the 30S, where the p130S exhibits a continuous increase in sedimentation to 30S (Fig. 2). Thus, assembly of the simpler 30S subunit is less likely to arrest, and higher concentrations of antibiotic would be needed to cause a similar precursor accumulation. These effects of erythromycin on the assembly of both subunits seemed unremarkable against the coherent backdrop of antibiotic effects observed since the earliest days of ribosome research.
Evidence for Direct Inhibition of Assembly
However, over the last 13 years, Scott Champney and colleagues (East Tennessee State University) have published an impressive body of work in support of an alternative theory (25-28). The experiments survey the effects of over 40 inhibitors of the 50S subunit and three 30S inhibitors on assembly in E. coli and five other bacterial species. This alternative view posits that antibiotics such as erythromycin bind to the assembling subunit and directly interfere with its assembly. The precursors are stalled long enough to be subjected to RNase action, resulting in the accumulation of rRNA fragments and a shortage of the completed subunit (25, 155, 177). In this view, while the assembly of both subunits may be slowed by protein limitation (Fig. 3), because of the specific effect of erythromycin on the 50S precursor, only this precursor accumulates to levels detectable in the steady-state 14C-labeled profile.
Methods Used To Detect Assembly Inhibition
To examine the speed of subunit assembly, Champney et al. pulse-label cultures growing with or without antibiotic for 1 to 3 min with [3H]uridine and then withdraw samples during a chase period (lasting 0 to 90 min). Samples are lysed and centrifuged on sucrose density gradients, and the arrival of counts in 30S and 50S peaks versus time of chase is presented, rather than the raw traces shown here (Fig. 3). Labeling in uninhibited cultures increases rapidly to a plateau as subunits are completed. Some of the inhibited cultures show a much more gradual and continuous increase, perhaps as a result of degradation followed by reincorporation, so that completion of assembly is less easily defined. Inhibitors of the 30S slow the assembly of both subunits. Some 50S inhibitors appear to affect the assembly of 50S only, although a summary of results (see Table 1 in reference 28) shows that most slow the assembly of both subunits, often to a similar degree.
To measure the accumulation of subunits, cultures growing with or without antibiotic are labeled with [3H]uridine for two generations, followed by a 30-min chase with cold uridine before harvest (25). Lysates are centrifuged on sucrose density gradients, and amounts of label in the 30S and 50S peaks are recorded. In these experiments, inhibitors of the 50S subunit reduce the accumulation of 50S with minimal effects on 30S accumulation except at high concentrations of antibiotic that inhibit translation by about 80% or more. Results are thus broadly similar to those shown in Fig. 3, with a slowed assembly of both subunits and altered accumulation of the 50S subunit in particular. More pronounced effects on 30S assembly, seen with chloramphenicol and streptogramins B in several species (35, 102) and some other 50S inhibitors in Haemophilus influenzae, are taken to be exceptions (28).
Both methods used by Champney employ a much lower magnesium ion concentration in the gradients than that used here (0.25 mM versus 10 mM) (Fig. 3). This dissociates 70S couples into subunits so that they can be tracked separately as 30S or 50S. However, precursors akin to the p150S, such as that accumulated as shown in Fig. 3, would now sediment with the 30S peak. Champney et al. do detect such a stalled precursor to 50S in the 30S peak during inhibition under their conditions. Its presence was revealed by the detection of L proteins, 23S rRNA, and binding of [14C]erythromycin or azithromycin (28, 30, 177, 178). Degradation would reduce this accumulation, yet measurements of [14C]erythromycin binding under conditions of degradation (177) still showed a significant presence of the precursor in the 30S peak, even compared to the total concentration of 50S subunits (also detected by binding). As a result, the assembly rate and accumulation of 30S would seem higher (see, e.g., reference 30), masking the effects of 50S inhibitors on 30S assembly. Sedimentation at higher magnesium concentrations would allow both an unambiguous quantitation of 30S subunits and a clearer definition of subunit completion by monitoring the arrival of 16S and 23S rRNA in 70S ribosomes (see, e.g., reference 117).
Comparison of IC50s for Inhibition of Translation and Assembly
Subunit accumulation measurements performed at a range of antibiotic concentrations allow determinations of the antibiotic concentration that inhibits subunit assembly by 50% (the assembly IC50). The same cultures used to determine assembly IC50s are also used to determine translation IC50s (25). Translation measurements are usually made by Champney et al. during the chase period with cold uridine (i.e., after at least two generations of growth with antibiotic). After growth of a normal culture (without antibiotic) for two doublings, only 25% of the total ribosomes are those present at the start of growth, with the other 75% being newly synthesized. Thus, after two generations of growth with antibiotic, the assembly of most of the ribosomes in the cell (75%) would have been impaired by the antibiotic so that the translation IC50s as measured reflect both the inhibition of ribosome function and the fact that fewer ribosomes have been made in the presence of antibiotic.
The contribution of assembly effects to the “translation” IC50 can be estimated as follows: for a culture treated with antibiotic at the translation IC50, translation is inhibited by 50%. If we assume that this inhibits assembly by 50%, then only half of the ribosomes made in the control will be made, and this (37.5%), together with the preexisting ones (25%), will be roughly half (62.5%) that of the uninhibited control. The activity of this reduced population is inhibited by 50% to give a combined effect that is approximately double that on translation alone so that the IC50 for translation is half that for assembly when measured in this way (25, 28).
As expected, IC50s for translation that are half that for assembly have been found in most cases (28), indicating that the inhibition of assembly is equivalent to that of translation. The same ratio would also be expected if assembly inhibition were caused simply by the inhibition of translation. Depending on which underlying cause is assumed, assembly inhibition can therefore be viewed either as a separate target of equal importance to translation or as an inevitable secondary consequence of translation inhibition.
IC50s that are the same for both translation and assembly have been recorded in some species for azithromycin, clarithromycin (29), cethromycin, and telithromycin (31). This is hard to reconcile with either model: since the translation IC50s as measured include assembly effects, these standard inhibitors of protein synthesis would have to inhibit only assembly in order to give this result, an unlikely scenario.
To resolve these uncertainties, effects on translation alone could be measured a few minutes after the addition of antibiotic to the growing culture before appreciable effects on subunit accumulation. Antibiotics such as kirromycin or fusidic acid inhibit translation by binding elongation factors EF-Tu and EF-G rather than the ribosome itself and therefore cannot directly influence the assembly of subunits by binding to their precursors. As such, they could provide baseline studies to show the nonspecific effects on assembly of inhibiting translation alone. Certain amino acid analogs that inhibit translation but allow rRNA synthesis to continue (175) might also be useful in this regard.
Resistance and the Binding of Antibiotics to Precursors
An important finding is that erythromycin binds to the stalled 50S precursor in inhibited cells, so direct interference with assembly is at least possible (177). However, the antibiotic could equally bind but have no influence on assembly. Perversely, perhaps, ketolide antibiotics can promote assembly under suboptimal conditions in vitro, (91, 149), so the inhibition of assembly should not be assumed.
The methylation of a single nucleotide in 23S rRNA by erm methylases confers erythromycin resistance on the 50S subunit, and this methylation abolishes erythromycin binding to both precursor and completed 50S, suggesting that at least some of the critical binding determinants are common to both. Champney attributed the loss of assembly inhibition in such resistant strains to the prevention of binding to the precursor (28), yet a simpler explanation is that since translation is not inhibited, neither is assembly.
These studies underline the difficulties in divorcing effects on translation from direct effects on assembly. Since translation depends on ribosome assembly, and ribosome assembly requires translation, we are faced with a chicken-and-egg situation. However, the erm methylases also provide a means to separate these effects since they can methylate only naked or partially assembled 23S rRNA and not the completed subunit (134). This means that if the production of the methylase were induced in an otherwise erythromycin-sensitive strain, the precursor pool would become resistant before the mature subunits. It would take a generation of growth to achieve resistance in half of the mature subunits, while changes in the susceptibility of assembly to direct interference could be detected on a time scale of seconds to minutes (using the techniques described in the legend of Fig. 3).
