Abstract
The human t(3;21)(q26;q22) translocation is found as a secondary mutation in some cases of chronic myelogenous leukemia during the blast phase and in therapy-related myelodysplasia and acute myelogenous leukemia. One result of this translocation is a fusion between the AML1, MDS1, and EVI1 genes, which encodes a transcription factor of approximately 200 kDa. The role of the AML1/MDS1/EVI1 (AME) fusion gene in leukemogenesis is largely unknown. In this study, we analyzed the effect of the AME fusion gene in vivo by expressing it in mouse bone marrow cells via retroviral transduction. We found that mice transplanted with AME-transduced bone marrow cells suffered from an acute myelogenous leukemia (AML) 5–13 mo after transplantation. The disease could be readily transferred into secondary recipients with a much shorter latency. Morphological analysis of peripheral blood and bone marrow smears demonstrated the presence of myeloid blast cells and differentiated but immature cells of both myelocytic and monocytic lineages. Cytochemical and flow cytometric analysis confirmed that these mice had a disease similar to the human acute myelomonocytic leukemia. This murine model for AME-induced AML will help dissect the molecular mechanism of AML and the molecular biology of the AML1, MDS1, and EVI1 genes.
Chromosomal abnormalities are common genetic bases of leukemias. Many of these cytogenetic changes result in the creation of fusion proteins that contain transcription factors. Studying the role of these fusion transcription factors in leukemogenesis is important for understanding the molecular mechanism of leukemias. A reciprocal translocation between chromosomes 3, band q26, and 21, band q22, has been found in certain patients with chronic myelogenous leukemia during blast phase (CML-BC), in patients with therapy-related myelodysplasia/acute myelogenous leukemia (t-MDS/t-AML), and on rare occasions in de novo acute myelogenous leukemia (AML) (1, 2). The t(3;21) translocation can result in fusion of the AML1 gene on chromosome 21 to several genes on chromosome 3, namely EAP, MDS1, and MDS1/EVI1 (reviewed in refs. 3 and 4).
The AML1 gene found on chromosome 21, band q22, encodes a transcription factor containing an N-terminal DNA-binding domain that is homologous to the Drosophila pair-rule gene runt. Normal AML1 protein (also known as CBFA2) is the DNA-binding subunit of the enhancer core-binding factor (CBF) and functions as a heterodimer with the non-DNA-binding subunit CBFβ that enhances AML1's DNA-binding affinity. Both AML1 and CBFβ play a crucial role in definitive hematopoiesis and blood vessel development (5, 6) and are targets of multiple chromosomal abnormalities in human leukemias and myelodysplasia. Besides the t(3;21) translocations mentioned above, the AML1 gene is also found fused to the ETO gene, a zinc-finger-containing transcription factor, in t(8;21)(q22;q22)-associated de novo AML (M2 subtype) (7, 8), fused to the TEL gene, which encodes an ETS family transcription factor, in t(12;21)(p13;q22)-associated de novo acute lymphocytic leukemia (9, 10) and in several other rare translocations (3). CBFβ is found fused to the smooth muscle myosin heavy chain gene (SMMHC) in inv(16)(p13;q22)-associated AML (M4 with eosinophilia subtype) (11).
EVI1 encodes a zinc-finger-containing transcription factor on chromosome 3 and was originally identified as a frequent site of retroviral integration in murine myeloid tumor cells (12, 13). EVI1 is normally expressed in nonhematopoietic tissues but abnormally expressed in hematopoietic cells in retrovirus-induced myeloid leukemias in mice and in human MDS, AML, and CML-BC that are associated with the t(3;3)(q21q26) and inv(3)(q21q26) (reviewed in ref. 4). MDS1 is a separate gene of unknown function located upstream of EVI1 on chromosome 3, although the fusion protein MDS1/EVI1 is also expressed normally through intergenic splicing (14, 15). The AME fusion protein retains the DNA-binding domains of AML1 and EVI1. The other possible AML1 partners on chromosome 3 are EAP and the above-mentioned MDS1. The EAP gene encodes the abundant ribosomal protein L22. The MDS1 gene is fused to AML1 in frame, which produces the chimeric protein AML1/MDS1, whereas fusion of AML1 to EAP is out of frame, resulting in a protein similar to a form of AML1 without the transactivation domain (AML1a) (2, 14).
The possible roles of the CBF subunit-containing fusion genes in leukemogenesis have been investigated by using cultured cells. The transforming activity of the AME and AML1/MDS1 fusion proteins has been demonstrated in Rat-1 fibroblast cells (16, 17), as has that of AML1 and AML1/ETO in NIH 3T3 fibroblast cells (18, 19). AME also blocks granulocytic differentiation of the IL-3-dependent 32D cell line when stimulated with granulocyte colony-stimulating factor (G-CSF) (20, 21). Similar results were observed in previous studies with EVI-1 (22). Interestingly, MDS1/EVI1 did not induce a differentiation blockade in the same system (21). In addition, it has been shown that AML1/ETO can block differentiation of mouse bone marrow cells in vitro (18).
