Abstract
Multilocus sequence types of 163 human Streptococcus agalactiae strains isolated in Bangui and Dakar were analyzed. We identified local specificities in the distribution of sequence types and capsular serotypes. However, the overall population structure is similar to that in the United States and Europe, suggesting that few specific clones colonize humans.
Streptococcus agalactiae, also referred to as group B streptococcus, is a leading cause of neonatal mortality and morbidity in Europe and in the United States (6). Infections caused by S. agalactiae have also been described as emerging among older adults, particularly among immunocompromised patients (25). However, S. agalactiae is primarily a commensal bacterium colonizing the gastrointestinal and genitourinary tracts of up to 30% of healthy adults (25). To improve the prevention of S. agalactiae infections, efforts have been directed toward identifying surface components that could be used in a universal vaccine (3, 13, 17, 22). A precise characterization of the genetic diversity at the population level is needed to evaluate the global efficiency of such a vaccine, as surface components have been shown to be highly variable within the population (2, 12, 14).
Capsular serotyping has been a classical method used in the descriptive epidemiology of S. agalactiae. Based on the immunologic reactivity of a capsular polysaccharide, 10 serotypes are distinguished: Ia, Ib, and II through IX (23, 26). Investigations in the United States and western Europe have shown that four serotypes (Ia, II, III, and V) account for 86 to 90% of the isolates colonizing and infecting humans in these countries. However, the serotype distribution may differ with respect to the geographical regions: in Japan, for instance, serotypes IV and VIII are described as predominant in vaginal isolates from colonized pregnant women (11). Temporal variations with respect to the predominant serotype were also noticed: e.g., serotype V strains emerged in the 1990s in the United States (8). The development of multilocus sequence typing (MLST), a nucleotide-sequence-based typing strategy, has allowed unambiguous comparison of the population structures of S. agalactiae among different geographical areas (9). Up to now, MLST has been used to investigate the population structure of S. agalactiae in different regions of the United Kingdom (9, 10), the United States (1), Sweden (15), Portugal (19), France (12), and Israel (18). In these regions, most isolates that colonize and infect humans belong to the same five clonal complexes (CCs), which are referred to as CC1, CC17, CC19, CC23, and CC9/10/12. By combining MLST and serotyping, these studies have shown that a given serotype is usually distributed among several CCs and that CCs contain several serotypes, suggesting recombinational exchanges (15). These different studies showed that the emerging serotype V isolates belong mostly to CC1 in the countries investigated. However, the population structure of S. agalactiae in other geographic areas such as Africa remains poorly documented. Epidemiological studies from Kenya (5), South Africa (16), and Malawi (7) suggest that S. agalactiae is also emerging in Africa as an important cause of neonatal sepsis and that serotypes III and Ia are predominant whereas the remaining strains belong to serotypes II, Ib, and V. In contrast, serotype V was predominant in large Gambian and Zimbabwean studies of maternal colonization (20, 29).
Complete knowledge about the clonal distribution of S. agalactiae in Africa does not exist. In the present work, we used MLST and molecular serotyping to characterize strains isolated from maternal carriage from 2005 to 2006 in two different African geographical regions: Dakar (Senegal) and Bangui (Central African Republic). The maternal carriage rates of 20.0% (n = 797 pregnant women) in Dakar and 17.5% (n = 1,000 pregnant women) in Bangui (J. M. Sire and R. Bercion, unpublished results) are similar to what was previously reported in studies of African countries (28). We aimed at inferring whether the same well-characterized CCs known to be dominant in Europe and the United States are also prevalent in these African countries.
Two sets of 88 and 75 isolates each were collected in clinical laboratories located at the Pasteur Institutes of Bangui and Dakar, respectively. These carriage isolates were recovered from genital swabs of healthy asymptomatic women in their last term of pregnancy, by the use of the recommended procedures for enhanced recovery of S. agalactiae (4). Isolates were identified to the species level by Gram staining, colony morphology determinations, catalase testing, and the use of a commercial latex agglutination kit (Slidex Strepto B; bioMérieux, Marcy l'Etoile, France). MLST was carried out as previously described by analyzing seven housekeeping genes (9). The 163 isolates were resolved into 32 unique sequence types (STs) (Fig. 1). In Bangui, 23 STs were identified. More than half of the strains (47 isolates [53% of the data set]) belonged to one of the four following STs: ST28 (22%), ST23 (14%), ST26 (10%), and ST196 (8%). Among the isolates from Dakar, 17 STs were identified and four major STs were distinguished: ST26 (20%), ST1 (20%), ST23 (15%), and ST17 (13%). Eight STs were present in both areas, whereas 15 STs were found only in Bangui and 9 only in Dakar. Eleven STs identified in Bangui and six STs identified in Dakar have not previously been reported to the MLST database.