The induction of ermC methylase production by the addition of erythromycin to cultures of Staphylococcus aureus (30) showed that methylation of the precursor increases in the first 15 min so that erythromycin can no longer bind to it (30). Champney et al. commented that at least 1 h is needed for a substantial fraction of the completed ribosomes to become methylated and that the rate of 50S subunit formation was reduced during this time period. They acknowledged that this assembly inhibition results from antibiotic binding to the 50S subunit rather than to the precursor, but they maintained that a direct effect on the precursor might still play a role. To obtain a definitive answer, the timing of assembly inhibition relative to translation inhibition during the induction of erm methylation by erythromycin should be directly observed on pulse-labeled gradients as shown in Fig. 3.
Inhibitors of the 30S Subunit
Effects of three aminoglycoside antibiotics have also been studied. These are neomycin and paromomycin in both E. coli and S. aureus (116) and hygromycin B in E. coli alone (114). The data showed a reduced accumulation of both subunits in all cases, with an effect on the 50S subunit that is proportionately greater than that on the 30S subunit in some instances (114, 115). A putative coupling between the assembly of the 50S subunit and that of the 30S subunit, rather than a restriction of translation, is proposed to account for this finding (28), but such a coupling has not been established.
Although aminoglycosides have normally been thought of as inhibitors of the 30S subunit and can be seen in X-ray crystallographic structures bound to the 30S of Thermus thermophilus, either on its own (14, 23) or as part of the 70S ribosome (148), recent crystal structures of the E. coli 70S subunit unexpectedly showed neomycin and paromomycin bound not just to the 30S subunit but also to a site in helix 69 of the large subunit 23S rRNA, at the subunit interface (9). Studies of ribosomal mutations that confer resistance to aminoglycosides are expected to shed light on the relevance of this interaction. If correct, it may explain an observed labeling of 50S subunits (as well as 30S subunits and their precursor) with [3H]neomycin or [3H]paromomycin during growth with these ligands (60). If 50S precursors can also bind aminoglycosides, then a direct effect on the assembly of both subunits is possible. More work will be needed to clarify possible effects of aminoglycosides on the 50S.
In Vitro Studies
A 21S precursor to the 30S subunit accumulated during assembly both in vivo and in vitro in the presence of neomycin or paromomycin (60). The in vitro study is particularly interesting, as it offers a rare glimpse of assembly divorced from synthesis and so could potentially resolve the issue of whether binding to precursors actually inhibits their assembly. Earlier work also reported an inhibition of in vitro assembly by streptomycin (8). However, particular caution is required with the aminoglycoside class of antibiotics. Due to their unusually polycationic nature, their interaction with RNA is dominated by electrostatics (180): they bind to various other RNAs and ribonucleoproteins in addition to ribosomes (147), and the binding of up to 100 streptomycin molecules per ribosome has been observed (46). Neomycin and paromomycin are particularly promiscuous in this respect (170) and could therefore quench assembly in vitro by binding to rRNA nonspecifically. By analogy, the polyanionic dye cibacron blue F3GA has been used to quench in vitro assembly reactions for kinetic studies, as it binds to ribosomal proteins and prevents further binding to rRNA (48). In fact, the antibiotic concentrations described by Champney seem quite moderate, but it would nevertheless be prudent to eliminate this caveat by assembling the 30S in vitro from rRNA or proteins harboring aminoglycoside resistance mutations. If the antibiotic effect on assembly is specific, these components should assemble in vitro without hindrance.
rRNA Degradation and Lethality
Mutants defective in certain RNase activities have been shown to be more sensitive to the inhibition of assembly by antibiotics, suggesting an involvement of the RNases in the degradation of abnormal precursors (155, 177). Sensitivity to degradation is expected for precursors regardless of how they are generated (69, 78, 166). Nucleases are often involved in both the degradation and biogenesis of RNA (53), and at least one of the implicated enzymes (RNase E) processes rRNA during assembly. Strains lacking RNases may have subtle assembly defects that sensitize them to antibiotic inhibition, while the inability to break down and recycle misfolded precursors will further stress the cells.
It has been suggested that the inhibition of 50S assembly is lethal for bacteria because of RNase degradation of the ribosomal RNAs (27, 30, 32-34), but no causal link has been established, and inhibitors of 50S assembly that are merely bacteriostatic also cause the degradation of rRNA (155).
Prospects
It is inevitable that if translation is inhibited enough, it will affect assembly. What is unclear is whether the direct binding to precursors affects assembly and, if so, whether this effect is more significant than the indirect one. If the direct effect on assembly is significant only at low levels of translation inhibition, as suggested previously (26), it may be less relevant given the significant levels of inhibition required for clinical effectiveness. The extensive studies of Champney et al. have highlighted the sometimes-overlooked effect of the inhibition of ribosome assembly by translation inhibitors, revealing many previously unknown consequences. However, different experiments will be required to understand how this assembly inhibition arises; for now, the ideas that binding to precursors affects assembly or that the assembly defects cause death remain speculative.
To differentiate between specific and nonspecific effects requires additional kinetic studies during the induction of methylases that confer resistance (or sensitivity) to antibiotics that affect assembly, studies of in vitro assembly using resistant components, monitoring of translation rates during the course of inhibition, and studies with translation factor inhibitors. These studies may resolve whether ribosome assembly is a direct and specific target of current antibiotics. A more practical question, addressed next, is whether assembly inhibition is worth pursuing as a drug target in its own right and, if so, whether we have the means to prosecute the target.
Effects of Inhibiting Assembly Alone
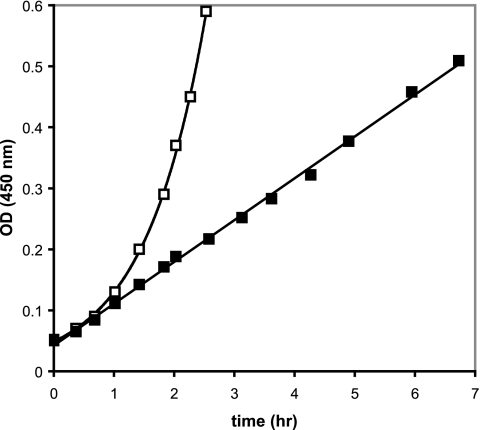
What would the effect of a direct and complete inhibition of assembly be on a growing culture? An example is provided by the conditional repression of synthesis of proteins L28 and L33 in vivo. Assembly of the 50S subunit ceases abruptly when these proteins are no longer made (106), and an abnormal precursor accumulates instead. Growth is limited by the existing ribosome concentration and so is arithmetic rather than logarithmic (Fig. 4). Assembly and growth recover if the synthesis of L28 is recommenced (106).
FIG. 4.
Growth of strain BM108 with isopropyl-β-thiogalactopyranoside (expressing protein L28 but not protein L33) or without isopropyl-β-thiogalactopyranoside (neither protein expressed). OD, optical density. (Based on data from reference 106.)
In this case, at least, the inhibition of assembly in the absence of direct inhibition of translation is effectively bacteriostatic, although growth cessation takes much longer than if translation were directly inhibited by an antibiotic. Assembly mutants generally have normal colony-forming abilities and are not prone to lysis in liquid culture, although in one isolated case (a cold-sensitive L6 gentamicin-resistant mutant), it was speculated that mutant ribosomes or their precursors might interact with the cytoplasmic membrane to cause lysis (11). The inability to synthesize ribosomes means that cells must rely on preexisting ones, but provided that the mature ribosomes are not degraded (a rare occurrence) (56, 90, 129), growth can continue at a low rate and recover when conditions for assembly improve. Under the conditions described in the legend of Fig. 4, recovery is swift, perhaps because assembly is arrested at a relatively late stage here, and the precursor is stable. Degradation of the precursors would certainly have slowed this recovery.
Given the persistent nature of some infections, bactericidal inhibitors are often preferred in order to limit the emergence of resistance during the course of treatment. Inhibition of assembly is a useful adjunct for any inhibitor of translation and doubtless accounts for some of the efficacy of currently used antibiotics. However, as an end in itself, more direct, rapid, and lethal mechanisms would be preferred.