Although these in vitro studies revealed the oncogenic potential of some of the CBF subunit-containing fusion genes, they do not model the pathogenesis of AML. Development of leukemia is a complex process that may involve both the effect of the fusion genes in correct target cells and interactions of the target cells with the rest of the in vivo environment. An in vivo experimental system is required to assess the direct role of these fusion genes in leukemogenesis. However, establishing that the CBF subunit-containing fusion proteins directly induce myelogenous leukemia in vivo has been difficult because of the essential nature of the genes involved. Knock-in of AML1/ETO or CBFβ/SMMHC fusion genes in mice resulted in embryonic lethality similar to the AML1−/− and CBFβ−/− mice, suggesting that the fusion genes may function as dominant negative alleles of normal AML1 (23–26). To circumvent the embryonic lethality problem and to target the human oncogene into hematopoietic stem/progenitor cells, here we used a bone marrow retroviral transduction/transplantation approach to examine the effects of the t(3;21) AML1/MDS1/EVI1 (AME) fusion gene in leukemogenesis. We found that expression of AME in mouse bone marrow cells induces a disease in mice similar to the human acute myelomonocytic leukemia.
Materials and Methods
DNA Constructs.
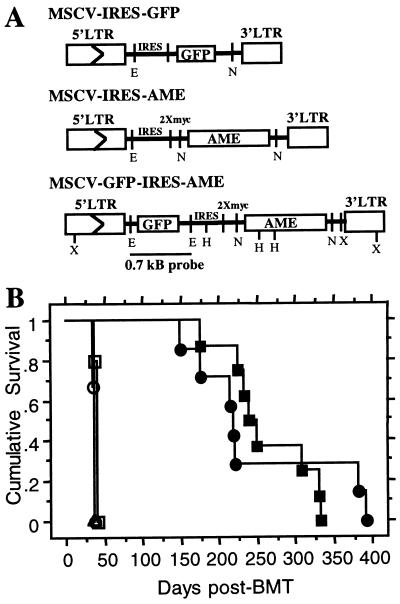
The 4.4-kB coding sequence of the human AME (14) was transferred from the pBK-CMV expression vector (Stratagene) to the retroviral vector MSCV (27, 28). We used a modified MSCV vector that contains an internal ribosomal entry site (IRES) followed by two myc tag sequences (Fig. 1A). The 5′ end of AME was amplified by PCR, removing 154 bp of 5′ untranslated sequence and adding a 5′ NotI site. This PCR fragment was digested with NotI and AvrII and used in a three-way ligation with a fragment containing the rest of AME cut with AvrII and NotI and the MSCV vector digested with NotI. This results in fusion of the myc tags at the N-terminal end of AME (Fig. 1A). The amplified DNA fragment was sequenced to confirm that no errors had been introduced. A second AME-containing construct was made by adding the 0.7-kb modified GFP gene into the EcoRI site upstream of the IRES of MSCV (Fig. 1A). The vector control for this study is MSCV-IRES-GFP (27).
Figure 1.
Retroviral DNA constructs used to transduce GFP and/or AME and cumulative survival of AME or GFP-AME mice (A) LTR, long terminal repeat; E, EcoRI; N, NotI; H, HindIII; X, XbaI. (B) Primary AME (closed circle, seven mice) and GFP-AME (closed square, eight mice) recipient mice; secondary AME (open circle, three from mouse no. 9) and GFP-AME (open square, five from mouse no. 6 and open triangle, three from mouse no. 2) recipient mice. Curves were generated by Kaplan–Meier survival analysis. All mice died or were killed because of disease conditions.
Cell Culture and Virus Preparation.
BOSC-23 cells and NIH 3T3 mouse fibroblasts were grown as previously described (27, 28). Helper-free retroviruses were generated by transiently transfecting retroviral vectors into BOSC-23 cells as described (27, 28).
Bone Marrow Infection and Transplantation.
Bone marrow cell isolation, infection, and transplantation were performed as described (27, 28). Peripheral blood counts of recipient mice were monitored starting from 2 wk after transplantation, every 2 wk for 3 mo, then changed to once a week for 9 mo. Secondary recipient mice received 1 × 106 bone marrow cells from the primary diseased mouse and 100,000 cells from a normal mouse of the same age.
Southern Blot Analysis.
Genomic DNA from 1 × 107 cells from the spleen or bone marrow of diseased mice was isolated by using the Qiagen (Chatsworth, CA) blood kit. Ten micrograms of DNA were digested with either HindIII or XbaI and subjected to Southern blot analysis as previously described (27), by using a 32P-labeled 0.7-kB GFP DNA fragment as probe.
Western Blot Analysis.
Twenty million cells from peripheral blood, bone marrow, and spleen of diseased mice were treated with solution ACK (0.15 M NH4Cl, 1.0 mM KHCO3, 0.1 mM Na2EDTA, pH 7.3) to lyse red blood cells, washed twice in PBS, resuspended in Laemmli sample buffer, and boiled for 10 min. Equal volumes of lysates were run on a 6–15% gradient polyacrylamide gel and transferred to nitrocellulose. The membrane was incubated with either a primary mouse anti-myc antibody (9E10, 1:100) or mouse anti-dynamin (1:1,000, Transduction Laboratories, Lexington, KY), followed by a secondary anti-mouse horseradish peroxidase antibody (1:2,000, Southern Biotechnology Associates). Signals were detected by chemiluminescence.
Blood Cell Count and Hematocrit Measurements.
Three microliters of whole blood from each mouse, obtained by tail bleed, was diluted in 3 ml of Isoton II (Fisher). Total blood counts were measured by using the Coulter Counter Z1 (Coulter). White blood cells (WBCs) were counted similarly after adding of 1 μl ZAP-O-Globin (Coulter), which lyses red blood cells. Peripheral blood and bone marrow smears were stained with Hema 3 (Fisher) for routine identification of cell morphology. Percentage of blasts in the peripheral blood was calculated by counting the number of blast cells in a total of at least 200 cells. Hematocrit was measured by capillary centrifugation.