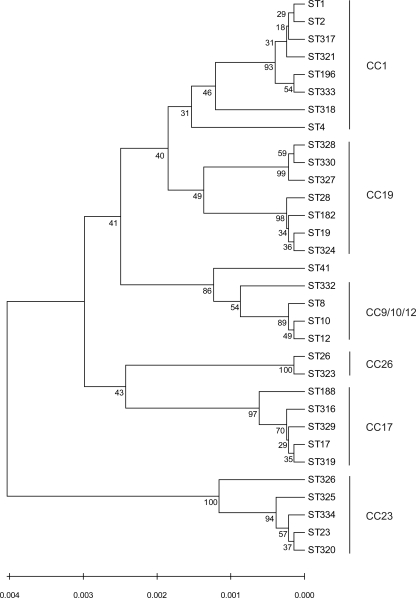
FIG. 1.
Dendrogram of the STs of human S. agalactiae isolates characterized in this study. The dendrogram was created by the unweighted-pair group method using average linkages. Numbers at the nodes represent bootstrap support values.
Strains were grouped into CCs by use of eBURST software by relaxing the group definition to five out of seven shared alleles (27). Thirty-one STs from both cities were clustered in six CCs, and one was a singleton ST. Out of the six CCs, five correspond to the major CCs identified in Europe and in the United States: CC1, CC9/10/12, CC17, CC19, and CC23. Eighty-three percent of the collected strains belonged to one of these five CCs. All new STs identified in Dakar and Bangui were single-locus or double-locus variants of the previously described CCs. This variability probably reflects local evolution within these CCs. Interestingly, 15% of the isolates belonged to CC26, which was rarely detected in North America and Europe. According to both nucleotide polymorphism and allelic combination results, isolates from CC26 are only distantly related to those from all other CCs. Similarly, one subgroup within CC19 was described in Bangui for the first time. This subgroup includes 11 strains belonging to ST327, ST328, and ST330 (12% of the isolates from Bangui). Furthermore, ST28 isolates were prevalent within CC19 in both Dakar and Bangui, while ST19 isolates were described as prevalent within this CC in Europe and the United States.
The capsular serotype of each strain was determined by a sensitive two-multiplex-PCR assay (24) (Tables 1 and 2). Five different serotypes were detected in both Dakar and Bangui: Ia, Ib, II, III, and V. In similarity to what has been previously reported in the United States and in Europe, we observed an overall correlation between the genotype defined by the ST and the serotype. CC23 and CC17 were strictly associated with serotypes Ia and III, respectively. Similarly, CC26 was mainly associated with serotype V (23/25), with the two remaining strains expressing serotype III and Ia. For CC9/10/12, serotype Ib was predominant, although the ST10 isolate expressed a type II capsule. CC1 and CC19 showed a higher degree of serotype variation. CC1 mainly contained serotype V isolates (18/33) but also contained serotypes Ia (9/33), Ib (3/33), and II (3/33), whereas CC19 was constituted of serotypes II (25/47), III (11/47), and V (11/47). The predominance of serotype V within CC1 was mainly due to the prevalence of ST1, while serotype Ia was associated with ST196. Similarly, serotypes II, III, and V within CC19 were associated with ST28, ST19, and subgroup ST328, respectively.
TABLE 1.