Two further observations are worth noting. The first is that in the original genetic background in which the mutation conferring loss of L28 and L33 was isolated, lethality was suppressed, apparently by an erythromycin resistance mutation in protein L4 that slightly alters the early kinetics of 50S assembly (40, 105). The finding that a complete block of assembly can be circumvented by compensating mutations in other components augurs badly for inhibitors that interfere with a specific assembly interaction, as a high frequency of mutation to resistance is likely. Second, if synthesis of the two proteins is not abolished but only halved, as in another mutant, then ribosomes are assembled, albeit slowly (21). The same abnormal precursor lacking proteins accumulates (18, 19), and the missing proteins (including L28 and L33) are just added later in the assembly process, as if an afterthought, to produce a subunit that functions well enough for growth and viability. Thus, if the inhibition of synthesis is less than absolute, the subunit simply assembles by a different route. Although these observations refer to a specific case, they provide instructive and cautionary examples of the pitfalls to be expected in targeting assembly. While there is essentially one way to make a protein, there are apparently many ways to make a ribosome. This inherent flexibility of assembly will make it a difficult target to constrain as effectively as translation itself.
Targeting Assembly
As described above, the assembly of ribosomes begins with transcription of the rRNA. In many species, transcription is from multiple copies of rRNA operons (seven in E. coli, six in H. influenzae, five in S. aureus, and four in Streptococcus pneumoniae and Pseudomonas aeruginosa) (144). Unlike mutations in the singly encoded ribosomal proteins, mutation of the rRNA to resistance is disfavored by the need to mutate multiple copies. Antibiotics that target the rRNA are attractive for this reason and could in theory target assembly just as they do translation. Although evidence for direct interference in assembly by current antibiotics remains inconclusive, the idea has inspired many attempts to develop such inhibitors. Antibiotics such as spectinomycin inhibit ribosome function by blocking structural transitions of the completed ribosome that are required for function (10). The smaller molecule may prevail in such David-versus-Goliath encounters by exploiting a restricted range of motion in the larger structure, rather like a small block of wood inserted into a door jamb to prevent closure. It could be harder to block such transitions in a largely unfolded rRNA during assembly, and the inhibitor might be shrugged off, circumventing the steric block. Since assembly can progress by multiple pathways, the blockage might also be cooperatively overcome by the completion of other steps of assembly that are not directly inhibited.
Alternatively, rather than inhibiting structural transitions, an inhibitor could block the binding of a ligand to the rRNA. This could be the binding of either a ribosomal protein, an auxiliary factor, or another rRNA element. The binding of the small molecule would have to be avid enough to prevent exchange with the natural ligand that might otherwise allow assembly to resume. In vitro studies showed that some proteins (such as L20 and L24) can be removed from subunits by high salt yet do not need to be added back to reassemble active subunits (128, 158); these proteins are required only for assembly and are not needed once they have fulfilled this role. For such proteins, even a fleeting interaction might bypass the blockage and allow assembly to complete. Effective inhibitors that do not allow sufficient escape for resistance to arise may thus be difficult to obtain. However, the degradation of stalled precursors could play an important part in hindering the reversibility of the inhibition.
The next issue is how to detect compounds that affect assembly. Assays using whole cells suffer from the same problems that attended selections for assembly mutants in early studies: the inhibition of translation itself is hard to distinguish from the inhibition of assembly, as both result in an accumulation of ribosome precursors and impaired translation. Some assembly mutants are cold sensitive, so increased sensitivity to inhibition at low temperatures could be considered, but this is a very crude measure. Inhibitors of a given function can sometimes be revealed by screening for compounds that preferentially inhibit mutants that are already somewhat impaired in that function, but as we have seen, mutations that impair assembly can radically redirect its course from normal assembly.
In light of this finding, in vitro assays might seem tempting. Although RNA-protein interactions can be a challenge for high-throughput screening, an attractive assay that uses fluorescent resonance energy transfer to monitor the interaction of protein S15 with a fragment of 16S rRNA has been developed (92). In another approach, a peptide derivative called a helix-threading peptide was found to intercalate with an RNA helix that is part of the S15 binding site. The peptide interferes with S15 binding, but whether results with this minimal system can be reproduced in vitro with the whole binding site or even the whole ribosome remains to be seen (64). More specifically, protein S15, although important for assembly in vitro, has since been found to be dispensable in vivo (17), confirming the disconnect between the processes in vitro and in vivo.
Another problem is that in vitro assembly is sensitive to the ionic conditions chosen. For instance, protein L15 was long considered to be essential in vitro yet became dispensable if different in vitro ionic conditions were used (61, 74). Failure to translate in vitro inhibition to efficacy against intact bacteria in vivo is a common cause of attrition for antibacterial drug candidates, and the pursuit of inhibitors of an in vitro assay so obviously divergent from assembly in vivo would render the odds of success prohibitively low. The challenge therefore remains to find or construct an in vivo reporter specific to assembly inhibition.
Another possibility is to target auxiliary factors (4, 41). Since these are single gene targets, resistance may be a problem. However, the GTPases are small, soluble, and amenable to X-ray crystallography to determine their structure (38, 137) and cryo-electron microscopy to determine their interaction with the ribosome (49, 150). This offers hope that structure-based drug design might guide the development of inhibitors that interact with residues of such functional importance that mutation to resistance is disfavored. The fact that some of these GTPases are essential for viability is encouraging, but many of them are involved in multiple processes in the cell, so this essentiality may not relate to their role in assembly (discussed in references 83, 86, and 176). Nevertheless, the pleiotropic effects of their inhibition may in fact offer multiple ways to inhibit cell growth through a single target, an attractive feature that might limit the emergence of resistance. Ironically, then, the best targets that affect assembly may be successful through their effects on other functions. In the meantime, the development of improved translational inhibitors remains a reliable option, albeit with concomitant effects on assembly that are now much better appreciated due to extensive recent studies.
ADDENDUM
During review of the manuscript, Siibak et al. published a provocative series of experiments that directly addressed the issue of specificity for the inhibition of assembly (153). The translation of a short “E peptide” was previously shown to confer cis-acting erythromycin resistance upon the translating ribosome (169). Siiback et al. now show that the overexpression of this peptide simultaneously relieves the erythromycin inhibition of assembly, with the interpretation that the inhibition of assembly is merely a result of translation inhibition. However, further studies may be required, since earlier unpublished experiments apparently suggested that at low levels of peptide expression, its effects on assembly and translation inhibition can be separable so that only the latter is relieved (26). A second experiment examined the erythromycin resistance or sensitivity of the 23S rRNA detected in assembly intermediates that accumulate during the inhibition of assembly by erythromycin. This was done by using a strain of S. aureus that contains two genes encoding wild-type 23S rRNA and three genes encoding 23S that has a point mutation conferring erythromycin resistance. Both sensitive and resistant 23S rRNA were found to accumulate in the precursor particles, suggesting that erythromycin binding to the precursors is not required for their accumulation. These elegant experiments may be the first of many experiments to shed new light on existing data by directly addressing the issue of mechanism.
Acknowledgments
I give special thanks to Don Wild (Oxford) and Matt Kidd (Pfizer Inc.) for data retrieval.
