Abstract
Papillomatous digital dermatitis (PDD) is an infectious foot disease of cattle that is prevalent throughout the world. Although it has been prevalent in Japan since the first case was reported in 1992, full epidemiological and bacteriological examinations have not been conducted. We collected 91 lesions of PDD from 80 dairy cattle on 12 farms in eight regions of Japan to isolate the spirochetes that are frequently detected in lesions. We isolated 40 strains of spirochetes from 24 cattle (30.0%) by a simple two-step culture technique, in which the biopsy samples were incubated at 4°C for 48 to 72 h in an enrichment broth supplemented with antibiotics, which improved the rate of isolation, and then inoculated on selective agar plates. All spirochetes examined were catalase positive and oxidase negative and showed weak beta-hemolytic activity. Enzyme activities were identical to those of Treponema phagedenis ATCC 27087. Sequencing of the 16S rRNA gene showed that all strains isolated had >99% identity to those of the T. phagedenis type strain and of T. phagedenis-like strains isolated from PDD lesions in the United States and Europe. Pulsed-field gel electrophoresis and PCR-based random amplified polymorphism DNA methods revealed considerable diversity among strains isolated not only from different cattle but also from the same individuals. These findings may provide further evidence for the role of these treponemes in the pathogenesis of persistent PDD.
Papillomatous digital dermatitis (PDD) is an infectious foot disease in cattle that was first reported in Italy in 1974 (4) and is now recognized throughout the world (3, 8, 31). In Japan, PDD in dairy cattle is now found in most regions (24, 26, 37) since the first case was reported in 1992 (20). PDD begins as a superficial dermatitis with an erosive lesion and later forms a hyperkeratotic papillomatous lesion with long hairlike projections (12). These lesions are usually located on the rear of the foot between the bulbs of the heel (3). PDD often leads to lameness and decreases in body weight and milk production, causing economic loss and animal welfare problems (30).
It is suspected that PDD is caused by bacteria, since treatment with antibiotics such as oxytetracycline and penicillin G results in rapid resolution of the lesions (19, 29, 46). Microscopic examinations of biopsy samples or direct stamp preparations of the lesions show various bacterial morphotypes, including long filaments, rods, coccoids, and helices. Although several culturable bacteria have been isolated from PDD lesions (3, 7, 22, 30, 36), it remains unclear whether all are involved in the etiology.
Among the bacteria detected in PDD lesions, a large number of spirochetes are observed (6, 10, 25, 32, 39). Choi et al. detected five spirochetal phylotypes using 16S rRNA gene sequence analysis (6). Similarly, Moter et al. detected four spirochetal phylotypes by using in situ hybridization with oligonucleotide probes of the 16S rRNA gene from treponemes (23). Indeed, several Treponema species have been isolated from dairy cattle with PDD, and are closely related to oral Treponema species in humans, including Treponema phagedenis, T. denticola, T. vincentii, and T. medium (6, 16, 21). Furthermore, Schrank et al. isolated and characterized the newly proposed species Treponema brennaborense (35). These reports suggest that multiple Treponema species are present in PDD lesions. Their presence in both superficial lesions and deeper layers of the dermis (2, 8, 12, 25) suggests that they play an important role in the infection. However, it has not been proved that these organisms satisfy Koch's postulates.
One possible reason why the etiology of PDD is not yet fully understood is the difficulty of isolating and culturing Treponema species present in PDD lesions. Several research groups have isolated Treponema species by the use of methods for the isolation of oral treponemes in humans (9, 16, 21, 35, 43). An immunomagnetic bead method using polyclonal antibodies against T. denticola and T. vincentii was used to separate Treponema from other bacteria (9). The growth rate of many Treponema species is poor, and the presence of other fast-growing bacteria in PDD lesions often prevents their isolation (43). No selective media supplemented with suitable antimicrobial agents for the primary isolation of treponemes from PDD lesions have been developed. Here, we sought to isolate treponemes from cattle with PDD lesions for the first time in Japan. We developed a simple two-step culture method which improved the rate of isolation of spirochetes and examined the biochemical characteristics and 16S rRNA gene sequences for phylogenic analysis. Finally, we examined the genotypic heterogeneity among the strains by molecular typing methods.
MATERIALS AND METHODS
Bacterial strains.
