Abstract
The first case of canine endocarditis caused by “Bartonella rochalimae” is reported. By PCR-restriction fragment length polymorphism, sequence, and phylogenetic analyses, Bartonella isolates from a dog with endocarditis, 22 gray foxes, and three dogs, described as B. clarridgeiae like, were confirmed to belong to the new species “B. rochalimae,” suggesting canids as the natural reservoir.
The genus Bartonella comprises more than 20 species and subspecies, many of which are agents of zoonoses. Bartonella spp. are usually vector borne, with the vector and reservoir host varying depending on the Bartonella species involved. However, the vector has not been definitively identified for many recently described Bartonella species, nor has the mammalian reservoir (1). One such new species is “Bartonella rochalimae,” which was isolated from a human after her visit to Peru (10). It is believed that this human was an accidental host for “B. rochalimae,” with the reservoir host not yet identified. Domestic dogs, like humans, appear to be accidental hosts of several Bartonella species (1, 3, 7) and may serve as sentinels for human exposure to Bartonella. The first Bartonella species isolated from a dog was B. vinsonii subsp. berkhoffii (5), which causes most canine Bartonella endocarditis cases (2, 18) and has been associated with at least one human case of endocarditis (25). Infection of dogs with other Bartonella species, including B. clarridgeiae (8), B. washoensis (9), B. henselae (15), B. elizabethae (19), and B. quintana (14), has also been documented. Infection of dogs with Bartonella spp. has been associated with endocarditis (5, 8, 14, 16, 18), arrhythmias, myocarditis (16), peliosis hepatis (15), granulomatous lymphadenitis, granulomatous rhinitis (21), and granulomatous and lymphocytic hepatitis (12).
Case report.
A 9-year-old male, neutered shepherd mix from San Francisco was referred to the University of California, Davis, Veterinary Medical Teaching Hospital for evaluation of lameness and obtunded mentation in January 2000. Blood cultures performed at that time were negative. Echocardiography revealed a hyperechoic vegetative lesion on the right coronary cusp of the aortic valve and severe aortic insufficiency. The lesion was consistent with severe infective aortic valvular endocarditis. The dog (dog 318006) died in August 2000, and DNA was extracted from the damaged aortic valve following necropsy. The Bartonella strain infecting this dog was identified as closely related to B. clarridgeiae (designated B. clarridgeiae-like) based on the PCR-restriction fragment length polymorphism (RFLP) pattern and partial sequencing of the citrate synthase (gltA) gene. The B. clarridgeiae antibody titer was determined by immunofluorescence to be 1:1,024. Further identification of the infecting species was not undertaken at the time (18).
To further identify this canine strain, Bartonella DNA sequences amplified from the damaged valve were compared with DNA sequences from Bartonella isolates previously found to be closely related to B. clarridgeiae by sequence analysis (10, 13). Isolates used for comparison included B. clarridgeiae-like isolates from 22 gray foxes (Urocyon cinereoargenteus) and three dogs, described in a previous study (13), and an isolate from a woman with a history of travel to Peru before the onset of her febrile illness. The Bartonella species infecting the woman was recently described and designated “B. rochalimae” sp. nov. (10).
PCR-RFLP analysis of the internal transcribed spacer (ITS) region (24) and the gltA (20), rpoB (23), and ftsZ (26) genes was performed on DNA extracted from the damaged canine heart valve, as well as on the fox, dog, and human isolates. DNA extraction and PCR vials were set up as previously described (6, 13). The primers for the rpoB gene were 5′-CGCATTGGCTTACTTCGTATG-3′ and 5′-GTAGACTGATTAGAACGCTG-3′. The PCR conditions were 94°C for 10 min; 35 cycles of 94°C for 0.5 min, 53°C for 0.5 min, and 72°C for 1 min; and 72°C for 5 min. The primers for the ftsZ gene were 5′-ATTAATCTGCAYCGGCCAGA-3′ and 5′-ACVGADACACGAATAACACC-3′. The PCR conditions were 94°C for 10 min; 44 cycles of 94°C for 0.5 min, 55°C for 0.5 min, and 72°C for 1 min; and 72°C for 10 min. The primers and PCR conditions for the 16S-to-23S ITS region and the gltA gene were previously described (13). An approximately 400-bp fragment of the gltA gene, an 860-bp fragment of the rpoB gene, a 900-bp fragment of the ftsZ gene, and a 670-bp fragment of the ITS region were amplified and then verified by gel electrophoresis.