REFERENCES
- 1.Agalarov, S. C., O. M. Selivanova, E. N. Zheleznyakova, L. A. Zheleznaya, N. I. Matvienko, and A. S. Spirin. 1999. Independent in vitro assembly of all three major morphological parts of the 30S ribosomal subunit of Thermus thermophilus. Eur. J. Biochem. 266533-537. [DOI] [PubMed] [Google Scholar]
- 2.Alix, J.-H. 2004. The work of chaperones. Protein Synth. Ribosome Struct. 2004529-562. [Google Scholar]
- 3.Alix, J. H. 1993. Extrinsic factors in ribosome assembly. Transl. Appar. Proc. Int. Conf. 1993173-184. [Google Scholar]
- 4.Al Refaii, A., and J.-H. Alix. 2008. Inhibition of chaperone-dependent bacterial ribosome biogenesis. Methods Mol. Med. 14275-85. [DOI] [PubMed] [Google Scholar]
- 5.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289902-921. [DOI] [PubMed] [Google Scholar]
- 6.Bennett, P. M., and O. Maaloe. 1974. Effects of fusidic acid on growth, ribosome synthesis, and RNA metabolism in Escherichia coli. J. Mol. Biol. 90541-561. [DOI] [PubMed] [Google Scholar]
- 7.Besancon, W., and R. Wagner. 1999. Characterization of transient RNA-RNA interactions important for the facilitated structure formation of bacterial ribosomal 16S RNA. Nucleic Acids Res. 274353-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas, D. K., and L. Gorini. 1972. Attachment site of streptomycin to the 30S ribosomal subunit. Proc. Natl. Acad. Sci. USA 692141-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borovinskaya, M. A., R. D. Pai, W. Zhang, B. S. Schuwirth, J. M. Holton, G. Hirokawa, H. Kaji, A. Kaji, and J. H. D. Cate. 2007. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 14727-732. [DOI] [PubMed] [Google Scholar]
- 10.Borovinskaya, M. A., S. Shoji, J. M. Holton, K. Fredrick, and J. H. D. Cate. 2007. A steric block in translation caused by the antibiotic spectinomycin. ACS Chem. Biol. 2545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosl, A., and A. Bock. 1981. Ribosomal mutation in Escherichia coli affecting membrane stability. Mol. Gen. Genet. 182358-360. [DOI] [PubMed] [Google Scholar]
- 12.Bremer, H., and P. P. Dennis. 1987. Modulation of chemical composition and other parameters of the cell by growth rate. Escherichia coli Salmonella typhimurium 21527-1542. [Google Scholar]
- 13.Brock, T. D., and M. L. Brock. 1959. Similarity in mode of action of chloramphenicol and erythromycin. Biochim. Biophys. Acta 33274-275. [DOI] [PubMed] [Google Scholar]
- 14.Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 1031143-1154. [DOI] [PubMed] [Google Scholar]
- 15.Bryant, R. E., and P. S. Sypherd. 1974. Genetic analysis of cold-sensitive ribosome maturation mutants of Escherichia coli. J. Bacteriol. 1171082-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bubunenko, M., T. Baker, and D. L. Court. 2007. Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J. Bacteriol. 1892844-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bubunenko, M., A. Korepanov, D. L. Court, I. Jagannathan, D. Dickinson, B. R. Chaudhuri, M. B. Garber, and G. M. Culver. 2006. 30S ribosomal subunits can be assembled in vivo without primary binding ribosomal protein S15. RNA 121229-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler, P. D., P. F. G. Sims, and D. G. Wild. 1979. Abnormal ribosome assembly in a mutant of Escherichia coli. Biochem. J. 182493-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler, P. D., P. F. G. Sims, and D. G. Wild. 1980. Intermediates in the assembly of ribosomes by a mutant of Escherichia coli. Biochem. J. 190157-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler, P. D., and D. G. Wild. 1985. The location of a mutation affecting ribosomal protein synthesis by Escherichia coli. J. Gen. Microbiol. 131135-144. [DOI] [PubMed] [Google Scholar]
- 21.Butler, P. D., and D. G. Wild. 1984. Ribosomal protein synthesis by a mutant of Escherichia coli. Eur. J. Biochem. 144649-654. [DOI] [PubMed] [Google Scholar]
- 22.Cabezon, T., G. Delcuve, M. Faelen, L. Desmet, and A. Bollen. 1980. Polarity of amber mutations in ribosomal protein genes of Escherichia coli. J. Bacteriol. 14141-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature (London) 407340-348. [DOI] [PubMed] [Google Scholar]
- 24.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 25.Champney, W. S. 2003. Bacterial ribosomal subunit assembly is an antibiotic target. Curr. Top. Med. Chem. (Sharjah) 3929-947. [DOI] [PubMed] [Google Scholar]
- 26.Champney, W. S. 2001. Bacterial ribosomal subunit synthesis: a novel antibiotic target. Curr. Drug Targets Infect. Disord. 119-36. [DOI] [PubMed] [Google Scholar]
- 27.Champney, W. S. 1999. Macrolide antibiotic inhibition of 50S ribosomal subunit formation in bacterial cells. Recent Res. Dev. Antimicrob. Agents Chemother. 339-58. [Google Scholar]
- 28.Champney, W. S. 2006. The other target for ribosomal antibiotics: inhibition of bacterial ribosomal subunit formation. Infect. Disord. Drug Targets. 6377-390. [DOI] [PubMed] [Google Scholar]
- 29.Champney, W. S., and R. Burdine. 1998. Azithromycin and clarithromycin inhibition of 50 S ribosomal subunit formation in Staphylococcus aureus cells. Curr. Microbiol. 36119-123. [DOI] [PubMed] [Google Scholar]
- 30.Champney, W. S., H. S. Chittum, and C. L. Tober. 2003. A 50S ribosomal subunit precursor particle is a substrate for the ErmC methyltransferase in Staphylococcus aureus cells. Curr. Microbiol. 46453-460. [DOI] [PubMed] [Google Scholar]
- 31.Champney, W. S., N. Mentens, and K. Zurawick. 2004. An examination of the differential sensitivity to ketolide antibiotics in ermB strains of Streptococcus pyogenes and Streptococcus pneumoniae. Curr. Microbiol. 49239-247. [DOI] [PubMed] [Google Scholar]
- 32.Champney, W. S., and M. Miller. 2002. Inhibition of 50S ribosomal subunit assembly in Haemophilus influenzae cells by azithromycin and erythromycin. Curr. Microbiol. 44418-424. [DOI] [PubMed] [Google Scholar]
- 33.Champney, W. S., and J. Pelt. 2002. The ketolide antibiotic ABT-773 is a specific inhibitor of translation and 50S ribosomal subunit formation in Streptococcus pneumoniae cells. Curr. Microbiol. 45155-160. [DOI] [PubMed] [Google Scholar]
- 34.Champney, W. S., and C. L. Tober. 2000. Evernimicin (SCH27899) inhibits both translation and 50S ribosomal subunit formation in Staphylococcus aureus cells. Antimicrob. Agents Chemother. 441413-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Champney, W. S., and C. L. Tober. 2000. Specific inhibition of 50 S ribosomal subunit formation in Staphylococcus aureus cells by 16-membered macrolide, lincosamide, and streptogramin B antibiotics. Curr. Microbiol. 41126-135. [DOI] [PubMed] [Google Scholar]
- 36.Charollais, J., M. Dreyfus, and I. Iost. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 322751-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charollais, J., D. Pflieger, J. Vinh, M. Dreyfus, and I. Iost. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 481253-1265. [DOI] [PubMed] [Google Scholar]
- 38.Chen, X., D. L. Court, and X. Ji. 1999. Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif. Proc. Natl. Acad. Sci. USA 968396-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole, J. R., C. L. Olsson, J. W. B. Hershey, M. Grunberg-Manago, and M. Nomura. 1987. Feedback regulation of rRNA synthesis in Escherichia coli. Requirement for initiation factor IF2. J. Mol. Biol. 198383-392. [DOI] [PubMed] [Google Scholar]
