Abstract
Scrub typhus, caused by the intracellular bacterium Orientia tsutsugamushi, is a major cause of febrile illness in the Asia/Pacific region. Here, we implemented a novel real-time PCR and determined the relation of DNA target gene concentration with serum cytokine levels. The limit of detection of the novel real-time PCR was 1,062 DNA copies per ml of EDTA whole blood. Specificity was excellent as determined on a panel of blood- and skin-borne bacteria, including Rickettsia spp. as well as healthy Vietnamese blood donors. Bacterial DNA concentrations after 9 to 12 days from symptoms onset were significantly higher than in earlier or later periods (P < 0.05). Significantly higher concentrations of gamma interferon (IFN-γ) and interleukin-10 (IL-10) occurred during the acute phase of disease (<10 days from onset) as opposed to the convalescent phase (P < 0.05). No significant differences were observed between the acute and the convalescent phases for tumor necrosis factor alpha (TNF-α) and IL-1β concentrations. Regression analysis of DNA concentrations and cytokine levels identified a significant positive relationship for IL-10 (P < 0.0182) but not for IFN-γ, TNF-α, and IL-1β. In conclusion, proinflammatory cytokines and IL-10 were differentially related to human bacteremia. They may thus be induced by different constituents of O. tsutsugamushi. As a future prospect in a clinical diagnostic laboratory, quantitative real-time PCR may serve as a reliable tool to monitor therapy and to detect treatment failure.
Scrub typhus, caused by the intracellular bacterium Orientia tsutsugamushi, is a major cause of febrile illness in South-East Asia and the Pacific region (11). The clinical picture ranges from asymptomatic to fatal, but it is unclear whether severity is contributed by traits of the infecting strain, the infecting bacterial dose, the patient's immune response, or other host properties.
Because symptoms overlap with that of other conditions, laboratory confirmation is desirable for differential diagnosis and specific treatment. Indirect immunofluorescence assays (IFA) are generally used to detect anti-Orientia antibodies. Because reinfection is possible and IFA is not very sensitive, serological diagnosis of acute cases is sometimes demanding. As an alternative, molecular methods are now being investigated for confirmation of scrub typhus (4, 12, 13). These methods, in particular real-time PCR, provide additional benefits such as the determination of target gene concentrations as a surrogate of bacteremia.
In the present study we implemented a quantitative 5′ nuclease (TaqMan)-based real-time PCR assay for O. tsutsugamushi. Because cytokines are known to play a role in the pathogenesis of scrub typhus, we have determined the correlation of pathogen concentrations with serum cytokine levels in order to determine whether DNA target gene concentration could serve as a surrogate of disease status or immune response.
MATERIALS AND METHODS
Antibody detection.
Immunoglobulin G (IgG) and IgM titers were measured by IFA using Vero cells infected with a mixture of the Kato, Karp, and Gilliam strains of O. tsutsugamushi. Sera were absorbed prior to IgM testing with an IgG absorbing reagent (Mastsorb; Mast Diagnostica, United Kingdom).
DNA preparation.
DNA was prepared from serum or whole EDTA blood by using a DNA minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The input and elution volume was 200 μl.
Real-time PCR.
A 25-μl reaction contained 5 μl of prepared DNA, 4 mM MgCl2, 1× Platinum Taq polymerase reaction buffer (Invitrogen, Karlsruhe, Germany), 200 μM concentrations of each deoxynucleoside triphosphate, 0.6 μM primer TSUS1 (ACGTAAGCGGTTTAAACTTAC; TIB-Molbiol, Berlin, Germany), 0.6 μM primer TSUAS2 (AATATCAATCCCAAAGTCACGAT; TIB-Molbiol), 0.2 μM probe TSUP (CCTACTATAATGCCTATAAGTAT), 0.2 μM probe TSUmutant (ATCGTTCGTTGAGCGATTAGCAGTT), and 1 U of Taq DNA polymerase. Probe TSUP was labeled with 5′FAM and a 3′ nonfluorescent quencher (Applied Biosystems, Weiterstadt, Germany), probe TSUmutant was labeled with 5′VIC and 3′Black Hole Quencher (Eurogentec, Seraing, Belgium). The cycling conditions in an ABI Prism 7000 machine (Applied Biosystems) were as follows: 94°C for 2 min, and 40 cycles of 94°C for 15 s and 58°C for 30 s. The data were analyzed with the Sequence detector software V 2.1 (Applied Biosystems).