The amplified product of the gltA gene was digested with the restriction endonucleases TaqI (Promega, Madison, WI), HhaI (New England BioLabs), AciI (New England BioLabs), and MseI (New England BioLabs). The restriction endonucleases HhaI (New England BioLabs) and AluI (Promega, Madison, WI) were used for digestion of the amplified product of the rpoB gene. Finally, the amplified product of the 16S-to-23S ITS region was digested with the restriction endonuclease HaeIII (Promega, Madison, WI). Banding patterns were compared with B. vinsonii subsp. berkhoffii (ATCC 51672), B. henselae (strain U-4; University of California, Davis), and B. clarridgeiae (ATCC 51734).
PCR products from the gltA, rpoB, and ftsZ genes and the ITS region were purified with the QIAquick PCR purification kit (Qiagen Sciences, Germantown, MD), and both strands were sequenced with a fluorescence-based automated sequencing system (Davis Sequencing, Davis, CA). Raw sequence data were imported into Vector NTI Suite 9.0 software (Invitrogen Co.). A consensus sequence from both strands was obtained and compared with nucleic acid sequence entries in GenBank by using BLASTn (http://ncbi.nih.gov/BLAST/). Sequence variants for each separate gene and for a concatenated sequence of the four gene fragments were then aligned with each other and with relevant sequences from the GenBank database by using AlignX in Vector NTI. A percent similarity table comparing pairs of sequences was also generated after multiple alignments. Phylogenetic trees were constructed for each gene by using both the neighbor-joining and maximum parsimony methods in MEGA version 3.0 (http://www.megasoftware.net) (17). Bootstrap replicates were performed to estimate the node reliability of the phylogenetic trees, with values obtained from 1,000 randomly selected samples of the aligned sequence data.
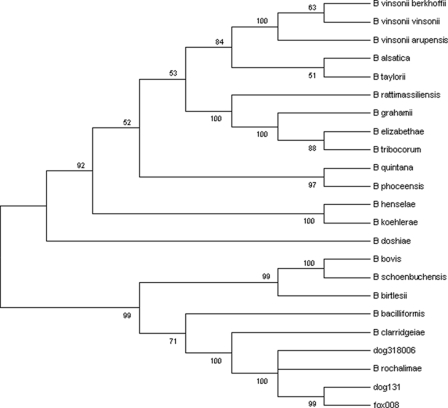
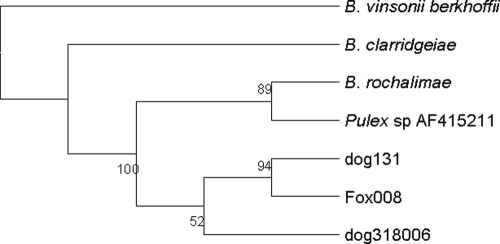
PCR amplification of the ITS region and the gltA, rpoB, and ftsZ genes produced fragments strongly suggestive of Bartonella. The molecular banding patterns of all of the isolates and the endocarditis dog strain were identical to each other based on RFLP analysis of the gltA gene, the rpoB gene, and the ITS region. Because the RFLP profiles were identical for the 22 gray fox isolates, only 6 isolates from this group were selected for sequencing of the gltA, rpoB, and ftsZ genes and the ITS region. Parts of these four sequences were also obtained from the three dogs, the canine endocarditis case, and the human “B. rochalimae” isolate. A BLAST search with partial sequences from the ITS region revealed a high degree of similarity with a GenBank entry (accession number AF415211.1) for a nucleic acid sequence from a flea (Pulex sp.) collected from a person in Peru (22), also reported to be the closest to the human isolate of “B. rochalimae” (10), followed by the sequence of B. clarridgeiae. Sequence data from the Peruvian flea are not available for the gltA, rpoB, and ftsZ genes. The levels of similarity between the Bartonella strains in this study and B. clarridgeiae ranged from 95.5% for the gltA gene to 92.3%, 94.3%, and 76.2% for the rpoB gene, the ftsZ gene, and the ITS region, respectively (Table 1). Small differences (1 to 4 bp) were observed between two or more genes when the northern California dog and fox isolates were compared with those from the dog with endocarditis and with the human “B. rochalimae” isolate. The ITS sequence from the human “B. rochalimae” isolate was identical to the sequence from the Peruvian flea that was also the closest match to the dog and gray fox Bartonella ITS sequences. No differences were found among or between the rural dog and gray fox isolates from northern California for the four gene fragments. Therefore, a representative sequence was chosen from one of the dog (dog 131) and one of the fox (fox 008) isolates for inclusion in the phylogenetic analysis (Fig. 1). The tree, constructed from a merged set of concatenated sequences, showed that the dog and gray fox strains cluster together in a distinct group that includes the human “B. rochalimae” isolate and is closely related to B. clarridgeiae (Fig. 1). A tree was also constructed by using only ITS sequences from the “B. rochalimae” strains, B. clarridgeiae, and B. vinsonii subsp. berkhoffii in order to demonstrate that the Pulex sp. sequence from Peru groups with the human “B. rochalimae” isolate, while the dog and gray fox samples from California cluster together (Fig. 2).