- 40.Coleman, S. H., B. A. Maguire, and D. G. Wild. 1993. Ribosome assembly in three strains of Escherichia coli with mutations in the rpmBG operon. J. Gen. Microbiol. 139707-716. [DOI] [PubMed] [Google Scholar]
- 41.Comartin, D. J., and E. D. Brown. 2006. Non-ribosomal factors in ribosome subunit assembly are emerging targets for new antibacterial drugs. Curr. Opin. Pharmacol. 6453-458. [DOI] [PubMed] [Google Scholar]
- 42.Condon, C., S. French, C. Squires, and C. L. Squires. 1993. Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. EMBO J. 124305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cowgill de Narvaez, C., and H. W. Schaup. 1979. In vivo transcriptionally coupled assembly of Escherichia coli ribosomal subunits. J. Mol. Biol. 1341-22. [DOI] [PubMed] [Google Scholar]
- 44.Culver, G. M. 2003. Assembly of the 30S ribosomal subunit. Biopolymers 68234-249. [DOI] [PubMed] [Google Scholar]
- 45.Dagley, S., G. Turnock, and D. G. Wild. 1963. The accumulation of ribonucleic acid (RNA) by a mutant of Escherichia coli. Biochem. J. 88555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dahlberg, A. E., F. Horodyski, and P. Keller. 1978. Interaction of neomycin with ribosomes and ribosomal ribonucleic acid. Antimicrob. Agents Chemother. 13331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dammel, C. S., and H. F. Noller. 1995. Suppression of a cold-sensitive mutation in 16S rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 9626-637. [DOI] [PubMed] [Google Scholar]
- 48.Datta, D., L. M. Changchien, and G. R. Craven. 1986. Studies on the kinetic sequence of in vitro ribosome assembly using cibacron blue F3GA as a general assembly inhibitor. Nucleic Acids Res. 144095-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Datta, P. P., D. N. Wilson, M. Kawazoe, N. K. Swami, T. Kaminishi, M. R. Sharma, T. M. Booth, C. Takemoto, P. Fucini, S. Yokoyama, and R. K. Agrawal. 2007. Structural aspects of RbfA action during small ribosomal subunit assembly. Mol. Cell 28434-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delcuve, G., T. Cabezon, A. Ghysen, A. Herzog, and A. Bollen. 1978. Amber mutations in Escherichia coli essential genes: isolation of mutants affected in the ribosomes. Mol. Gen. Genet. 157149-153. [DOI] [PubMed] [Google Scholar]
- 51.Dennis, P. P. 1976. Effects of chloramphenicol on the transcriptional activities of ribosomal RNA and ribosomal protein genes in Escherichia coli. J. Mol. Biol. 108535-546. [DOI] [PubMed] [Google Scholar]
- 52.Dennis, P. P., M. Ehrenberg, and H. Bremer. 2004. Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol. Mol. Biol. Rev. 68639-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deutscher, M. P. 2006. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 34659-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dodd, J., J. M. Kolb, and M. Nomura. 1991. Lack of complete cooperativity of ribosome assembly in vitro and its possible relevance to in vivo ribosome assembly and the regulation of ribosomal gene expression. Biochimie 73757-767. [DOI] [PubMed] [Google Scholar]
- 55.Dohme, F., and K. H. Nierhaus. 1976. Total reconstitution and assembly of 50 S subunits from Escherichia coli ribosomes in vitro. J. Mol. Biol. 107585-599. [DOI] [PubMed] [Google Scholar]
- 56.Dong, H., L. Nilsson, and C. G. Kurland. 1995. Gratuitous overexpression of genes in Escherichia coli leads to growth inhibition and ribosome destruction. J. Bacteriol. 1771497-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dubin, D. T. 1964. Some effects of streptomycin on RNA [ribonucleic acid] metabolism in Escherichia coli. J. Mol. Biol. 8749-767. [DOI] [PubMed] [Google Scholar]
- 58.El Hage, A., and J.-H. Alix. 2004. Authentic precursors to ribosomal subunits accumulate in Escherichia coli in the absence of functional DnaK chaperone. Mol. Microbiol. 51189-201. [DOI] [PubMed] [Google Scholar]
- 59.El Hage, A., M. Sbai, and J. H. Alix. 2001. The chaperonin GroEL and other heat-shock proteins, besides DnaK, participate in ribosome biogenesis in Escherichia coli. Mol. Gen. Genet. 264796-808. [DOI] [PubMed] [Google Scholar]
- 60.Foster, C., and W. Champney. 2007. Characterization of a 30S ribosomal subunit assembly intermediate found in Escherichia coli cells growing with neomycin or paromomycin. Arch. Microbiol. 189441-449. [DOI] [PubMed] [Google Scholar]
- 61.Franceschi, F. J., and K. H. Nierhaus. 1988. Ribosomal protein L20 can replace the assembly-initiator protein L24 at low temperatures. Biochemistry 277056-7059. [DOI] [PubMed] [Google Scholar]
- 62.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 2782092-2097. [DOI] [PubMed] [Google Scholar]
- 63.Gaal, T., and R. L. Gourse. 1990. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 875533-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gooch, B. D., M. Krishnamurthy, M. Shadid, and P. A. Beal. 2005. Binding of helix-threading peptides to E. coli 16S ribosomal RNA and inhibition of the S15-16S complex. ChemBioChem 62247-2254. [DOI] [PubMed] [Google Scholar]
- 65.Guthrie, C., H. Nashimoto, and M. Nomura. 1969. Studies on the assembly of ribosomes in vivo. Cold Spring Harb. Symp. Quant. Biol. 3469-75. [DOI] [PubMed] [Google Scholar]
- 66.Hager, J., B. L. Staker, H. Buegl, and U. Jakob. 2002. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 27741978-41986. [DOI] [PubMed] [Google Scholar]
- 67.Hardy, S. J. S. 1975. Stoichiometry of the ribosomal proteins of Escherichia coli. Mol. Gen. Genet. 140253-274. [DOI] [PubMed] [Google Scholar]
- 68.Haseltine, W. A., and R. Block. 1973. Synthesis of guanosine tetra- and pentaphosphate requires the presence of codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. USA 701564-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hayes, F., and D. H. Hayes. 1971. Biosynthesis of ribosomes in Escherichia coli. I. Properties of ribosomal precursor particles and their RNA components. Biochimie 53369-382. [DOI] [PubMed] [Google Scholar]
- 70.Held, W. A., B. Ballou, S. Mizushima, and M. Nomura. 1974. Structure and function of bacterial ribosomes. 23. Assembly mapping of 30S ribosomal proteins from Escherichia coli. J. Biol. Chem. 2493103-3111. [PubMed] [Google Scholar]
- 71.Held, W. A., and M. Nomura. 1973. Structure and function of bacterial ribosomes. XX. Rate-determining step in the reconstitution of Escherichia coli 30S ribosomal subunits. Biochemistry 123273-3281. [DOI] [PubMed] [Google Scholar]
- 72.Hernandez, V. J., and H. Bremer. 1993. Characterization of RNA and DNA synthesis in Escherichia coli strains devoid of ppGpp. J. Biol. Chem. 26810851-10862. [PubMed] [Google Scholar]
- 73.Herold, M., and K. H. Nierhaus. 1987. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J. Biol. Chem. 2628826-8833. [PubMed] [Google Scholar]
- 74.Herold, M., V. Nowotny, E. R. Dabbs, and K. H. Nierhaus. 1986. Assembly analysis of ribosomes from a mutant lacking the assembly-initiator protein L24: lack of L24 induces temperature sensitivity. Mol. Gen. Genet. 203281-287. [DOI] [PubMed] [Google Scholar]
- 75.Herzog, A., M. Yaguchi, T. Cabezon, M. C. Corchuelo, J. Petre, and A. Bollen. 1979. A missense mutation in the gene coding for ribosomal protein S17 (rpsQ) leading to ribosomal assembly defectivity in Escherichia coli. Mol. Gen. Genet. 17115-22. [DOI] [PubMed] [Google Scholar]