Characteristics of the 163 S. agalactiae isolates from Dakar and Bangui
| CC and ST | Allelic profile | Dakar resulta
|
Bangui resultb
|
||
|---|---|---|---|---|---|
| Serotype | No. of isolates | Serotype | No. of isolates | ||
| CC1 | |||||
| 1 | 1,1,2,1,1,2,2 | V | 15 | V | 1 |
| 2 | 1,1,3,1,1,2,2 | II | 1 | ||
| 4 | 1,1,4,1,1,3,4 | Ia | 1 | ||
| 196 | 1,1,3,1,1,12,2 | Ib | 3 | Ia | 7 |
| 317 | 1,1,2,1,33,2,2 | V | 1 | ||
| 318 | 1,1,2,3,1,1,2 | II | 2 | ||
| 321 | 64,1,2,1,1,2,2 | V | 1 | ||
| 333 | 1,1,3,1,1,12,5 | Ia | 1 | ||
| CC9/10/12 | |||||
| 8 | 4,1,4,1,3,3,2 | Ib | 6 | ||
| 10 | 9,1,4,1,3,3,2 | II | 1 | ||
| 12 | 10,1,4,1,1,3,2 | Ib | 1 | ||
| 332 | 4,1,3,1,3,3,2 | Ib | 1 | ||
| CC19 | |||||
| 19 | 1,1,3,2,2,2,2 | III | 3 | III | 3 |
| 28 | 1,1,3,5,2,2,2 | II | 6 | II | 19 |
| 182 | 1,1,3,2,18,2,2 | III | 3 | ||
| 324 | 1,1,40,2,2,2,2 | III | 2 | ||
| 327 | 1,1,4,2,20,3,2 | V | 6 | ||
| 328 | 1,1,4,2,2,3,2 | V | 4 | ||
| 330 | 1,1,27,2,2,3,2 | V | 1 | ||
| CC23 | |||||
| 23 | 5,4,6,3,2,1,3 | Ia | 11 | Ia | 12 |
| 320 | 5,4,6,3,2,1,23 | Ia | 1 | ||
| 325 | 5,4,6,4,2,1,3 | Ia | 1 | ||
| 326 | 5,4,41,3,2,3,3 | Ia | 1 | ||
| 334 | 5,4,42,3,2,1,3 | Ia | 1 | ||
| CC17 | |||||
| 17 | 2,1,1,2,1,1,1 | III | 10 | III | 4 |
| 188 | 2,1,1,2,1,1,2 | III | 1 | ||
| 316 | 63,1,1,2,1,1,1 | III | 1 | ||
| 319 | 2,1,1,2,1,1,22 | III | 2 | ||
| 329 | 2,1,1,38,1,1,1 | III | 1 | ||
| CC26 | |||||
| 26 | 1,1,5,4,1,4,6 | V | 15 | V/III/Ia | 7/1/1 |
| 323 | 1,1,39,4,1,4,6 | V | 1 | ||
| Singleton | |||||
| 41 | 10,1,12,1,3,2,2 | V | 1 | V | 2 |
For the Dakar isolates, n = 24, 1, 9, 12, 13, and 15 for CC1, CC9/10/12, CC19, CC23, CC17, and CC26, respectively.
For the Bangui isolates, n = 9, 8, 38, 15, 6, and 10 for CC1, CC9/10/12, CC19, CC23, CC17, and CC26, respectively.
TABLE 2.
Distribution of serotypes according to CCs
| Serotype | Dakar result
|
Bangui result
|
||
|---|---|---|---|---|
| No. of isolates | CC or singleton (no. of isolates) | No. of isolates | CC(s) or singleton (no. of isolates) | |
| Ia | 13 | CC1 (1), CC23 (12) | 24 | CC1 (8), CC23 (15), CC26 (1) |
| Ib | 4 | CC1 (3), CC10 (1) | 7 | CC10 (7) |
| II | 9 | CC1 (3), CC19 (6) | 20 | CC10 (1), CC19 (19) |
| III | 16 | CC19 (3), CC17 (13) | 15 | CC19 (8), CC17 (6), CC26 (1) |
| V | 33 | CC1 (17), CC26 (15), ST41 (1) | 22 | CC1 (1), CC19 (11), CC26 (8), ST41 (2) |
Although the six identified CCs are present in Dakar and Bangui, slight differences were observed in the local prevalences of CCs as well as the prevalences of STs within CCs. These differences were associated with different serotype distributions in both cities (Table 2). In Dakar, isolates from CC1 were prevalent (32%), whereas isolates from CC19 were prevalent in Bangui (43%). Interestingly, differences within CC1 and CC19 between the Dakar and Bangui data were observed. In Dakar, ST1/V isolates represented 63% of CC1 isolates, while in Bangui, seven out of the nine CC1 isolates corresponded to ST196/Ia isolates. In Bangui, the prevalence of CC19 was associated with 19 ST28/II isolates as well as with 11 isolates from the subgroup composed of ST327, ST328, and ST330 (serotype V) that were not detected in Dakar. In Bangui, eight strains belonged to CC10, among which six were ST8/Ib, while those latter were not detected in Dakar. Finally, CC26/V isolates were twice as frequent in Bangui whereas CC17/III strains were half as frequent. CC23 showed a homogeneous distribution between both areas.