Internal control.
The target sequence of above assay was cloned into plasmid pCR4 (Invitrogen). Using PCR extension technique (5), the hybridization sequence of probe TSUP was removed from the plasmid and the sequence of TSUmutant inserted at the same position.
Statistical procedures.
All calculations were done by using the Statgraphics 5.1 software package (Manugustics, Dresden, Germany).
RESULTS
Forty-eight serum samples from Vietnamese patients admitted to Hue Medical College were collected from October 2002 to October 2004. Median duration from onset of symptoms to admission was 10 days (95% confidence interval [CI] = 8.6 to 11.1, range 3 to 30 days). Patients were clinically diagnosed with scrub typhus (Table 1) and treated empirically with doxycycline upon admission. All responded to therapy within 48 h as assessed by defervescence. Before treatment a single blood sample was drawn from each patient.
TABLE 1.
Main symptoms of patients upon admission at Hue Medical Collegea
| Symptom | Frequency (%) | No. of patients (n = 48) | 95% CI |
|---|---|---|---|
| Fever (>38.5°C) | 85 | 41 | 73-94 |
| Eschar | 94 | 45 | 81-98 |
| Lymphadenopathy | 73 | 35 | 58-84 |
| Maculopapular skin rash | 25 | 12 | 9-32 |
| Headache | 88 | 42 | 75-96 |
| Hepatosplenomegaly | 31 | 15 | 17-44 |
Listed are the symptoms, frequency of occurrence, numbers of patients, and 95% CI.
In total, 45 (94%) patients showed IgG antibodies. All sera taken after day 8 of symptoms tested positive (Fig. 1). IgM antibodies were detected in 85.4% (n = 41) of all patients. In those sera taken after day 5 from onset, the IgM detection rate exceeded 75% (Fig. 1).
FIG. 1.
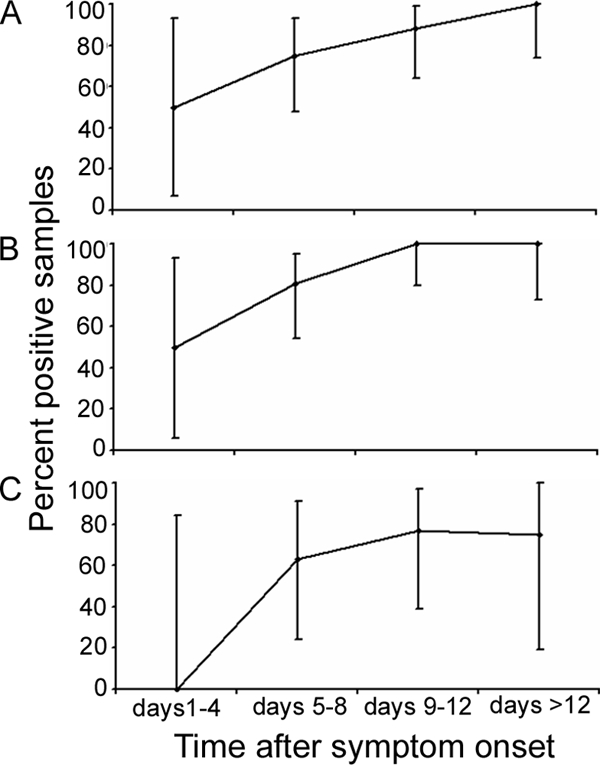
Percent positive samples for IgM antibody detection (A), IgG antibody detection (B), and real-time PCR (C) in 4-day intervals after symptom onset. Datum points are means. Range indicators show the 95% CI values of the means.
A real-time PCR assay with a target-derived internal control was designed for the 56-kDa gene of O. tsutsugamushi, based on public sequences and using procedures described earlier (3, 8). The analytical sensitivity was determined by probit analysis of multiple parallel amplifications of blood spiked with an Escherichia coli plasmid containing the assay's target region (data not shown). The statistically validated limit of detection was 1,062 (95% CI = 720 to 2,691) target gene copies per ml of EDTA blood. Specificity was confirmed on a comprehensive panel of blood- and skin-borne bacteria, including different Rickettsia spp. (3, 8), and on 50 sera from healthy Vietnamese blood donors. All of these samples tested negative. In all of these negative samples the internal control yielded positive results, demonstrating the technical robustness of the assay.