Histologic Examination and Cytochemistry.
Tissues were fixed with 4% paraformaldehyde in PBS and processed for paraffin-embedded sectioning at 5 μm in thickness, followed by staining with hematoxylin and eosin. For cytochemistry, smears and cytospin preparations of bone marrow cells were stained for myeloperoxidase (MPO), N-acetyl-chloroacetate esterase (NACE), and α-naphthol acetate esterase (ANAE) by using appropriate detection kits from Sigma.
Flow Cytometry.
Cells from peripheral blood, bone marrow, and spleen were treated with ACK, washed in PBS, then resuspended in FACS staining buffer (PBS/1% FBS/0.1% sodium azide). Cells were stained with the following mouse antibodies from PharMingen: phycoerythrin-conjugated Mac-1 (M1/70), Gr-1 (RB6–8C5), B220 (RA3–6B2), CD3 (145–2C11), CD4 (RM4–5), CD8a (53–6.7), Ter119, CD14 (rmC5–3); biotinylated c-Kit (2B8), CD34 (RAM34), CD38 (90) followed by incubation with streptavidin-allophycocyanin (APC). All cells were blocked with anti-mouse CD16-CD32 (Fc block), except for cells stained with CD14 antibodies. Cells were then washed twice in staining buffer and resuspended in 2 μg/ml propidium iodide in staining buffer. Staining intensities of viable cells were measured by using a FACSCalibur (Becton Dickinson). Viable green fluorescent protein (GFP) (+) and GFP (−) cells were sorted by using FACStar or FACS Vantage (Becton Dickinson).
Results
The Human AME Fusion Gene Associated with t(3;21) Induces a Myelogenous Leukemia in Mice.
To examine whether AME induces a hematopoietic neoplasm, we constructed retroviruses as shown in Fig. 1A. Flow cytometry analysis and Western blot by using the anti-myc monoclonal antibody 9E10 showed that NIH 3T3 cells infected with MSCV-GFP-IRES-myc-AME or MSCV-IRES-myc-AME express GFP and/or myc-tagged AME protein (data not shown). We then infected 5-fluorouracil-treated mouse bone marrow cells in vitro with retroviruses containing AME alone or GFP-AME and transplanted these cells into lethally irradiated syngeneic recipient mice. Five to thirteen months after bone marrow transplantation, all mice that received bone marrow cells infected with AME-retrovirus (AME mice) or GFP-AME-retrovirus (GFP-AME mice) developed the symptoms of cachexia, abnormal gait, pale veins, and labored breathing. Some of these diseased mice died (four of seven AME mice), and others were killed because of moribund condition (three of seven AME mice and four of eight GFP-AME mice) or for disease characterization. In contrast, mice that received bone marrow cells infected with vector alone were unaffected in the same period of time (Table 1 and data not shown). All the diseased mice suffered from anemia as shown by their pale veins, low total blood counts (Table 1), decreased red blood cells on peripheral blood smears (Fig. 2 Ad and Ap vs. Ah) and low hematocrit levels in two of seven AME mice analyzed (Table 1). Anemia seemed to be the cause of death or moribund conditions of most AME and GFP-AME mice. Variability of total blood counts in these mice (Table 1) may partially reflect different disease stages at the time they were examined. Analysis of the cumulative survival by using the Mantel–Cox (log-rank) test revealed no significant difference between the survival rate of mice that received AME or GFP-AME (Fig. 1B).
Table 1.
Hematologic parameters of diseased AME and GFP-AME mice
| Retroviral construct | Mouse no. | Latency, days | Total blood count, μl | WBC count, μl | Hematocrit, % | % Blast cells in PB | % GFP in PB | Spleen weight, mg |
|---|---|---|---|---|---|---|---|---|
| Vector | 1 | 224† | 3,400,000 | 24,000 | 49 | n/a | 9 | 90 |
| Vector | 2 | 224† | 3,900,000 | 34,000 | 44 | n/a | 23 | 70 |
| Vector | 3 | 224† | 3,200,000 | 26,000 | 45 | n/a | 18 | 60 |
| Vector | 4 | 224† | 3,500,000 | 22,000 | 47 | n/a | 32 | ND |
| AME | 14* | 148 | ND | 126,990 | ND | ND | n/a | ND |
| AME | 9‡ | 176 | ND | 313,260 | ND | 37 | n/a | 230 |
| AME | 15‡ | 215 | 518,150 | 146,620 | ND | ND | n/a | ND |
| AME | 16* | 219 | 1,240,000 | 30,450 | ND | ND | n/a | ND |
| AME | 10* | 221 | 2,700,000 | 13,680 | ND | ND | n/a | ND |
| AME | 18* | 382 | 1,500,000 | 69,680 | 21 | 49 | n/a | 190 |
| AME | 11*§ | 392 | 2,400,000 | 12,120 | 21 | ND | n/a | ND |
| GFP-AME | 4‡ | 177 | 1,400,000 | 252,710 | ND | 54 | 92 | 360 |
| GFP-AME | 6‡ | 225 | 932,180 | 46,680 | ND | 42 | 93 | ND |
| GFP-AME | 3‡ | 233 | 862,400 | 9,690 | ND | 44 | 29 | 290 |
| GFP-AME | 0¶ | 238 | 2,700,000 | 32,250 | ND | 53 | 67 | ND |
| GFP-AME | 8¶ | 248 | 3,100,000 | 32,490 | ND | 35 | 62 | ND |
| GFP-AME | 2‡ | 309 | 1,000,000 | 60,000 | ND | 65 | 83 | 233 |
| GFP-AME | 7¶ | 331 | 2,213,000 | 20,080 | ND | 59 | 79 | 280 |
| GFP-AME | 5¶ | 332 | 2,900,000 | 19,310 | ND | 46 | 74 | 290 |
ND, not determined; n/a, not applicable.