Here, we report for the first time the detailed characterization of 163 S. agalactiae strains isolated from vaginal carriage in two African areas. The overall population structures of isolates from Dakar and Bangui overlap those described for the United States and in Europe. However, two main specific features were identified in these African cities that are compatible with both global and local expansion. First, CC26 isolates represented 15% of all identified isolates in Dakar and Bangui whereas they were rarely detected elsewhere. Only two studies conducted in the United States and in England reported occasional ST26 strains: 2/899 and 3/369, respectively (1, 10). This ST26 was shown to be related to bovine ST256 strains isolated from a bovine udder in Brazil (1, 21) and thus likely corresponds to a clone adapted to both humans and animals. Second, in Bangui, the high prevalence of serotype V strains mainly corresponds to the high number of CC26 strains and to isolates from a CC19 subgroup (ST327, ST328, and ST330), whereas in Dakar, CC26 and CC1 contributed equally to serotype V. The high prevalence of serotype V isolates in the United States and in Europe was shown to be mainly associated with CC1 (1). These observations thus show that although serotype V isolates are disseminated in the United States, in Europe, and in Africa, they may correspond to distinct genetic lineages in Africa. In conclusion, the results of this study suggest that the worldwide genotypic diversity of human S. agalactiae isolates corresponds to a limited number of CCs, probably representing the ancestral population of S. agalactiae isolates colonizing humans, while local variations may reflect a more recent diversification.
Acknowledgments
We acknowledge Carmen Buchrieser, Claire Poyart, and Patrick Trieu-Cuot for fruitful discussions. We thank Nicola Jones for allocating new STs in the S. agalactiae MLST database.
Financial support was provided by the Institut Pasteur (ACIP A/2/2006). M.B. acknowledges support by the CNRSI (Caisse Nationale du Régime Social des Indépendants).
Footnotes
Published ahead of print on 24 December 2008.
REFERENCES
- 1.Bohnsack, J. F., A. Whiting, M. Gottschalk, D. M. Dunn, R. Weiss, P. H. Azimi, J. B. Philips 3rd, L. E. Weisman, G. G. Rhoads, and F. Y. Lin. 2008. Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. academic centers from 1995 to 1999. J. Clin. Microbiol. 461285-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brochet, M., E. Couvé, M. Zouine, T. Vallaeys, C. Rusniok, M. C. Lamy, C. Buchrieser, P. Trieu-Cuot, F. Kunst, C. Poyart, and P. Glaser. 2006. Genomic diversity and evolution within the species Streptococcus agalactiae. Microbes Infect. 81227-1243. [DOI] [PubMed] [Google Scholar]
- 3.Brodeur, B. R., M. Boyer, I. Charlebois, J. Hamel, F. Couture, C. R. Rioux, and D. Martin. 2000. Identification of group B streptococcal Sip protein, which elicits cross-protective immunity. Infect. Immun. 685610-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2002. Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC. MMWR Recommend. Rep. 55(RR)1-22. [Google Scholar]
- 5.English, M., M. Ngama, C. Musumba, B. Wamola, J. Bwika, S. Mohammed, M. Ahmed, S. Mwarumba, B. Ouma, K. McHugh, and C. Newton. 2003. Causes and outcome of young infant admissions to a Kenyan district hospital. Arch. Dis. Child. 88438-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibbs, R. S., S. Schrag, and A. Schuchat. 2004. Perinatal infections due to group B streptococci. Obstet. Gynecol. 1041062-1076. [DOI] [PubMed] [Google Scholar]
- 7.Gray, K. J., S. L. Bennett, N. French, A. J. Phiri, and S. M. Graham. 2007. Invasive group B streptococcal infection in infants, Malawi. Emerg. Infect. Dis. 13223-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison, L. H., D. M. Dwyer, and J. A. Johnson. 1995. Emergence of serotype V group B streptococcal infection among infants and adults. J. Infect. Dis. 171513. [DOI] [PubMed] [Google Scholar]
- 9.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M.-S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 412530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones, N., K. A. Oliver, J. Barry, R. M. Harding, N. Bisharat, B. G. Spratt, T. Peto, and D. W. Crook. 2006. Enhanced invasiveness of bovine-derived neonatal sequence type 17 group B Streptococcus is independent of capsular serotype. Clin. Infect. Dis. 42915-924. [DOI] [PubMed] [Google Scholar]