From the study cohort, 23 sera from 23 individual patients were available for PCR analysis (the remaining samples could not be tested because they had been used up for serology in the local hospital). The median duration of symptoms in these patients was 9 days (95% CI = 7.6 to 11, range 3 to 30 days). Sixty-five percent (n = 15) yielded a positive result by real-time PCR. Interestingly, no positive PCR was seen in any sample taken before day 5 (Fig. 1C) (n = eight samples). For confirmation, these eight samples were also tested by universal Rickettsia-specific PCR, with negative results.
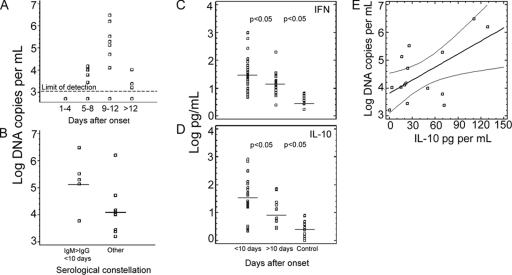
Quantitative analysis (Fig. 2) showed that bacterial DNA concentration between days 9 and 12 was significantly higher than in earlier or later periods (P < 0.05, one-way analysis of variance [ANOVA]).
FIG. 2.
(A) Log DNA copy numbers per ml of blood plotted in intervals of 4 days from symptom onset. The statistically validated limit of detection (95% probit point) is indicated by a dashed line. Highest concentrations were seen between days 9 to 12 after onset (P < 0.05, ANOVA). (B) DNA target gene concentration in samples (n = 5) representing acute disease with IgM > IgG and drawn within the first 10 days after onset were compared to all other PCR-positive samples (n = 10). Acute disease samples show significantly higher DNA target gene concentrations than other samples. Horizontal lines are means, squares are individual values. (C and D) IFN-γ and IL-10 concentrations, respectively, as measured in sera (n = 10) taken within the first 10 days after onset, sera taken after >10 days (n = 17), and healthy control sera (n = 20). Horizontal lines are means; squares are individual values. The levels of significance of the differences of means are shown between the groups. (E) IL-10 concentrations (pg per ml) plotted against level of bacteremia in 15 PCR-positive samples. A significant correlation could be demonstrated (P = 0.0182, linear regression model). The straight line is the regression line. Curved lines are continuous 95% CI values. Squares indicate individual datum values.
To assess whether target gene concentrations correlated with the course of disease, PCR-positive patients were grouped into two categories: first, patients with an IgM/IgG titer ratio >2 in a sample taken before day 10; second, all other patients with a combined positive result in serology and PCR (Fig. 2). It was assumed that group 1 was likely to represent primary infections, whereas group 2 would probably also contain secondary infections. One-way ANOVA identified significantly higher DNA target gene concentrations in group 1 (P < 0.016). However, there was no quantitative correlation in regression analysis between antibody titers and bacterial burden.
Concentrations of gamma interferon (IFN-γ), interleukin-10 (IL-10), tumor necrosis factor alpha (TNF-α), and IL-1β were measured in all samples by OptEIA II enzyme immunoassay (Becton Dickinson, San Diego, CA) as recommended by the manufacturer. Eighty-five percent (41/48) of patients displayed abnormally increased IFN-γ levels (mean, 66.5 pg/ml [7.5 to 985 pg/ml]); fifty-six percent (27/48) showed elevated IL-10 levels (mean, 47.2 pg/ml [13.3 to 682 pg/ml]). Peak concentrations of IFN-γ and IL-10 occurred during the acute phase of the disease (<10 days after onset) with significantly lower levels in the convalescent phase (P < 0.05) (Fig. 2). Elevated TNF-α and IL-1β levels were measured only in 5 of 48 (10.4%) and 12 of 48 (25%) patients, respectively (mean, 18 pg/ml [22 to 414 pg/ml]; mean, 58 pg/ml [11.7 to 2,190 pg/ml]). TNF-α level peaked within days 7 to 14, IL-1β within days 5 to 8. No significant differences (P > 0.05) were observed between acute and convalescent phases for TNF-α and IL-1β. A regression analysis was performed on bacteremia and cytokine levels in 15 PCR-positive samples. A significant relationship was seen for IL-10 (P < 0.0182) (Fig. 2) but not for IFN-γ, TNF-α, and IL-1β (P > 0.05).