T. phagedenis ATCC 27087 and T. denticola JCM8225 were used as type strains for comparing characteristics of treponemes from PDD lesions. These strains were cultured on the anaerobic medium described below. Isolated strains were suspended in brucella broth (BBL Becton and Dickinson, Sparks, MD) supplemented with 10% glycerol and were kept at −80°C until testing.
Sampling of PDD lesions.
We sampled 91 PDD lesions from 80 dairy cattle (Holstein) on 12 farms (A to L) in eight prefectures of Japan from October 2004 to January 2007 (Table 1). Lesions from 12 cattle on farms A, B, C, I, J, and L were taken from both rear feet. Cattle were restrained in a frame, and the affected foot was restrained with a rope. Before sampling, the lesions were washed with water using a soft brush to remove fecal material. Biopsy samples were obtained by using a sterile dermal biopsy punch with a 6-mm diameter (Kai Industries Co., Ltd., Gifu, Japan) and placed in the enrichment medium for anaerobes described below. The biopsy samples were kept on ice or at 4°C and transported to the laboratory within 48 h after sampling.
TABLE 1.
Isolation of treponemes from cattle with PDD
| Region | Farm | No. of cattle examined | No. of lesions sampleda | No. of cattle with isolated treponemes | Isolation rate in cattle (%) | No. of treponeme strains isolatedb |
|---|---|---|---|---|---|---|
| Hokkaido | A | 6 | 8 | 5 | 83.3 | 17 |
| Hokkaido | B | 8 | 12 | 6 | 75.0 | 10 |
| Yamagata | C | 2 | 3 | 2 | 100 | 2 |
| Chiba | D | 4 | 4 | 1 | 25.0 | 1 |
| Chiba | E | 2 | 2 | 1 | 50.0 | 1 |
| Hyougo | F | 22 | 22 | 2 | 9.1 | 2 |
| Hiroshima | G | 6 | 6 | 1 | 16.7 | 1 |
| Kumamoto | H | 2 | 2 | 1 | 50.0 | 1 |
| Miyazaki | I | 11 | 13 | 0 | 0 | 0 |
| Kagoshima | J | 5 | 6 | 0 | 0 | 0 |
| Kagoshima | K | 9 | 9 | 5 | 55.6 | 5 |
| Kagoshima | L | 3 | 4 | 0 | 0 | 0 |
| Total | 12 | 80 | 91 | 24 | 30.0 (mean) | 40 |
PDD lesions from 12 cattle in farms A, B, C, I, J, and L were taken from both rear feet.
The number of strains includes different colonies isolated from the same lesion and different foot lesions from identical cattle in farms A and B.
Media for transport and isolation.
To isolate treponemes from PDD lesions, we had to kill other bacteria present in the specimens with selective agents. We used rifampin and enrofloxacin, which have been used by others for the isolation of Treponema species from PDD lesions (9, 43). We modified an anaerobic medium, named PDDTp agar, to isolate the treponemes. PDDTp consists of GAM agar (Nissui Pharmaceutical Co., Tokyo, Japan) as a base medium for anaerobes supplemented with 0.8% brucella broth (BBL), 0.8% heart infusion broth (Nissui Pharmaceutical), 10% heat-inactivated fetal bovine serum (Invitrogen, San Diego, CA), 10% defibrinated horse blood, rifampin (1 μg/ml), and enrofloxacin (1 μg/ml). As a transport and enrichment medium, we used PDDTp broth: GAM broth (Nissui Pharmaceutical) for anaerobes supplemented with the same concentrations of brucella broth (BBL), heart infusion broth (Nissui), fetal bovine serum (Invitrogen), and antibiotics without horse blood.
Primary microbial isolation.
Biopsy specimens were kept in PDDTp broth at 4°C for 48 to 72 h in air after receipt at our laboratory. The tissue was cut with a sterile scalpel and was stamped onto PDDTp agar plates. Then, several stamped spots were streaked with a loop. The plates were incubated at 35°C for 14 days without oxygen in an AnaeroPack (Mitsubishi Gas Chemical Co., Tokyo, Japan). Film-like or swarming colonies were stained with crystal violet solution (used for Gram staining) for morphological observations. Spiral-shaped rods were analyzed for their biochemical characteristics and 16S rRNA gene sequences as described below. The isolates were subcultured on fresh PDDTp agar plates, and then the bacteria were suspended in brucella broth supplemented with 10% glycerol. These isolates were kept at −80°C until testing.