- 76.Himeno, H., K. Hanawa-Suetsugu, T. Kimura, K. Takagi, W. Sugiyama, S. Shirata, T. Mikami, F. Odagiri, Y. Osanai, D. Watanabe, S. Goto, L. Kalachnyuk, C. Ushida, and A. Muto. 2004. A novel GTPase activated by the small subunit of ribosome. Nucleic Acids Res. 325303-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holmes, I. A., and D. G. Wild. 1967. Inhibition of the growth of Escherichia coli by chlortetracycline. Biochem. J. 10469-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holmes, I. A., and D. G. Wild. 1965. The synthesis of ribonucleic acid during inhibition of Escherichia coli by chlortetracycline. Biochem. J. 97277-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holmes, K. L., and G. M. Culver. 2005. Analysis of conformational changes in 16S rRNA during the course of 30S subunit assembly. J. Mol. Biol. 354340-357. [DOI] [PubMed] [Google Scholar]
- 80.Homann, H. E., and K. H. Nierhaus. 1971. Ribosomal proteins. Protein compositions of biosynthetic precursors and artificial subparticles from ribosomal subunits in Escherichia coli K 12. Eur. J. Biochem. 20249-257. [DOI] [PubMed] [Google Scholar]
- 81.Hosokawa, K., and M. Nomura. 1965. Incomplete ribosomes produced in chloramphenicol- and puromycin-inhibited Escherichia coli. J. Mol. Biol. 12225-241. [DOI] [PubMed] [Google Scholar]
- 82.Hwang, J., and M. Inouye. 2006. The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 611660-1672. [DOI] [PubMed] [Google Scholar]
- 83.Inoue, K., J. Alsina, J. Chen, and M. Inouye. 2003. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol. Microbiol. 481005-1016. [DOI] [PubMed] [Google Scholar]
- 84.Jensen, K. F., and S. Pedersen. 1990. Metabolic growth rate control in Escherichia coli may be a consequence of subsaturation of the macromolecular biosynthetic apparatus with substrates and catalytic components. Microbiol. Rev. 5489-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jeong, J.-H., M. Kitakawa, S. Isono, and K. Isono. 1993. Cloning and nucleotide sequencing of the genes, rplU and rpmA, for ribosomal proteins L21 and L27 of Escherichia coli. DNA Seq. 459-67. [DOI] [PubMed] [Google Scholar]
- 86.Jiang, M., K. Datta, A. Walker, J. Strahler, P. Bagamasbad, P. C. Andrews, and J. R. Maddock. 2006. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 1886757-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jinks-Robertson, S., R. L. Gourse, and M. Nomura. 1983. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell 33865-876. [DOI] [PubMed] [Google Scholar]
- 88.Kaczanowska, M., and M. Ryden-Aulin. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71477-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaczanowska, M., and M. Ryden-Aulin. 2005. The YrdC protein—a putative ribosome maturation factor. Biochim. Biophys. Acta 172787-96. [DOI] [PubMed] [Google Scholar]
- 90.Kalpaxis, D. L., P. Karahalios, and M. Papapetropoulou. 1998. Changes in ribosomal activity of Escherichia coli cells during prolonged culture in sea salts medium. J. Bacteriol. 1803114-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khaitovich, P., and A. S. Mankin. 1999. Effect of antibiotics on large ribosomal subunit assembly reveals possible function of 5 S rRNA. J. Mol. Biol. 2911025-1034. [DOI] [PubMed] [Google Scholar]
- 92.Klostermeier, D., P. Sears, C.-H. Wong, D. P. Millar, and J. R. Williamson. 2004. A three-fluorophore FRET assay for high-throughput screening of small-molecule inhibitors of ribosome assembly. Nucleic Acids Res. 322707-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Korepanov, A. P., G. M. Gongadze, M. B. Garber, D. L. Court, and M. G. Bubunenko. 2007. Importance of the 5 S rRNA-binding ribosomal proteins for cell viability and translation in Escherichia coli. J. Mol. Biol. 3661199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kushner, S. R. 2004. Pre-tRNA and pre-rRNA processing in bacteria. Encycl. Biol. Chem. 3420-424. [Google Scholar]
- 95.Liang, S. T., M. Bipatnath, Y. C. Xu, S. L. Chen, P. Dennis, M. Ehrenberg, and H. Bremer. 1999. Activities of constitutive promoters in Escherichia coli. J. Mol. Biol. 29219-37. [DOI] [PubMed] [Google Scholar]
- 96.Liiv, A., and J. Remme. 2004. Importance of transient structures during post-transcriptional refolding of the pre-23S rRNA and ribosomal large subunit assembly. J. Mol. Biol. 342725-741. [DOI] [PubMed] [Google Scholar]
- 97.Lindahl, L. 1975. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J. Mol. Biol. 9215-37. [DOI] [PubMed] [Google Scholar]
- 98.Lindahl, L. 1973. Two new ribosomal precursor particles in Escherichia coli. Nature (London) 243170-172. [DOI] [PubMed] [Google Scholar]
- 99.Lindahl, L., and J. M. Zengel. 1986. Ribosomal genes in Escherichia coli. Annu. Rev. Genet. 20297-326. [DOI] [PubMed] [Google Scholar]
- 100.Lovgren, J. M., O. B. Goran, K. S. Manoj, L. A. C. Lundberg, P. P. Olof, G. Wingsle, and P. M. Wikstrom. 2004. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA 101798-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lund, E., and N. O. Kjeldgaard. 1972. Metabolism of guanosine tetraphosphate in Escherichia coli. Eur. J. Biochem. 28316-326. [DOI] [PubMed] [Google Scholar]
- 102.Mabe, S., and W. S. Champney. 2005. A comparison of a new oral streptogramin XRP 2868 with quinupristin-dalfopristin against antibiotic-resistant strains of Haemophilus influenzae, Staphylococcus aureus, and Streptococcus pneumoniae. Curr. Microbiol. 51363-366. [DOI] [PubMed] [Google Scholar]
- 103.Mackow, E. R., and F. N. Chang. 1985. Processing of precursor ribosomal RNA and the presence of a modified ribosome assembly scheme in Escherichia coli relaxed strain. FEBS Lett. 182407-412. [DOI] [PubMed] [Google Scholar]
- 104.Maguire, B. A., and D. G. Wild. 1997. The effects of mutations in the rpmB,G operon of Escherichia coli on ribosome assembly and ribosomal protein synthesis. Biochim. Biophys. Acta 1353137-147. [DOI] [PubMed] [Google Scholar]
- 105.Maguire, B. A., and D. G. Wild. 1997. Mutations in the rpmBG operon of Escherichia coli that affect ribosome assembly. J. Bacteriol. 1792486-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maguire, B. A., and D. G. Wild. 1997. The roles of proteins L28 and L33 in the assembly and function of Escherichia coli ribosomes in vivo. Mol. Microbiol. 23237-245. [DOI] [PubMed] [Google Scholar]
- 107.Maguire, B. A., and R. A. Zimmermann. 2001. The ribosome in focus. Cell 104813-816. [DOI] [PubMed] [Google Scholar]
- 108.Mandiyan, V., S. J. Tumminia, J. S. Wall, J. F. Hainfeld, and M. Boublik. 1991. Assembly of the Escherichia coli 30S ribosomal subunit reveals protein-dependent folding of the 16S rRNA domains. Proc. Natl. Acad. Sci. USA 888174-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mangiarotti, G., D. Apirion, D. Schlessinger, and L. Silengo. 1968. Biosynthetic precursors of 30S and 50S ribosomal particles in Escherichia coli. Biochemistry 7456-472. [DOI] [PubMed] [Google Scholar]
- 110.Mangiarotti, G., L. Silengo, G. Corte, and A. Bonsignore. 1970. Biogenesis of 50S ribosomes in E. coli: characterization of 43S precursors. Ital. J. Biochem. 1983-88. [PubMed] [Google Scholar]
- 111.Mangiarotti, G., E. Turco, A. Ponzetto, and F. Altruda. 1974. Precursor 16 S RNA in active 30 S ribosomes. Nature (London) 247147-148. [DOI] [PubMed] [Google Scholar]