Died of disease.
No disease.
Killed because of moribund conditions.
Blood count obtained 10 days before death.
Disease mice killed for characterization.
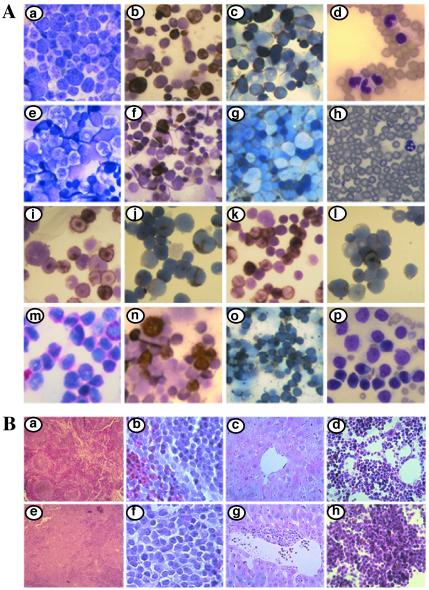
Figure 2.
Morphological, cytochemical, and histologic analysis of AME and GFP-AME mice. (A) Bone marrow smears of normal mouse (e–g) were compared with bone marrow smears from GFP-AME mouse no. 0 (a–c, i–l), mouse no. 6 (n–o), and mouse no. 4 (m). Comparison of MPO+ (b, n) and ANAE+ (c, o) cells from GFP-AME mice and MPO+ (f) and ANAE+ (g) cells from normal mouse. NACE staining of GFP-AME cells can be seen in m. Hema 3-stained peripheral blood smears of primary (mouse no. 6) (d) and secondary GFP-AME mouse (mouse no. 6.4) (p) were compared with smears from normal mouse (h). Cytospin preparations of c-Kit+/GFP+ (i–j) and Mac-1+/GFP+ (k–l)-sorted bone marrow cells from GFP-AME mice stained with MPO (i and k) and ANAE (j and l). (B) Comparison of histologic sections of spleen, liver and bone marrow of vector control mouse no. 2 (a–d), spleen and liver of AME mouse no. 9 (e–g) and bone marrow of GFP-AME mouse no. 6 (h). All photos are at ×600 except for Ao, Bc, Bd, Bg, and Bh, which are at ×200; Bb and Bf, which are at ×630; and Ba and Be which are at ×25).
WBC counts in AME and GFP-AME mice varied: some contained very high counts (up to ≈300,000 cells/μl), and others were comparable to the vector mice (Table 1). But invariably all the diseased mice contained greater than 30% blasts in the peripheral blood (Table 1 and Fig. 2 Ad and Ap). Consistently, bone marrow smears from the diseased mice also revealed the presence of large numbers of myeloid blast cells (Fig. 2Aa). Differentiated but immature myelocytic and monocytic cells were also found in bone marrow smears (Fig. 2Aa). In contrast, normal blood and bone marrow smears consisted predominantly of lymphoid cells and mature myeloid cells (Fig. 2 Ae and Ah).
Splenomegaly was seen in all diseased AME and GFP-AME mice (Table 1). Histologic examination revealed extensive growth of blast cells of myeloid lineage, which resulted in partial to complete destruction of the normal splenic architecture (Fig. 2 Be and Bf vs. Ba and Bb). No significant enlargement of liver, lung, thymus, and lymph nodes was observed in the diseased mice. The livers of all of the diseased mice were very pale because of the anemia. Histologic examination showed that liver tissues were nearly normal except for moderate perivascular infiltration of tumor cells (Fig. 2 Bg vs. Bc). No significant extramedullary hematopoiesis was observed in liver. The bone marrow was hypercellular, and the extensive tumor growth led to the destruction of normal marrow architecture (Fig. 2 Bh vs. Bd). Impairment of normal hematopoiesis in bone marrow is consistent with the severe anemia of diseased mice.
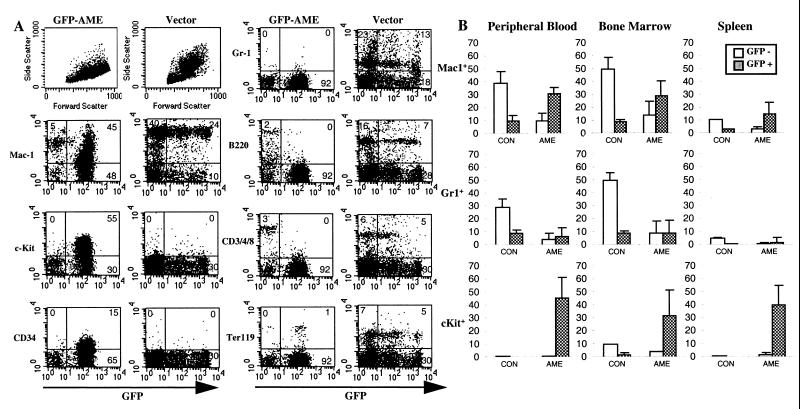
To identify and quantify further various types of WBCs, we analyzed peripheral WBCs by flow cytometry for cell size and granularity and for expression of lineage-specific antigens and GFP. Fig. 3A presents results from a representative diseased GFP-AME mouse (no. 6) and one of the vector mouse. Compared with vector control, the majority of WBCs in the diseased mouse were GFP+. Flow cytometry analysis based on the forward scatter (cell size) and side scatter (cell complexity) showed that the proportion of large granulated cells in GFP-AME mice was greatly reduced. Consistent with this result, very few Gr-1+ cells (differentiated granulocytes) were detected in the GFP-AME mice (Fig. 3 A and B). Almost no GFP+ B-cells (B220+) or T-cells (CD3/4/8+) were detected, although there are still some GFP− B and T lymphocytes present in the circulation. The proportion of GFP+/Ter119+ erythroid cells also decreased when compared with that of the vector mice. Most of the GFP+ cells in the GFP-AME mice express Mac-1, indicating that they are of myeloid lineage. Low to medium expression of Mac-1 suggests that these cells are at an immature stage of differentiation. Interestingly, a large proportion (55%) of WBCs in this mouse are c-Kit+ and significant amounts of CD34+ cells are also present. In contrast, very little c-Kit+ and CD34+ cells can be seen in the peripheral blood of vector mice (Fig. 3A). These results further confirm that early hematopoietic progenitors and immature myeloid cells are substantially increased in the peripheral blood of the GFP-AME mice.