Electron microscopic observations.
Bacteria grown on PDDTp agar plates at 35°C for 14 days were suspended in distilled water: 10 μl of bacterial suspension was mixed with an equal volume of 1% phosphotungstate for negative staining. One drop was placed on a collodion-carbon-coated grid and examined with a Hitachi H-800MU transmission electron microscope (Hitachi Seisakusho, Tokyo, Japan). Three spirochetes were examined for determination of the size and the numbers of periplasmic flagella per cell.
Enzymatic activities of the isolates.
Bacteria grown on PDDTp agar plates at 35°C for 14 days were examined for hemolytic activity and for catalase and oxidase production. Other enzymatic activities were tested by using an API-ZYM strip system (Biomérieux Japan, Tokyo, Japan). Bacteria were suspended in saline and washed twice with phosphate-buffered saline (PBS) by centrifugation of 6,000 × g for 20 min at 4°C. After a washing step, cells were resuspended with PBS and adjusted to a density equivalent to McFarland turbidity standard 5. The suspensions were then inoculated into wells of the API-ZYM strip system, which contains substrates for detecting 19 enzymes. The strips were incubated and interpreted according to the manufacturer's instructions.
Amplification of 16S rRNA gene.
Forty isolates were grown on PDDTp agar plates at 35°C for 10 to 14 days, and chromosomal DNA was prepared with CTAB (cetyltrimethyl ammonium bromide), as described elsewhere (44). The near complete 16S rRNA gene (∼1,500 bp) was PCR amplified with universal primers 8F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 15R (5′-AAGGAGGTGATCCARCCGCA-3′), which were based on the Escherichia coli 16S rRNA numbering system (14). PCR was performed in a final volume of 20 μl containing 20 pM concentrations of forward and reverse primers, a 200 μM concentration of each deoxynucleoside triphosphate, 0.5 U of Taq DNA polymerase (Qiagen, Inc., Tokyo, Japan), 1× PCR buffer, and 20 ng of DNA template. Thermal cycle conditions were 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C, followed by a final 72°C for 10 min. After amplification, the resulting amplicons were electrophoresed in 1% agarose gel.
16S rRNA sequence data analysis.
The fragments for sequencing were amplified by PCR as described above, and the products were purified by using a QIAquick PCR purification kit (Qiagen). The nucleotide sequences of 40 isolates were determined directly from the PCR fragment in a PCR-based reaction by using a ABI Prism BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems, Tokyo, Japan) and analyzed with a 3130 DNA sequencer (Applied Biosystems). An ∼800-bp fragment was sequenced from each 5′ and 3′ end of the amplicon, and then they were assembled to determine the nucleotide sequence of the nearly complete 16S rRNA gene (∼1,500 bp) using GENETYX-WIN 5.4 software. Database similarity searches were performed through the National Center for Biotechnology Information by use of the BLASTN algorithm. To align the 16S rRNA gene sequences, CLUSTAL W software was used (5). Phylogenetic trees were drawn by using TreeView software (28).
Molecular typing of isolates by PFGE.
Treponema strains were grown on PDDTp agar plates without antibiotics for 2 weeks. Bacteria were suspended in 10 mM PBS (pH 7.2), washed twice, and then used to construct plugs with a CHEF Genomic DNA plug kit (Japan Bio-Rad Laboratories, Tokyo, Japan) according to the manufacturer's instructions. Encapsulated DNA was digested at 37°C with 20 U of XbaI (New England Biolabs, Beverly, MA) for 16 h and then separated in 1.2% agarose gel by pulsed-field gel electrophoresis (PFGE) in 1:5-diluted Tris-borate-EDTA (45 mM Tris, 45 mM borate, 1 mM EDTA) buffer (pH 8.3).
The running conditions were as follows: initial switch time, 5 s; final switch time, 50 s; field strength, 6 V/cm with a 120° field angle; and 22 h at 14°C. Restriction fragments were stained with ethidium bromide and visualized under UV light.
RAPD DNA analysis.