TABLE 1.
Levels of similarity between B. clarridgeiae and new “B. rochalimae” strains in the citrate synthase (gltA), the RNA polymerase beta subunit (rpoB), the cell division protein (ftsZ), and the 16S-to-23S intergenic spacer (ITS) region
| Origin(s) | % Similarity to B. clarridgeiae
|
|||
|---|---|---|---|---|
| gltA (270 bp) | rpoB (784 bp) | ftsZ (789 bp) | ITS (593 bp) | |
| Humboldt County dogs, gray foxes | 95.9 | 92.6 | 94.6 | 76.2 |
| Humana | 95.9 | 92.3 | 94.6 | 76.2 |
| Dog 318006b | 95.5 | 92.3 | 94.3 | 76.2 |
Isolated designated “B. rochalimae.”
Endocarditis case.
FIG. 1.
Phylogenetic tree of Bartonella species based on the combined gltA, rpoB, ftsZ, and ITS sequence alignments. The cluster with dog 318006, the human-associated Bartonella sp., dog 131, and Fox 008 represents the “B. rochalimae” group. The tree shown is a neighbor-joining tree based on the Kimura two-parameter model of nucleotide substitution. Bootstrap values are based on 1,000 replicates. The human-associated Bartonella sp. has been designated “B. rochalimae.” The analysis provided tree topology only, and the lengths of the vertical and horizontal lines are not significant.
FIG. 2.
Phylogenetic tree based on the 16S-to-23S intergenic spacer region showing the grouping of “B. rochalimae” strains from domestic dogs, gray foxes, a human, and a Pulex sp. flea in relation to B. clarridgeiae. The human-associated Bartonella sp. has been designated “B. rochalimae.” The tree shown is a neighbor-joining tree based on the Kimura two-parameter model of nucleotide substitution. Bootstrap values are based on 1,000 replicates. The analysis provided tree topology only, and the lengths of the vertical and horizontal lines are not significant.
Discussion.
We recently reported the isolation of B. clarridgeiae-like strains from 22 gray foxes and three domestic dogs from northern California (13). However, PCR-RFLP and sequence analyses of the gltA, rpoB, and ftsZ genes and the 16S-to-23S ITS region of these B. clarridgeiae-like isolates and from the DNA extracted from the damaged cardiac valve of a dog with endocarditis confirmed that all of these strains belong to the new species “B. rochalimae,” when compared to the only human isolate of “B. rochalimae” (≥99.5% similarity). “B. rochalimae” was isolated from an American woman who presented with fever, rash, and splenomegaly (10). She had a history of travel to Peru, where she sustained multiple insect bites. This is the first time that “B. rochalimae” has been identified in domestic and wild animals and the first report of “B. rochalimae” isolation from mammals in North America. The results of this study suggest that this new species of Bartonella is zoonotic and that it could occur in both wildlife species and domestic dogs in areas within and outside of California. It is also the first time that this new Bartonella species has been associated with a case of endocarditis, as previously reported in humans and dogs for many other Bartonella species (7). It is not unexpected that this new species of Bartonella is associated with disease in both dogs and humans, because dogs have been reported to become infected mainly by species of Bartonella known to be human pathogens and exhibit many of the same clinical manifestations (4). Endocarditis appears to be the most dramatic clinical expression of Bartonella infection in dogs (1, 2, 5, 7, 16), and the spectrum of Bartonella species or subspecies involved is widening (8, 9, 18). The canine endocarditis case included in this study is strongly suspected to have been caused by this newly identified Bartonella species. Factors supporting causation include a failure to isolate other bacteria by conventional blood culture and PCR amplification of the Bartonella DNA characterized above from the abnormal aortic valve, as well as a high Bartonella antibody titer (>1:512 in the present case) commonly observed in cases of endocarditis (18).