Figure 3.
Flow cytometry analysis of hematopoietic tissues from GFP-AME and vector alone mice. (A) Comparison of cell size and granularities (Top Left) and expression of GFP and surface markers Mac-1, c-Kit, CD34, Gr-1, B220, CD3/4/8, and Ter119 in peripheral blood. (B) Average percent of GFP+ and GFP− Mac-1, Gr-1, and c-Kit positive cells in the peripheral blood, bone marrow, and spleen of eight GFP-AME mice (AME) compared with three vector mice (CON).
We also analyzed bone marrow cells and splenocytes from the diseased GFP-AME mice. Fig. 3B displays the mean levels of Mac-1+, Gr1+, and c-Kit+ cells in the peripheral blood, bone marrow, and spleen. In all three tissues, there is a general decrease in Gr-1+ cells and a dramatic increase in GFP+/Mac-1+ and GFP+/c-Kit+ cells in GFP-AME mice vs. vector mice. Double staining of these cells with Mac-1 and c-Kit demonstrate that c-Kit+ cells were Mac-1lo/−. Mice that received AME alone also contain large numbers of cells expressing Mac-1lo/med, c-Kit, and CD34 in the peripheral blood, bone marrow, and spleen (data not shown).
In summary, we found that AME induces a fatal hematopoietic malignancy resembling an AML with a latency of 5–13 mo. This murine AML, like human AML, is characterized by a marked elevation of early hematopoietic progenitors and immature myeloid cells in the bone marrow, spleen, and peripheral blood, and by a severe impairment of normal hematopoiesis.
AME Mice Contain Both Elevated Immature Myelocytic and Monocytic Cells.
To identify further the various AME-transduced tumor cells and assess similarities to human disease subtypes, we performed cytochemical staining on the bone marrow smears and cytospin preparations. These staining techniques are similar to those used by the French, American, and British (FAB) Cooperative Group to classify human AML subtypes. Staining for MPO activity differentiates strongly positive cells in the granulocytic lineage vs. the weakly positive cells in the monocytic lineage. A large number of marrow cells from the diseased mice are MPO+ (Fig. 2Ab), whereas normal bone marrow contains much fewer MPO+ cells (Fig. 2Af).
Stains for NACE and ANAE or nonspecific esterase are also useful for differentiating these cell lineages. NACE strongly stains differentiated cells of the granulocytic lineage and very weakly or negatively stains cells of the monocytic lineage, whereas ANAE is present in monocytes, T-cells, and megakaryocytes. A large fraction of marrow cells from the diseased mice were ANAE+ (Fig. 2Ac), whereas normal bone marrow contained very few ANAE+ cells except for some weakly positive lymphocytes (Fig. 2Ag). Weakly positive NACE-stained cells from the diseased mice can be seen in Fig. 2Am.
To verify that MPO- and ANAE-containing cells are AME-transduced cells, we sorted the GFP+/c-Kit+ and GFP+/Mac-1+ cells from the GFP-AME mice by flow cytometry and performed cytochemical analysis on the cytospin preparations of these cells. We found both MPO+ and ANAE+ cells in the sorted GFP+ cell population (Fig. 2 Ai vs. Al). There were more MPO+ cells from the Mac-1-sorted than the c-Kit-sorted populations (Fig. 2 Ai vs. Ak), whereas more ANAE+ cells were detected in c-Kit-sorted cells (Fig. 2 Aj vs. Al). All of the diseased mice contained significant amounts of both MPO+ and ANAE+ cells, although some had more MPO+ and less ANAE+ (Fig. 2 Ab and Ac), and others had more ANAE+ and less MPO+ (Fig. 2 An and Ao). Despite this variation, the presence of both monocytic and granulocytic cell lineages in the GFP-AME or AME mice indicate that AME induces a disease resembling the human acute myelomonocytic leukemia (AML FAB-M4).
Rapid Development of AML in the Secondary Recipient Mice.
To examine whether tumor cells from diseased primary mice are transplantable, we transferred bone marrow cells from the diseased AME (mouse no. 9) or GFP-AME (mice nos. 6 and 2) mice into groups of three to five lethally irradiated secondary recipients. All of the secondary recipient mice became progressively moribund or died within 40 days after bone marrow transplantation (Fig. 1B). Analysis of the bone marrow and blood smears, flow cytometry, and cytochemistry showed that the secondary AME recipient mice developed the same disease as primary recipients. The drastic difference in disease latency between the primary and secondary AME mice suggests that additional genetic abnormalities may have been acquired in the AME-transduced cells during the development of AML.