For RAPD [random(ly) amplified polymorphic DNA] analysis of Treponema strains, reaction mixtures were prepared in a total volume of 25 μl, containing 20 ng of DNA, 2.5 μl of 10× PCR buffer, 3 mM MgCl2, 0.625 U of Taq DNA polymerase (Qiagen), 250 μM concentrations of each deoxynucleoside triphosphate (Invitrogen), and a 160 pM concentration of random primer 1254 (5′-CCGCAGCCAA-3′) (1). The PCR program comprised 4 cycles of 94°C for 5 min, 36°C for 5 min, and 72°C for 5 min; followed by 30 cycles of 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min; with a final extension at 72°C for 10 min. The resulting amplicons were separated by electrophoresis in 1% agarose gel and visualized by ethidium bromide. This analysis was repeated at least twice to confirm its reproducibility.
Dendrogram analysis of PFGE results and 16S rRNA gene sequences.
To examine the relationships between strains, we calculated banding patterns in PFGE by UPGMA (unweighted pair-group method with arithmetic averages) analysis (38) using the Statistica clustering program (Design Technologies, Tokyo, Japan). 16S rRNA gene sequences among strains isolated in the present study and spirochetes submitted to GenBank were compared, and phylogenetic trees were constructed by using the neighbor-joining method of Saitou and Nei (33).
Nucleotide sequence accession number.
The partial 16S rRNA gene sequence of strain YG3903R was submitted to GenBank and assigned accession number FJ004921.
RESULTS
Isolation of helical bacteria from PDD biopsy samples.
The PDDTp culture plates were examined every 7 days after inoculation. Treponemes formed diffuse film-like colonies at the stamped spots after 14 days at 35°C. Although bacterial colonies other than treponemes also grew on the selective agar plates, treponeme-like colonies were easily differentiated from them. Suspected colonies were stained with crystal violet for morphological observations. Helix-shaped bacteria were taken from the edge of their colonies and subcultured on fresh PDDTp agar plates to obtain pure cultures. A total of 40 spirochetes were isolated from 24 of 80 cattle with PDD (30.0%) and from 25 of 91 PDD lesions (27.5%) from nine farms in seven prefectures (Table 1). Multiple strains were isolated from a single lesion or from both feet of six cattle on farms A and B. The isolation rate ranged from 0 to 100% of cattle (Table 1).
Electron microscopic observation.
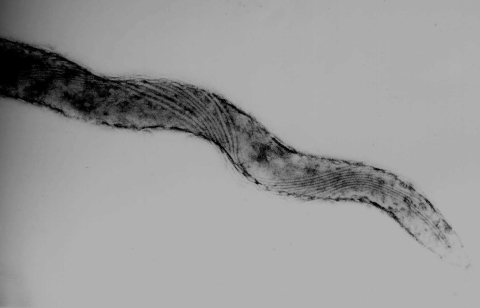
Electron microscopy of strain HT201 showed helical shapes 6 to 15 μm long and 0.2 to 0.3 μm in diameter (Fig. 1). These bacteria had eight axial flagella attached to each end.
FIG. 1.
Transmission electron microscopy of T. phagedenis-like spirochete strain HT201 isolated from a PDD lesion. The spirochete has eight axial flagella at each end.
Biochemical characteristics.
We examined 12 better-growing strains among the 40 isolates since enough amount of bacterial cells were necessary for the tests. The strains which did not recover one loopful of bacteria from one PDDTp agar plate after 14 days of incubation were not subjected to the tests. They showed weak beta-hemolytic activity on PDDTp agar plates supplemented with horse blood. T. phagedenis ATCC 27087 and all PDD isolates were catalase positive and oxidase negative. The enzyme activities are shown in Table 2. PDD isolates showed results similar to those of T. phagedenis ATCC 27087 but not to those of T. denticola (Table 2).
TABLE 2.
Enzyme activities of T. phagedenis-like spirochetes isolated from PDD lesions
| Strain | API-ZYM resultb
|
Other analysesc
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | H | C | O | |
| T. denticola JCM8152 | − | + | + | − | + | − | − | + | − | + | − | + | + | − | − | − | − | − | − | + | + | − |
| T. phagedenis ATCC 27087 | + | + | + | − | − | − | − | − | − | + | − | − | + | + | − | − | + | − | + | + | + | − |
| T. phagedenis-like spirochetes from PDDa | + | + | + | − | − | − | − | − | − | + | − | − | + | + | − | − | + | − | + | + | + | − |
Twelve strains of T. phagedenis-like spirochetes isolated from PDD were examined for the biochemical tests.