DISCUSSION
We implemented a novel real-time PCR and determined the relation of DNA target gene concentration with serum cytokine levels. Our assay provides enhanced features, such as an internal control and a clearly defined limit of detection, making it suitable for clinical diagnostic laboratories. Although only a small number of patients were tested, it can be projected from the high analytical sensitivity (1,062 DNA copies/ml) that clinical sensitivity with our PCR assay is at least equivalent to results in earlier studies (7, 13, 15). Importantly, among other factors the time point of sampling seems to be relevant for the clinical sensitivity of PCR assays. We could show that no PCR-positive sample was seen within the first 5 days after symptom onset. In this respect, despite for the first time a thoroughly evaluated and highly sensitive real-time PCR assay was used a significant improvement in the rapid diagnosis of acute scrub typhus could not be demonstrated. The capability of quantifying bacteremia is a novel feature that improves the selection of diagnostic methods. Interestingly, bacteremia in our patients was at its peak concentration only after a comparatively long period of time (9 to 12 days). Caution is thus necessary when acute disease is to be ruled out in acute cases by PCR or by any other pathogen detection test that is based on blood. On the other hand, molecular bacterial quantification may serve in therapy monitoring to detect treatment failure. Cases of antibiotic resistance have been published and may increase in affected regions (10).
Apart from its diagnostic usefulness, quantitative PCR also provides novel insights into pathogenesis. Interestingly, we found in the present study that peak bacteremia occurred relatively late in the course of disease. In the early phase when O. tsutsugamushi multiplies at the bite site and in endothelium, shedding of organisms via the bloodstream therefore seems unlikely. In the later phase of the disease, however, the organism replicates in macrophages (14). Symptoms may be triggered mostly through immune cells in the skin and in local lymph nodes, rather than resulting from immediate systemic distribution of bacteria. This would make it less likely that the infecting dose of O. tsutsugamushi influences measured bacteremia (2). The virulence of the infecting strain may also be an important contribution.
It is suggested for O. tsutsugamushi that reinfection with a heterologous strain is associated with more severe disease (1). However, it is difficult to discriminate upon serological constellation at the time of diagnosis whether a patient suffers from primary or secondary infection. Our study shows that patients with a serological constellation typical of primary infection (i.e., a high IgM/IgG ratio) present higher DNA target gene concentrations in general. However, the bandwidth of DNA target gene concentrations in these patients seems to be too large to propose that quantitative PCR could predict primary infection. Interestingly, however, in the patient group that likely contained cases of secondary infection, some patients with very high bacteremia were observed, while in general these patients had lower concentrations. One could speculate that this might be a reflection of nonprotective, infection-enhancing immunity. Along these lines it may be rewarding in future studies to determine whether secondary cases can be identified with a combination of serology and PCR results.
Our study determined cytokine responses in combination with O. tsutsugamushi bacteremia for the first time. The combined upregulation of IFN-γ and IL-10 confirms findings in a mouse model (6). It is interesting that an association with the level of bacteremia could only be shown for IL-10 in our patients. IL-10 is a regulatory cytokine that ensures homeostasis within the host. Its antagonistic action against proinflammatory cytokines exerts an inhibitory effect on the immune response that helps surviving in an intracellular environment, in a similar way as in Legionella pneumophila infection (9).
For scrub typhus Kim et al. recently reported that O. tsutsugamushi induced IL-10 in mice, which then inhibited TNF-α production by murine macrophages (5). They postulated hereupon a specific IL-10-inducing component in O. tsutsugamushi. Our data in humans match these observations. By measuring bacterial DNA concentrations we could show that proinflammatory cytokines and IL-10 were differentially related to human bacteremia. Proinflammatory cytokines and IL-10 may thus be induced by different constituents of O. tsutsugamushi.
Acknowledgments
The study was supported by the European Commission (contract SSPE-CT-2005-022639) and the Bundesamt für Bevölkerungsschutz und Katastrophenhilfe (contract BBK-F-440-00-1).
We are grateful to Britta Liedigk and Sabine Koehler for excellent technical assistance.
Footnotes
Published ahead of print on 14 January 2009.