Detection of AME Provirus in Tumor Cells of Diseased Mice.
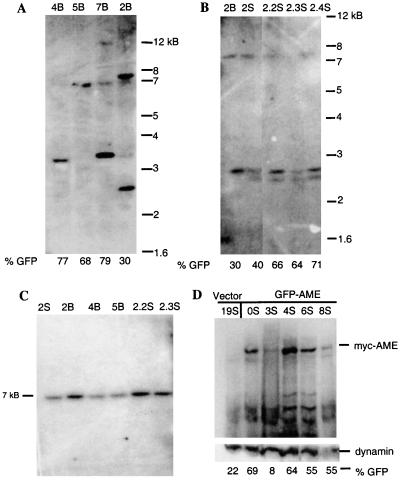
To demonstrate integration of the AME provirus and to establish the number of AME provirus-containing clones in the diseased mice, genomic DNA isolated from the bone marrow or spleen of primary and secondary GFP-AME mice was digested with HindIII (Fig. 1A) and analyzed by Southern blot by using a 0.7-kB GFP probe. One to three prominent bands were observed, suggesting that only a few clones of cells are responsible for disease manifestation (Fig. 4 A and B). Fig. 4B also shows that different clones can arise independently in the bone marrow and spleen (lanes 2B vs. 2S), and that most of these clones persist in the spleen of secondary recipient mice (last three lanes).
Figure 4.
Detection of the AME provirus and expression of the AME protein in affected tissues. Southern blot analysis of genomic DNA from (A) bone marrow of GFP-AME mice digested with HindIII; (B) from bone marrow (2B) and spleen (2S) of primary GFP-AME mouse no. 2, and spleen of its secondary recipients (2.2S, 2.3S, 2.4S) digested with HindIII; and (C) from spleen (lanes 1, 5–6) and bone marrow (lanes 2–4) of primary (lanes 1–4) and secondary recipient (lanes 5–6) GFP-AME mice digested with XbaI. (D). Western blot analysis of the expression of the myc-tagged AME in total spleen cell lysate of GFP-AME mice by using anti-myc antibody (9E10). (Lower) Reprobing of the same membrane with anti-dynamin antibody. The percentages of GFP+ cells are indicated under each lane.
To confirm that the AME provirus did not undergo rearrangement during the long disease latency, we digested genomic DNA with XbaI, which releases nearly the entire GFP-AME provirus (Fig. 1A) and probed with the same GFP DNA fragment. Fig. 4C shows one prominent 7-kB band from primary (first four lanes) and secondary recipient mice (last two lanes) that corresponds to the GFP-AME provirus. These results demonstrate that the AML seen in the primary and secondary recipient mice is a clonal disease.
Expression of the AME Fusion Protein in Tumor Cells of the Diseased Mice.
To establish that the AME fusion protein is expressed in the diseased mice, we performed Western blot analysis on spleen cells. Fig. 4D shows that the myc-tagged-AME is detectable in total lysate of splenocytes derived from several diseased GFP-AME mice (lanes 2–6) but not in splenocytes derived from the vector control mouse (lane 1). The proportion of GFP+ cells (indicated at the bottom of each lane) generally correlated with the AME expression levels. Reprobing with anti-dynamin antibody confirmed that all lanes except lane 6 contained equal amounts of proteins. Expression of the myc-tagged AME can also be seen in total cell lysate of cells from the peripheral blood and bone marrow of GFP-AME mice (data not shown). Mice that received AME alone cannot be monitored by using GFP, but we found that cells from the peripheral blood, bone marrow, and spleen of these mice all express the myc-tagged-AME by Western blot (data not shown). These results confirm the association between expression of the AME fusion protein and the induction of AML.
Discussion
In this study, we established that the human AME fusion gene associated with t(3;21) can induce a disease in mice similar to human acute myelomonocytic leukemia. Common features of the disease include: (i) anemia; (ii) large numbers of myeloid blast cells as well as differentiated but immature granulocytic and monocytic cells accumulate in the bone marrow, spleen, and peripheral blood; (iii) the presence of the AME provirus and the expression of the AME protein in affected tissues; and (iv) efficient transfer of the leukemia to secondary recipient mice, which developed the disease with a much shorter latency.
The difference in the latency of the disease between the primary and secondary AME mice (Fig. 1B) suggests that additional genetic abnormalities may have been acquired in the AME-transduced hematopoietic progenitor cells during the development of the murine acute myelomonocytic leukemia. Consistent with this notion of multistep leukemogenesis in the AME mice, only one or a few clones of AME provirus-containing cells was detected by Southern blot (Fig. 4 A and B). The presence of more than one clone of tumor cells in the diseased mice suggests that the frequency of secondary mutations may have been increased in AME-expressing cells. The murine AML model established here will be useful in studying the molecular mechanisms of multistep leukemogenesis.
The t(3;21) fusion proteins have not been preferentially linked to AML with a particular AML-FAB classification, unlike that of the t(8;21) that is typically associated with AML-M2 and the inv(16) with AML-M4Eo. However, both AML-M4 and M5 subtypes have been found in cases of t(3;21)-associated AML (1, 29). In this study, expression of GFP helped us demonstrate that both myelocytic and monocytic leukemic cells exist in AME mice (Fig. 2A and Fig. 3). Although variations in the amount of MPO+ and ANAE+ cells were detected, the AML-M4 diagnosis was consistent in each of the diseased mice. These variations may be influenced by different secondary mutations acquired during leukemogenesis, which may possibly also account for variations in WBC counts and proportion of GFP+ cells in hematopoietic tissues observed in AME and/or GFP-AME mice (Table 1).