Enzymes are represented by numbers above each column as follows: 1, alkaline phosphatase; 2, C4 esterase; 3, C8 esterase lipase; 4, C14 lipase; 5, leucine arylamidase; 6, valine arylamidase; 7, cystine arylamidase; 8, trypsin; 9, chymotrypin; 10, acid phosphatase; 11, naphtholphosphohydrolase; 12, α-galactosidase; 13, β-galactosidase; 14, β-glucuronidase; 15, α-gulucosidase; 16, β-gulucosidase; 17, N-acetyl-β-glucosaminidase; 18, α-mannosidase; 19, α-fucosidase.
H, hemolytic activity; C, catalase; O, oxidase.
16S rRNA gene sequence analysis.
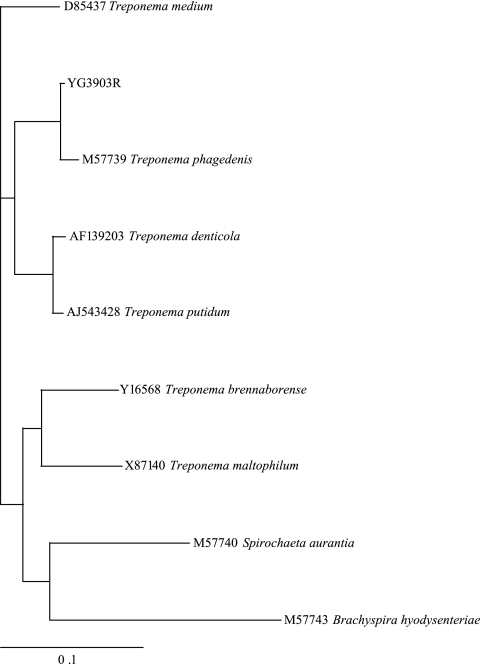
A fragment of the 16S rRNA gene (∼1,500 bp) of each 40 strains was amplified by using universal primers. BLAST searches of the sequence data showed 99 to 100% identity of all strains examined with each other, as well as with PDD strains isolated in the United States, the United Kingdom, and Denmark (accession numbers L78126, EF645248, and AM942450, respectively). The phylogenic tree (Fig. 2) placed our PDD isolates (only strain YG3903R is shown in the figure) in a single cluster closely related to T. phagedenis ATCC 27087.
FIG. 2.
Phylogenic tree based on nearly complete 16S rRNA gene sequences of strain YG3903R isolated from PDD in the present study and from other Treponema species. Brachyspira hyodysenteriae was added as an outgroup.
PFGE.
We compared 26 strains (Table 3) and type strains of T. denticola and T. phagedenis. They included multiple colonies isolated from the same lesion or from different lesions on the same cattle on farms A and B. The results revealed considerable diversity among strains isolated not only from different cattle but also from the same individuals (Fig. 3).
TABLE 3.
Strains from 19 cattle used for PFGE and RAPD types
| Cattle no.a | Strain | Farm | Lesion sourceb | Yr of isolation | Region |
|---|---|---|---|---|---|
| 1 | HT201 | H | R-L | 2004 | Kumamoto |
| 2 | YG3093R | C | R-R | 2005 | Yamagata |
| 3 | YG5618 | C | R-R | 2005 | Yamagata |
| 4 | HG42 | F | NA | 2005 | Hyogo |
| 5 | IZ6-2 | K | NA | 2005 | Kagoshima |
| 6 | IZ7-2 | K | R-L | 2005 | Kagoshima |
| 7 | CH6 | D | R-R | 2005 | Chiba |
| 8 | CH9 | E | R-L | 2006 | Chiba |
| 9* | HD21-66 | A | R-L | 2007 | Hokkaido |
| HD21R-R7 | A | R-R | 2007 | Hokkaido | |
| 10* | HD22-63 | A | R-L | 2007 | Hokkaido |
| HD22R-16 | A | R-R | 2007 | Hokkaido | |
| 11 | HD23-1 | A | R-L | 2007 | Hokkaido |
| 12 | HD24-2 | A | R-R | 2007 | Hokkaido |
| 13† | HD26-43 | A | R-R | 2007 | Hokkaido |
| HD26-67 | A | R-R | 2007 | Hokkaido | |
| HD26-R4 | A | R-R | 2007 | Hokkaido | |
| HD26-R5 | A | R-R | 2007 | Hokkaido | |
| 14† | HD27-4 | B | R-L | 2007 | Hokkaido |
| HD27-24 | B | R-L | 2007 | Hokkaido | |
| HD27-R6 | B | R-L | 2007 | Hokkaido | |
| 15 | HD30R-16 | B | R-R | 2007 | Hokkaido |
| 16 | HD31-84 | B | R-L | 2007 | Hokkaido |
| 17 | HD32-R10 | B | R-L | 2007 | Hokkaido |
| 18 | HD33-86 | B | R-R | 2007 | Hokkaido |
| 19 | HD34-27 | B | R-L | 2007 | Hokkaido |
*, Strains isolated from different lesions on the same cow; †, strains isolated from the same lesion on the same cow.