REFERENCES
- 1.Bourgeois, A. L., J. G. Olson, R. C. Fang, J. Huang, C. L. Wang, L. Chow, D. Bechthold, D. T. Dennis, J. C. Coolbaugh, and E. Weiss. 1982. Humoral and cellular responses in scrub typhus patients reflecting primary infection and reinfection with Rickettsia tsutsugamushi. Am. J. Trop. Med. Hyg. 31532-540. [DOI] [PubMed] [Google Scholar]
- 2.Chattopadhyay, S., J. Jiang, T. C. Chan, T. S. Manetz, C. C. Chao, W. M. Ching, and A. L. Richards. 2005. Scrub typhus vaccine candidate Kp r56 induces humoral and cellular immune responses in cynomolgus monkeys. Infect. Immun. 735039-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten, C., M. Weber, E. Seifried, and W. K. Roth. 2000. Evaluation of a new PCR assay with competitive internal control sequence for blood donor screening. Transfusion 40718-724. [DOI] [PubMed] [Google Scholar]
- 4.Jiang, J., T. C. Chan, J. J. Temenak, G. A. Dasch, W. M. Ching, and A. L. Richards. 2004. Development of a quantitative real-time polymerase chain reaction assay specific for Orientia tsutsugamushi. Am. J. Trop. Med. Hyg. 70351-356. [PubMed] [Google Scholar]
- 5.Kim, M. J., M. K. Kim, and J. S. Kang. 2006. Orientia tsutsugamushi inhibits tumor necrosis factor alpha production by inducing interleukin 10 secretion in murine macrophages. Microb. Pathog. 401-7. [DOI] [PubMed] [Google Scholar]
- 6.Koh, Y. S., J. H. Yun, S. Y. Seong, M. S. Choi, and I. S. Kim. 2004. Chemokine and cytokine production during Orientia tsutsugamushi infection in mice. Microb. Pathog. 3651-57. [DOI] [PubMed] [Google Scholar]
- 7.Kramme, S., G. Bretzel, M. Panning, J. Kawuma, and C. Drosten. 2004. Detection and quantification of Mycobacterium leprae in tissue samples by real-time PCR. Med. Microbiol. Immunol. 193189-193. [DOI] [PubMed] [Google Scholar]
- 8.Panning, M., M. Asper, S. Kramme, H. Schmitz, and C. Drosten. 2004. Rapid detection and differentiation of human pathogenic orthopox viruses by a fluorescence resonance energy transfer real-time PCR assay. Clin. Chem. 50702-708. [DOI] [PubMed] [Google Scholar]
- 9.Park, D. R., and S. J. Skerrett. 1996. IL-10 enhances the growth of Legionella pneumophila in human mononuclear phagocytes and reverses the protective effect of IFN-gamma: differential responses of blood monocytes and alveolar macrophages. J. Immunol. 1572528-2538. [PubMed] [Google Scholar]
- 10.Rosenberg, R. 1997. Drug-resistant scrub typhus: paradigm and paradox. Parasitol. Today 13131-132. [DOI] [PubMed] [Google Scholar]
- 11.Silpapojakul, K. 1997. Scrub typhus in the Western Pacific region. Ann. Acad. Med. Singapore 26794-800. [PubMed] [Google Scholar]
- 12.Singhsilarak, T., W. Leowattana, S. Looareesuwan, V. Wongchotigul, J. Jiang, A. L. Richards, and G. Watt. 2005. Short report: detection of Orientia tsutsugamushi in clinical samples by quantitative real-time polymerase chain reaction. Am. J. Trop. Med. Hyg. 72640-641. [PubMed] [Google Scholar]
- 13.Sonthayanon, P., W. Chierakul, V. Wuthiekanun, S. D. Blacksell, K. Pimda, Y. Suputtamongkol, S. Pukrittayakamee, N. J. White, N. P. Day, and S. J. Peacock. 2006. Rapid diagnosis of scrub typhus in rural Thailand using polymerase chain reaction. Am. J. Trop. Med. Hyg. 751099-1102. [PubMed] [Google Scholar]
- 14.Walsh, D. S., K. S. Myint, P. Kantipong, K. Jongsakul, and G. Watt. 2001. Orientia tsutsugamushi in peripheral white blood cells of patients with acute scrub typhus. Am. J. Trop. Med. Hyg. 65899-901. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida, Y., Y. Furuya, T. Katayama, I. Kaiho, and S. Yamamoto. 1994. Serotype-specific amplification of Rickettsia tsutsugamushi DNA from clinical specimens by nested polymerase chain reaction. Kansenshogaku Zasshi 68601-606. (In Japanese.) [DOI] [PubMed] [Google Scholar]