- 11.Lachenauer, C. S., D. L. Kasper, J. Shimada, Y. Ichiman, H. Ohtsuka, M. Kaku, L. C. Paoletti, P. Ferrieri, and L. C. Madoff. 1999. Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese women. J. Infect. Dis. 1791030-1033. [DOI] [PubMed] [Google Scholar]
- 12.Lamy, M. C., S. Dramsi, A. Billoet, H. Reglier-Poupet, A. Tazi, J. Raymond, F. Guerin, E. Couve, F. Kunst, P. Glaser, P. Trieu-Cuot, and C. Poyart. 2006. Rapid detection of the highly virulent group B Streptococcus ST-17 clone. Microbes Infect. 81714-1722. [DOI] [PubMed] [Google Scholar]
- 13.Larsson, C., J. Holmgren, G. Lindahl, and C. Bergquist. 2004. Intranasal immunization of mice with group B streptococcal protein Rib and cholera toxin B subunit confers protection against lethal infection. Infect. Immun. 721184-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindahl, G., M. Stalhammar-Carlemalm, and T. Areschoug. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18102-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luan, S. L., M. Granlund, M. Sellin, T. Lagergard, B. G. Spratt, and M. Norgren. 2005. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 433727-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madhi, S. A., K. Radebe, H. Crewe-Brown, C. E. Frasch, G. Arakere, M. Mokhachane, and A. Kimura. 2003. High burden of invasive Streptococcus agalactiae disease in South African infants. Ann. Trop. Paediatr. 2315-23. [DOI] [PubMed] [Google Scholar]
- 17.Maione, D., I. Margarit, C. D. Rinaudo, V. Masignani, M. Mora, M. Scarselli, H. Tettelin, C. Brettoni, E. T. Iacobini, R. Rosini, N. D'Agostino, L. Miorin, S. Buccato, M. Mariani, G. Galli, R. Nogarotto, V. N. Dei, F. Vegni, C. Fraser, G. Mancuso, G. Teti, L. C. Madoff, L. C. Paoletti, R. Rappuoli, D. L. Kasper, J. L. Telford, and G. Grandi. 2005. Identification of a universal group B streptococcus vaccine by multiple genome screen. Science 309148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchaim, D., S. Efrati, R. Melamed, L. Gortzak-Uzan, K. Riesenberg, R. Zaidenstein, and F. Schlaeffer. 2006. Clonal variability of group B Streptococcus among different groups of carriers in southern Israel. Eur. J. Clin. Microbiol. Infect. Dis. 25443-448. [DOI] [PubMed] [Google Scholar]
- 19.Martins, E. R., M. A. Pessanha, M. Ramirez, and J. Melo-Cristino. 2007. Analysis of group B streptococcal isolates from infants and pregnant women in Portugal revealing two lineages with enhanced invasiveness. J. Clin. Microbiol. 453224-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moyo, S. R., J. A. Maeland, and K. Bergh. 2002. Typing of human isolates of Streptococcus agalactiae (group B Streptococcus, GBS) strains from Zimbabwe. J. Med. Microbiol. 51595-600. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira, I. C., M. C. de Mattos, T. A. Pinto, B. T. Ferreira-Carvalho, L. C. Benchetrit, A. A. Whiting, J. F. Bohnsack, and A. M. Figueiredo. 2006. Genetic relatedness between group B streptococci originating from bovine mastitis and a human group B Streptococcus type V cluster displaying an identical pulsed-field gel electrophoresis pattern. Clin. Microbiol. Infect. 12887-893. [DOI] [PubMed] [Google Scholar]
- 22.Paoletti, L. C., and L. C. Madoff. 2002. Vaccines to prevent neonatal GBS infection. Semin. Neonatol. 7315-323. [DOI] [PubMed] [Google Scholar]
- 23.Paoletti, L. C., L. C. Madoff, and D. L. Kasper. 2000. Surface structures of Group B Streptococcus important in human immunity, p. 137-153. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, DC.
- 24.Poyart, C., A. Tazi, H. Reglier-Poupet, A. Billoet, N. Tavares, J. Raymond, and P. Trieu-Cuot. 2007. Multiplex PCR assay for rapid and accurate capsular typing of group B streptococci. J. Clin. Microbiol. 451985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slotved, H. C., F. Kong, L. Lambertsen, S. Sauer, and G. L. Gilbert. 2007. A proposed new Streptococcus agalactiae serotype, serotype IX. J. Clin. Microbiol. 452929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spratt, B. G., W. P. Hanage, B. Li, D. M. Aanensen, and E. J. Feil. 2004. Displaying the relatedness among isolates of bacterial species—the eBURST approach. FEMS Microbiol. Lett. 241129-134. [DOI] [PubMed] [Google Scholar]
- 28.Stoll, B. J., and A. Schuchat. 1998. Maternal carriage of group B streptococci in developing countries. Pediatr. Infect. Dis. J. 17499-503. [DOI] [PubMed] [Google Scholar]
- 29.Suara, R. O., R. A. Adegbola, C. J. Baker, O. Secka, E. K. Mulholland, and B. M. Greenwood. 1994. Carriage of group B streptococci in pregnant Gambian mothers and their infants. J. Infect. Dis. 1701316-1319. [DOI] [PubMed] [Google Scholar]