- 112.Markey, F., and D. G. Wild. 1975. 30-S precursor of 30-S ribosomes in a mutant of Escherichia coli. Biochem. J. 151463-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Markey, F., and D. G. Wild. 1976. An unusual precursor of 50-S ribosomes in a mutant of Escherichia coli. Biochem. J. 154311-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McGaha, S. M., and W. S. Champney. 2007. Hygromycin B inhibition of protein synthesis and ribosome biogenesis in Escherichia coli. Antimicrob. Agents Chemother. 51591-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mehta, R., and W. S. Champney. 2002. 30S ribosomal subunit assembly is a target for inhibition by aminoglycosides in Escherichia coli. Antimicrob. Agents Chemother. 461546-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mehta, R., and W. S. Champney. 2003. Neomycin and paromomycin inhibit 30S ribosomal subunit assembly in Staphylococcus aureus. Curr. Microbiol. 47237-243. [DOI] [PubMed] [Google Scholar]
- 117.Michaels, G. A. 1972. Ribosome maturation of Escherichia coli growing at different growth rates. J. Bacteriol. 110889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mikkola, R., and C. G. Kurland. 1991. Evidence for demand-regulation of ribosome accumulation in E. coli. Biochimie 731551-1556. [DOI] [PubMed] [Google Scholar]
- 119.Mogridge, J., and J. Greenblatt. 1998. Specific binding of Escherichia coli ribosomal protein S1 to boxA transcriptional antiterminator RNA. J. Bacteriol. 1802248-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Murray, H. D., and R. L. Gourse. 2004. Unique roles of the rrn P2 rRNA promoters in Escherichia coli. Mol. Microbiol. 521375-1387. [DOI] [PubMed] [Google Scholar]
- 121.Murray, K. D., and H. Bremer. 1996. Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli. J. Mol. Biol. 25941-57. [DOI] [PubMed] [Google Scholar]
- 122.Nashimoto, H., and M. Nomura. 1970. Structure and function of bacterial ribosomes. XI. Dependence of 50S ribosomal assembly on simultaneous assembly of 30S subunits. Proc. Natl. Acad. Sci. USA 671440-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nierhaus, K. H. 2004. Ribosome assembly: assembly of the prokaryotic ribosome. Protein Synth. Ribosome Struct. 200485-105. [Google Scholar]
- 124.Nierhaus, K. H., and F. Dohme. 1974. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl. Acad. Sci. USA 714713-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nilsson, L., A. Vanet, E. Vijgenboom, and L. Bosch. 1990. The role of FIS in trans activation of stable RNA operons of E. coli. EMBO J. 9727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ninnemann, O., C. Koch, and R. Kahmann. 1992. The E. coli fis promoter is subject to stringent control and autoregulation. EMBO J. 111075-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nodwell, J. R., and J. Greenblatt. 1993. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell 72261-268. [DOI] [PubMed] [Google Scholar]
- 128.Nowotny, V., and K. H. Nierhaus. 1980. Protein L20 from the large subunit of Escherichia coli ribosomes is an assembly protein. J. Mol. Biol. 137391-399. [DOI] [PubMed] [Google Scholar]
- 129.Okamura, S., H. B. Maruyama, and T. Yanagita. 1973. Ribosome degradation and degradation products in starved Escherichia coli. VI. Prolonged culture during glucose starvation. J. Biochem. (Tokyo) 73915-922. [DOI] [PubMed] [Google Scholar]
- 130.Olsson, M., L. Isaksson, and C. G. Kurland. 1974. Pleiotropic effects of ribosomal protein S4 studied in Escherichia coli mutants. Mol. Gen. Genet. 135191-202. [DOI] [PubMed] [Google Scholar]
- 131.Olsson, M. O. 1979. Analysis of rpsD mutations in Escherichia coli. II. Physiology of some representative mutants. Mol. Gen. Genet. 169259-269. [DOI] [PubMed] [Google Scholar]
- 132.Pardo, D., C. Vola, and R. Rosset. 1979. Assembly of ribosomal subunits affected in a ribosomal mutant of E. coli having an altered L22 protein. Mol. Gen. Genet. 17453-58. [DOI] [PubMed] [Google Scholar]
- 133.Pichon, J., J. Marvaldi, and G. Marchis-Mouren. 1975. In vivo order of protein addition in the course of Escherichia coli 30 S and 50 S subunit biogenesis. J. Mol. Biol. 96125-137. [DOI] [PubMed] [Google Scholar]
- 134.Pokkunuri, I., and W. S. Champney. 2007. Characteristics of a 50S ribosomal subunit precursor particle as a substrate for ermE methyltransferase activity and erythromycin binding in Staphylococcus aureus. RNA Biol. 4147-153. [DOI] [PubMed] [Google Scholar]
- 135.Powers, T., G. Daubresse, and H. F. Noller. 1993. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J. Mol. Biol. 232362-374. [DOI] [PubMed] [Google Scholar]
- 136.Pul, U., R. Wurm, and R. Wagner. 2007. The role of LRP and H-NS in transcription regulation: involvement of synergism, allostery and macromolecular crowding. J. Mol. Biol. 366900-915. [DOI] [PubMed] [Google Scholar]
- 137.Robinson, V. L., J. Hwang, E. Fox, M. Inouye, and A. M. Stock. 2002. Domain arrangement of Der, a switch protein containing two GTPase domains. Structure 101649-1658. [DOI] [PubMed] [Google Scholar]
- 138.Roehl, R., and K. H. Nierhaus. 1982. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc. Natl. Acad. Sci. USA 79729-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ross, W., J. F. Thompson, J. T. Newlands, and R. L. Gourse. 1990. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 93733-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Roth, H. E., and K. H. Nierhaus. 1980. Assembly map of the 50S subunit from Escherichia coli ribosomes, covering the proteins present in the first reconstitution intermediate particle. Eur. J. Biochem. 10395-98. [DOI] [PubMed] [Google Scholar]
- 141.Samaha, R. R., B. O'Brien, T. W. O'Brien, and H. F. Noller. 1994. Independent in vitro assembly of a ribonucleoprotein particle containing the 3′ domain of 16S rRNA. Proc. Natl. Acad. Sci. USA 917884-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sato, A., G. Kobayashi, H. Hayashi, H. Yoshida, A. Wada, M. Maeda, S. Hiraga, K. Takeyasu, and C. Wada. 2005. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells 10393-408. [DOI] [PubMed] [Google Scholar]
- 143.Sayed, A., S.-I. Matsuyama, and M. Inouye. 1999. Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem. Biophys. Res. Commun. 26451-54. [DOI] [PubMed] [Google Scholar]
- 144.Schmidt, T. M. 1998. Multiplicity of ribosomal RNA operons in prokaryotic genomes. Bact. Genomes 1998221-229. [Google Scholar]
- 145.Schneider, D. A., T. Gaal, and R. L. Gourse. 2002. NTP-sensing by rRNA promoters in Escherichia coli is direct. Proc. Natl. Acad. Sci. USA 998602-8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schneider, D. A., W. Ross, and R. L. Gourse. 2003. Control of rRNA expression in Escherichia coli. Curr. Opin. Microbiol. 6151-156. [DOI] [PubMed] [Google Scholar]
- 147.Schroeder, R., C. Waldsich, and H. Wank. 2000. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Selmer, M., C. M. Dunham, F. V. Murphy IV, A. Weixlbaumer, S. Petry, A. C. Kelley, J. R. Weir, and V. Ramakrishnan. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 3131935-1942. [DOI] [PubMed] [Google Scholar]
- 149.Semrad, K., and R. Green. 2002. Osmolytes stimulate the reconstitution of functional 50S ribosomes from in vitro transcripts of Escherichia coli 23S rRNA. RNA 8401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sharma, M. R., C. Barat, D. N. Wilson, T. M. Booth, M. Kawazoe, C. Hori-Takemoto, M. Shirouzu, S. Yokoyama, P. Fucini, and R. K. Agrawal. 2005. Interaction of Era with the 30S ribosomal subunit: implications for 30S subunit assembly. Mol. Cell 18319-329. [DOI] [PubMed] [Google Scholar]