DNA sequences from the human “B. rochalimae” isolate and the Peruvian flea PCR product paired closely together in our phylogenetic analysis, as did sequences from the dog from rural northern California and the gray fox isolates from Humboldt County, reflecting small differences in gene sequences by geographical origin. The vector responsible for the transmission of this new species of Bartonella is unknown. However, the finding that DNA sequenced from a Pulex sp. flea collected in Peru (22) is a close match with ITS sequences from isolates in California supports a role for fleas as a potential vector. “B. rochalimae” and B. vinsonii subsp. berkhoffii DNAs have been identified in fleas (Pulex spp.) collected from the foxes sampled as part of the present study (11), but further studies are needed to demonstrate vector competence.
Nucleotide sequence accession numbers.
Representative sequences from strains analyzed in this study were submitted to GenBank under the following accession numbers: gltA, (DQ676484 (dog 131) and DQ676488 (dog 318006); rpoB, DQ676485 (dog 131) and DQ676489 (dog 318006); ftsZ, DQ676486 (dog 131) and DQ676490 (dog 318006); 16S-to-23S ITS, DQ676487 (dog 131) and DQ676491 (dog 318006).
Acknowledgments
This work was supported by a grant from the Center for Companion Animal Health at the University of California, Davis, and by the American Kennel Club Canine Health Foundation. J.E.K. was supported by NIH R01 AI43703, R01 AI52813, and a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research.
We thank Jerry Theis, the Hoopa Tribe, and the Rural Area Veterinary Service for their contributions to this research.
Footnotes
Published ahead of print on 24 December 2008.
REFERENCES
- 1.Boulouis, H. J., C. C. Chang, J. B. Henn, R. W Kasten, and B. B. Chomel. 2005. Factors associated with rapid emergence of zoonotic Bartonella infections. Vet. Res. 36383-410. [DOI] [PubMed] [Google Scholar]
- 2.Breitschwerdt, E. B., C. E. Atkins, T. T. Brown, D. L. Kordick, and P. S. Snyder. 1999. Bartonella vinsonii subsp. berkhoffii and related members of the alpha subdivision of the Proteobacteria in dogs with cardiac arrhythmias, endocarditis, or myocarditis. J. Clin. Microbiol. 373618-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitschwerdt, E. B., K. R. Blann, M. E. Stebbins, K. R. Munana, M. G. Davidson, H. A. Jackson, and M. D. Willard. 2004. Clinicopathological abnormalities and treatment response in 24 dogs seroreactive to Bartonella vinsonii (berkhoffii) antigens. J. Am. Anim. Hosp. Assoc. 4092-101. [DOI] [PubMed] [Google Scholar]
- 4.Breitschwerdt, E. B., B. C. Hegarty, R. Maggi, E. Hawkins, and P. Dyer. 2005. Bartonella species as a potential cause of epistaxis in dogs. J. Clin. Microbiol. 432529-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitschwerdt, E. B., D. L. Kordick, D. E. Malarkey, B. Keene, T. L. Hadfield, and K. Wilson. 1995. Endocarditis in a dog due to infection with a novel Bartonella subspecies. J. Clin. Microbiol. 33154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, C. C., B. B. Chomel, R. W. Kasten, R. Heller, M. Kocan, H. Ueno, K. Yamamoto, V. C. Bleich, B. M. Pierce, B. J. Gonzales, P. K. Swift, W. M. Boyce, S. S. Jang, H. J. Boulouis, and Y. Piémont. 2000. Bartonella spp. isolated from wild and domestic ruminants in North America. Emerg. Infect. Dis. 6306-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomel, B. B., H. J. Boulouis, S. Maruyama, and E. B. Breitschwerdt. 2006. Bartonella spp. in pets and effect on human health. Emerg. Infect. Dis. 12389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomel, B. B., K. A. MacDonald, R. W. Kasten, C. C. Chang, A. C. Wey, J. E. Foley, W. P. Thomas, and M. D. Kittleson. 2001. Aortic valve endocarditis in a dog due to Bartonella clarridgeiae. J. Clin. Microbiol. 393548-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomel, B. B., A. C. Wey, and R. W. Kasten. 2003. Isolation of Bartonella washoensis from a dog with mitral valve endocarditis. J. Clin. Microbiol. 415327-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eremeeva, M. E., H. L. Gerns, S. L. Lydy, J. S. Goo, E. T. Ryan, S. S. Mathew, M. J. Ferraro, J. M. Holden, W. L. Nicholson, G. A. Dasch, and J. E. Koehler. 2007. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N. Engl. J. Med. 3562381-2387. [DOI] [PubMed] [Google Scholar]