The majority of diseased AME and GFP-AME mice contained large number of c-Kit+ cells (Fig. 3). Many of the c-Kit+ cells were Mac-1lo/−, suggesting that differentiation of hematopoietic cells is blocked at early developmental stages in AME cells. Increases in c-Kit+ cells have also been found in many AML patients, although the specificity and reliability of this as a prognostic marker for AML are still controversial (34–37). Despite this, the predominance of c-Kit+ cells in the AME mice suggests that c-Kit may be a useful marker to characterize t(3;21)-associated AML.
AME is the first CBF subunit-containing translocation protein shown to induce AML in mice. Previous studies have demonstrated that AML1-ETO and CBFβ/SMMHC act as dominant negative alleles of normal AML1. However, expression of these fusion genes in hematopoietic cells either by knock-in (23–26) or by transgenic technology by using a myeloid-specific gene hMPR8 promoter (30) did not induce leukemia in mice. The apparent difference in the leukemogenic potential of AME vs. AML1-ETO and CBFβ/SMMHC may lie in the function of AML1's fusion partner: truncated ETO or SMMHC or MDS1/EVI1. Among these three, EVI1 is the only one that has been implicated in the development of myeloid leukemia in mice (through its overexpression after retroviral integration) (13). It is possible that the role of AML1-ETO and CBFβ/SMMHC in the development of AML may involve only blocking differentiation of hematopoietic cells, but establishment of a neoplasia requires mutations that activate other oncogenes. On the other hand, AME may also block differentiation in hematopoietic cells but in addition have intrinsic transforming properties that allow development of a neoplasia.
Alternatively, all of the CBF subunit-containing fusion proteins may have the potential to induce leukemia when they are targeted into the correct hematopoietic progenitors. Indeed, in vitro studies have already demonstrated that EVI1, AME, and AML1-ETO can block differentiation of a mouse myeloid cell line or mouse bone marrow cells in vitro (20–22, 25, 31), and that AME, AML1/MDS1, AML1/ETO, and AML1 can transform mouse fibroblast cells (16–19). Therefore, it is possible that most, if not all, CBF subunit-containing translocation proteins can block differentiation of hematopoietic progenitor cells at a certain developmental stage and may even stimulate their growth to some extent. However, malignant transformation of these targeted progenitors may require acquisition of additional genetic abnormalities. In knock-in experiments, leukemia may not be realized because AML1-ETO and CBFβ/SMMHC were expressed in cells that were in a stage that is too early in hematopoietic development. Conversely, using the hMPR8 promoter to drive expression of CBFβ/SMMHC in transgenic mice may have allowed targeting into progenitor cells that are too late in hematopoietic development to induce leukemia.
It has been shown that the induction of a CML-like disease in mice by BCR-ABL, which is associated with 95% of human CML, relies on targeting the gene into multipotential hematopoietic stem/progenitor cells by using an efficient retroviral transduction system (27, 32, 33). In this report, we used the same retroviral transduction system to show that AME induces AML in mice. This in vivo experimental system will be useful in assessing the role of the domains of AME as well as the role of other AML1-containing fusion genes in leukemogenesis.
Acknowledgments
We thank E. Leonard and J. Joyce for technical assistance and Dr. H. Dong for pathological consultation. This work was supported by National Cancer Institute grant CA68008 (to R.R.). G.M.C. is supported by National Institutes of Health Training Grant GM07122. G.N. and R.R. are recipients of a Leukemia Society of America Scholar Award.
Abbreviations
- AML
acute myelogenous leukemia
- CML-BC
chronic myelogenous leukemia during blast phase
- t-MDS
therapy-related myelodysplasia
- t-AML
therapy-related AML
- CBF
core-binding factor
- G-CSF
granulocyte colony-stimulating factor
- IRES
internal ribosomal entry site
- WBC
white blood cell
- MPO
myeloperoxidase
- NACE
N-acetyl-chloroacetate esterase
- ANAE
α-naphthol acetate esterase
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.030421197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.030421197
References
- 1.Johansson B, Fioretos T, Garwicz S, Heim S, Mitelman F. Br J Haematol. 1996;92:429–431. doi: 10.1046/j.1365-2141.1996.d01-1468.x. [DOI] [PubMed] [Google Scholar]
- 2.Nucifora G, Begy C R, Erickson P, Drabkin H A, Rowley J D. Proc Natl Acad Sci USA. 1993;90:7784–7788. doi: 10.1073/pnas.90.16.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nucifora G, Rowley J D. Blood. 1995;86:1–14. [PubMed] [Google Scholar]