- 151.Sharrock, R. A., R. L. Gourse, and M. Nomura. 1985. Defective antitermination of rRNA transcription and derepression of rRNA and tRNA synthesis in the nusB5 mutant of Escherichia coli. Proc. Natl. Acad. Sci. USA 825275-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shen, V., and H. Bremer. 1977. Chloramphenicol-induced changes in the synthesis of ribosomal, transfer, and messenger ribonucleic acids in Escherichia coli B/r. J. Bacteriol. 1301098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Siibak, T., L. Peil, L. Xiong, A. Mankin, J. Remme, and T. Tenson. 24 November 2008. Erythromycin- and chloramphenicol-induced ribosomal assembly defects are secondary effects of protein synthesis inhibition. Antimicrob. Agents Chemother. doi: 10.1128/AAC.00870-08. [DOI] [PMC free article] [PubMed]
- 154.Silengo, L., F. Altruda, G. P. Dotto, F. Lacquaniti, C. Perlo, E. Turco, and G. Mangiarotti. 1977. Ribosome maturation in E. coli. Ital. J. Biochem. 26133-143. [PubMed] [Google Scholar]
- 155.Silvers, J. A., and W. S. Champney. 2005. Accumulation and turnover of 23S ribosomal RNA in azithromycin-inhibited ribonuclease mutant strains of Escherichia coli. Arch. Microbiol. 18466-77. [DOI] [PubMed] [Google Scholar]
- 156.Singer, P., and M. Nomura. 1985. Stability of ribosomal protein mRNA and translational feedback regulation in Escherichia coli. Mol. Gen. Genet. 199543-546. [DOI] [PubMed] [Google Scholar]
- 157.Slechta, L. 1967. Sparsomycin. Antibiotics 1410-414, 756. [Google Scholar]
- 158.Spillmann, S., and K. H. Nierhaus. 1978. The ribosomal protein L24 of Escherichia coli is an assembly protein. J. Biol. Chem. 2537047-7050. [PubMed] [Google Scholar]
- 159.Squires, C. L., J. Greenblatt, J. Li, C. Condon, and C. L. Squires. 1993. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl. Acad. Sci. USA 90970-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Srivastava, A. K., and D. Schlessinger. 1988. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc. Natl. Acad. Sci. USA 857144-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Srivastava, A. K., and D. Schlessinger. 1989. Escherichia coli 16S rRNA 3′-end formation requires a distal transfer RNA sequence at a proper distance. EMBO J. 83159-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Srivastava, A. K., and D. Schlessinger. 1990. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 44105-129. [DOI] [PubMed] [Google Scholar]
- 163.Suthers, P. F., R. L. Gourse, and J. Yin. 2007. Rapid responses of ribosomal RNA synthesis to nutrient shifts. Biotechnol. Bioeng. 971230-1245. [DOI] [PubMed] [Google Scholar]
- 164.Sykes, J., E. Metcalf, and J. D. Pickering. 1977. The nature of the proteins in ‘chloramphenicol particles’ from Escherichia coli A19 (Hfr rel met rns). J. Gen. Microbiol. 981-16. [DOI] [PubMed] [Google Scholar]
- 165.Sykes, J., E. Metcalf, and J. D. Pickering. 1977. The nature of the proteins present in the ‘relaxed particles’ from methionine-starved Escherichia coli A19 (Hfr rel met rns). J. Gen. Microbiol. 9817-27. [DOI] [PubMed] [Google Scholar]
- 166.Sypherd, P. S. 1965. Accumulation of ribonucleoprotein particles in a relaxed mutant of Escherichia coli. J. Bacteriol. 90403-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Takebe, Y., A. Miura, D. M. Bedwell, M. Tam, and M. Nomura. 1985. Increased expression of ribosomal genes during inhibition of ribosome assembly in Escherichia coli. J. Mol. Biol. 18423-30. [DOI] [PubMed] [Google Scholar]
- 168.Talkington, M. W. T., G. Siuzdak, and J. R. Williamson. 2005. An assembly landscape for the 30S ribosomal subunit. Nature 438628-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tenson, T., T. DeBlasio, and A. Mankin. 1996. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc. Natl. Acad. Sci. USA 938796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Thomas, J. R., and P. J. Hergenrother. 2008. Targeting RNA with small molecules. Chem. Rev. 1081171-1224. [DOI] [PubMed] [Google Scholar]
- 171.Torres, M., C. Condon, J.-M. Balada, C. Squires, and C. L. Squires. 2001. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 203811-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Traub, P., and M. Nomura. 1968. Structure and function of Escherichia coli ribosomes. V. Reconstitution of functionally active 30 S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. USA 59777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Traub, P., and M. Nomura. 1969. Structure and function of Escherichia coli ribosomes. VI. Mechanism of assembly of 30 S ribosomes. J. Mol. Biol. 40391-413. [DOI] [PubMed] [Google Scholar]
- 174.Tumminia, S. J., W. Hellmann, J. S. Wall, and M. Boublik. 1994. Visualization of protein-nucleic acid interactions involved in the in vitro assembly of the Escherichia coli 50 S ribosomal subunit. J. Mol. Biol. 2351239-1250. [DOI] [PubMed] [Google Scholar]
- 175.Turnock, G., and D. G. Wild. 1966. Synthesis of ribonucleic acid and protein during inhibition of Escherichia coli by analogs of amino acids. Biochim. Biophys. Acta 123402-415. [DOI] [PubMed] [Google Scholar]
- 176.Uicker, W. C., L. Schaefer, and R. A. Britton. 2006. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 59528-540. [DOI] [PubMed] [Google Scholar]
- 177.Usary, J., and W. S. Champney. 2001. Erythromycin inhibition of 50S ribosomal subunit formation in Escherichia coli cells. Mol. Microbiol. 40951-962. [DOI] [PubMed] [Google Scholar]
- 178.Usary, J. E., and W. S. Champney. 1999. Accumulation of a 50S ribosomal subunit precursor in erythromycin-treated Escherichia coli cells. Nucleic Acids Symp. Ser. 41169-172. [Google Scholar]
- 179.Wagner, R. 2002. Regulation of ribosomal RNA synthesis in E. coli: effects of the global regulator guanosine tetraphosphate (ppGpp). J. Mol. Microbiol. Biotechnol. 4331-340. [PubMed] [Google Scholar]
- 180.Walter, F., Q. Vicens, and E. Westhof. 1999. Aminoglycoside-RNA interactions. Curr. Opin. Chem. Biol. 3694-704. [DOI] [PubMed] [Google Scholar]
- 181.Weitzmann, C. J., P. R. Cunningham, K. Nurse, and J. Ofengand. 1993. Chemical evidence for domain assembly of the Escherichia coli 30S ribosome. FASEB J. 7177-180. [DOI] [PubMed] [Google Scholar]
- 182.Williamson, J. R. 2005. Assembly of the 30S ribosomal subunit. Q. Rev. Biophys. 38397-403. [DOI] [PubMed] [Google Scholar]
- 183.Wilson, D. N., and K. H. Nierhaus. 2007. The weird and wonderful world of bacterial ribosome regulation. Crit. Rev. Biochem. Mol. Biol. 42187-219. [DOI] [PubMed] [Google Scholar]
- 184.Wower, I. K., J. Wower, and R. A. Zimmermann. 1998. Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. J. Biol. Chem. 27319847-19852. [DOI] [PubMed] [Google Scholar]
- 185.Yamagishi, M., H. A. De Boer, and M. Nomura. 1987. Feedback regulation of rRNA synthesis. A mutational alteration in the anti-Shine-Dalgarno region of the 16 S rRNA gene abolishes regulation. J. Mol. Biol. 198547-550. [DOI] [PubMed] [Google Scholar]
- 186.Yamagishi, M., and M. Nomura. 1988. Effects of induction of rRNA overproduction on ribosomal protein synthesis and ribosome subunit assembly in Escherichia coli. J. Bacteriol. 1705042-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zengel, J. M., and L. Lindahl. 1994. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol. 47331-370. [DOI] [PubMed] [Google Scholar]
- 188.Zhang, X., S.-T. Liang, and H. Bremer. 2006. Feedback control of ribosome synthesis in Escherichia coli is dependent on eight critical amino acids. Biochimie 881145-1155. [DOI] [PubMed] [Google Scholar]