- 11.Gabriel, M. W., J. B. Henn, J. E. Foley, R. N. Brown, R. W. Kasten, P. Foley, P., and B. B. Chomel. 6 January 2009. Zoonotic Bartonella species in fleas collected on gray foxes (Urocyon cinereoargenteus). Vector Borne Zoonotic Dis. [Epub ahead of print.] doi: 10.1089/vbz.2008.0134. [DOI] [PubMed]
- 12.Gillespie, T. N., R. J. Washabau, M. H. Goldschmidt, J. M. Cullen, A. R. Rogala, and E. B. Breitschwerdt. 2003. Detection of Bartonella henselae and Bartonella clarridgeiae DNA in hepatic specimens from two dogs with hepatic disease. J. Am. Vet. Med. Assoc. 22247-51. [DOI] [PubMed] [Google Scholar]
- 13.Henn, J. B., M. W. Gabriel, R. W. Kasten, R. N. Brown, J. H. Theis, J. E. Foley, and B. B. Chomel. 2007. Gray foxes (Urocyon cinereoargenteus) as a potential reservoir of a Bartonella clarridgeiae-like bacterium and domestic dogs as sentinels for zoonotic arthropod-borne pathogens in northern California. J. Clin. Microbiol. 452411-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly, P., J. M. Rolain, R. Maggi, S. Sontakke, B. Keene, S. Hunter, H. Lepidi, K. T. Breitschwerdt, and E. B. Breitschwerdt. 2006. Bartonella quintana endocarditis in dogs. Emerg. Infect. Dis. 121869-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitchell, B. E., T. M. Fan, D. Kordick, E. B. Breitschwerdt, G. Wollenberg, and C. A. Lichtensteiger. 2000. Peliosis hepatis in a dog infected with Bartonella henselae. J. Am. Vet. Med. Assoc. 216519-523. [DOI] [PubMed] [Google Scholar]
- 16.Kordick, D. L., B. Swaminathan, C. E. Greene, K. H. Wilson, A. M. Whitney, S. O'Connor, D. G. Hollis, G. M. Matar, A. G. Steigerwalt, G. B. Malcolm, P. S. Hayes, T. L. Hadfield, E. B. Breitschwerdt, and D. J. Brenner. 1997. Bartonella vinsonii subsp. berkhoffii subsp. nov., isolated from dogs; Bartonella vinsonii subsp. vinsonii; and emended description of Bartonella vinsonii. Int. J. Syst. Bacteriol. 46704-709. [DOI] [PubMed] [Google Scholar]
- 17.Kumar, S., K. Tamura, and M. Nei. 2004. Mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 18.MacDonald, K. A., B. B. Chomel, M. D. Kittleson, R. W. Kasten, W. P. Thomas, and P. Pesavento. 2004. A prospective study of canine infective endocarditis in northern California (1999-2001): emergence of Bartonella as a prevalent etiologic agent. J. Vet. Intern. Med. 1856-64. [DOI] [PubMed] [Google Scholar]
- 19.Mexas, A. M., S. I. Hancock, and E. B. Breitschwerdt. 2002. Bartonella henselae and Bartonella elizabethae as potential canine pathogens. J. Clin. Microbiol. 404670-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norman, A. F., R. Regnery, P. Jameson, C. Greene, and D. C. Krause. 1995. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 331797-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pappalardo, B. L., T. Brown, J. L. Gookin, C. L. Morrill, and E. B. Breitschwerdt. 2000. Granulomatous disease associated with Bartonella infection in 2 dogs. J. Vet. Intern. Med. 1437-42. [DOI] [PubMed] [Google Scholar]
- 22.Parola, P., S. Shpynov, M. Montoya, M. Lopez, P. Houpikian, Z. Zeaiter, H. Guerra, and D. Raoult. 2002. First molecular evidence of new Bartonella spp. in fleas and a tick from Peru. Am. J. Trop. Med. Hyg. 67135-136. [DOI] [PubMed] [Google Scholar]
- 23.Renesto, P., J. Gouvernet, M. Drancourt, V. Roux, and D. Raoult. 2001. Use of rpoB gene analysis for detection and identification of Bartonella species. J. Clin. Microbiol. 39430-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rolain, J. M., F. Gouriet, M. Enea, M. Aboud, and D. Raoult. 2003. Detection by immunofluorescence assay of Bartonella henselae in lymph nodes from patients with cat scratch disease. Clin. Diagn. Lab. Immunol. 10686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roux, V., S. J. Eykyn, S. Wyllie, and D. Raoult. 2000. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J. Clin. Microbiol. 381698-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeaiter, Z., Z. Liang, and D. Raoult. 2002. Genetic classification and differentiation of Bartonella species based on comparison of partial ftsZ gene sequences. J. Clin. Microbiol. 403641-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]