- 4.Nucifora G. Leukemia. 1997;11:2022–2031. doi: 10.1038/sj.leu.2400880. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Stacy T, Miller J D, Lewis A F, Gu T L, Huang X, Bushweller J H, Bories J C, Alt F W, Ryan G, et al. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 6.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bitter M A, Le Beau M M, Rowley J D, Larson R A, Golomb H M, Vardiman J W. Hum Pathol. 1987;18:211–225. doi: 10.1016/s0046-8177(87)80002-3. [DOI] [PubMed] [Google Scholar]
- 9.Golub T R, Barker G F, Bohlander S K, Hiebert S W, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romana S P, Poirel H, Leconiat M, Flexor M A, Mauchauffe M, Jonveaux P, Macintyre E A, Berger R, Bernard O A. Blood. 1995;86:4263–4269. [PubMed] [Google Scholar]
- 11.Liu P, Tarle S A, Hajra A, Claxton D F, Marlton P, Freedman M, Siciliano M J, Collins F S. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 12.Mucenski M L, Taylor B A, Ihle J N, Hartley J W, Morse H C, 3d, Jenkins N A, Copeland N G. Mol Cell Biol. 1988;8:301–308. doi: 10.1128/mcb.8.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morishita K, Parker D S, Mucenski M L, Jenkins N A, Copeland N G, Ihle J N. Cell. 1988;54:831–840. doi: 10.1016/s0092-8674(88)91175-0. [DOI] [PubMed] [Google Scholar]
- 14.Nucifora G, Begy C R, Kobayashi H, Roulston D, Claxton D, Pedersen-Bjergaard J, Parganas E, Ihle J N, Rowley J D. Proc Natl Acad Sci USA. 1994;91:4004–4008. doi: 10.1073/pnas.91.9.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fears S, Mathieu C, Zeleznik-Le N, Huang S, Rowley J D, Nucifora G. Proc Natl Acad Sci USA. 1996;93:1642–1647. doi: 10.1073/pnas.93.4.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurokawa M, Ogawa S, Tanaka T, Mitani K, Yazaki Y, Witte O N, Hirai H. Oncogene. 1995;11:833–840. [PubMed] [Google Scholar]
- 17.Zent C S, Mathieu C, Claxton D F, Zhang D E, Tenen D G, Rowley J D, Nucifora G. Proc Natl Acad Sci USA. 1996;93:1044–1048. doi: 10.1073/pnas.93.3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank R C, Sun X, Berguido F J, Jakubowiak A, Nimer S D. Oncogene. 1999;18:1701–1710. doi: 10.1038/sj.onc.1202459. [DOI] [PubMed] [Google Scholar]
- 19.Kurokawa M, Tanaka T, Tanaka K, Ogawa S, Mitani K, Yazaki Y, Hirai H. Oncogene. 1996;12:883–892. [PubMed] [Google Scholar]
- 20.Tanaka T, Mitani K, Kurokawa M, Ogawa S, Tanaka K, Nishida J, Yazaki Y, Shibata Y, Hirai H. Mol Cell Biol. 1995;15:2383–2392. doi: 10.1128/mcb.15.5.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sood R, Talwar-Trikha A, Chakrabarti S R, Nucifora G. Leukemia. 1999;13:348–357. doi: 10.1038/sj.leu.2401360. [DOI] [PubMed] [Google Scholar]
- 22.Morishita K, Parganas E, Matsugi T, Ihle J N. Mol Cell Biol. 1992;12:183–189. doi: 10.1128/mcb.12.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castilla L H, Wijmenga C, Wang Q, Stacy T, Speck N A, Eckhaus M, Marin-Padilla M, Collins F S, Wynshaw-Boris A, Liu P P. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 24.Niki M, Okada H, Takano H, Kuno J, Tani K, Hibino H, Asano S, Ito Y, Satake M, Noda T. Proc Natl Acad Sci, USA. 1997;94:5697–5702. doi: 10.1073/pnas.94.11.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okuda T, Cai Z, Yang S, Lenny N, Lyu C J, van Deursen J M, Harada H, Downing J R. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- 26.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A H, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Ren R. Blood. 1998;92:3829–3840. [PubMed] [Google Scholar]
- 28.Gross A W, Zhang X, Ren R. Mol Cell Biol. 1999;19:6918–6928. doi: 10.1128/mcb.19.10.6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitelman F. Catalog of Chromosomal Aberrations in Cancer. New York: Wiley-Liss; 1994. pp. 373–376. [Google Scholar]
- 30.Kogan S C, Lagasse E, Atwater S, Bae S C, Weissman I, Ito Y, Bishop J M. Proc Natl Acad Sci USA. 1998;95:11863–11868. doi: 10.1073/pnas.95.20.11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreider B L, Orkin S H, Ihle J N. Proc Natl Acad Sci USA. 1993;90:6454–6458. doi: 10.1073/pnas.90.14.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pear W S, Miller J P, Xu L, Pui J C, Soffer B, Quackenbush R C, Pendergast A M, Bronson R, Aster J C, Scott M L, et al. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 33.Li S, Ilaria R L, Jr, Million R P, Daley G Q, Van Etten R A. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cascavilla N, Musto P, D'Arena G, Melillo L, Carella A M, Petrilli M P, Sanpaolo G, Carotenuto M. Haematologica. 1998;83:392–397. [PubMed] [Google Scholar]
- 35.Schwartz S, Heinecke A, Zimmermann M, Creutzig U, Schoch C, Harbott J, Fonatsch C, Loffler H, Buchner T, Ludwig W D, et al. Leuk Lymphoma. 1999;34:85–94. doi: 10.3109/10428199909083383. [DOI] [PubMed] [Google Scholar]
- 36.Bene M C, Bernier M, Casasnovas R O, Castoldi G, Knapp W, Lanza F, Ludwig W D, Matutes E, Orfao A, Sperling C, et al. Blood. 1998;92:596–599. [PubMed] [Google Scholar]
- 37.Nomdedeu J F, Mateu R, Altes A, Llorente A, Rio C, Estivill C, Lopez O, Ubeda J, Rubiol E. Leuk Res. 1999;23:341–347. doi: 10.1016/s0145-2126(98)00185-4. [DOI] [PubMed] [Google Scholar]