R-R, rear right foot; R-L, rear left foot; NA, not available.
FIG. 3.
PFGE analysis of PDD isolates. We examined 26 strains isolated from PDD lesions and two type strains of Treponema species. Multiple colonies from individual PDD lesions on the same host are included. Lanes and strains are as follows: 1, T. denticola JCM8225; 2, T. phagedenis ATCC 27087; 3, HT201; 4, YG3903R; 5, YG5618; 6, HG42; 7, IZ6-2; 8, IZ7-2; 9, CH6; 10, CH9; 11, HD21-66; 12, HD21R-R7; 13, HD22-63; 14, HD22R-16; 15, HD23-1; 16, HD24-2; 17, HD26-43; 18, HD26-67; 19, HD26-R4; 20, HD26-R5; 21, HD27-4; 22, HD27-24; 23, HD27-R6; 24, HD30-R16; 25, HD31-84; 26, HD32-R10; 27, HD33-86; and 28, HD34-27. Lanes M contain the ladder markers for PFGE.
RAPD analysis.
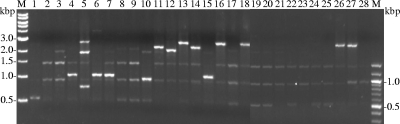
The same strains used for PFGE (Table 3) were tested by RAPD analysis. The RAPD profiles of the isolates appeared to be divergent, as seen in the PFGE analysis. Even the colonies from the same host revealed different banding patterns, confirming that the result corresponded to that of the PFGE analysis (Fig. 4).
FIG. 4.
RAPD fingerprints of 26 PDD isolates and 2 type strains. Lanes and strains are as follows: 1, T. denticola JCM8225; 2, T. phagedenis ATCC 27087; 3, HT201; 4, YG3903R; 5, YG5618; 6, HG42; 7, IZ6-2; 8, IZ7-2; 9, CH6; 10, CH9; 11, HD21-66; 12, HD21R-R7; 13, HD22-63; 14, HD22R-16; 15, HD23-1; 16, HD24-2; 17, HD26-43; 18, HD26-67; 19, HD26-R4; 20, HD26-R5; 21, HD27-4; 22, HD27-24; 23, HD27-R6; 24, HD30-R16; 25, HD31-84; 26, HD32-R10; 27, HD33-86; and 28, HD34-27. Lanes M contain 1.0-kb (left) and 0.1-kb (right) ladder markers.
Dendrograms by UPGMA.
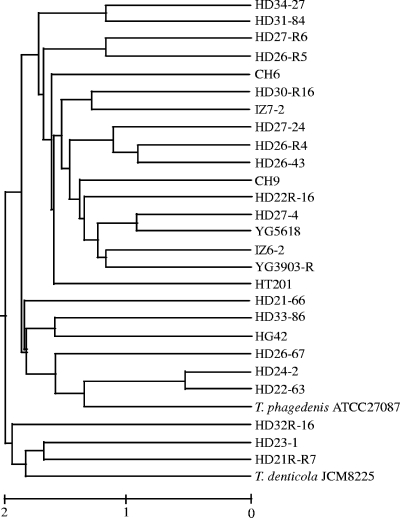
A total of 36 banding patterns were detected in PFGE of the 26 strains examined. Cluster analysis indicated that they formed three major clusters, and each strain showed multiple branching (Fig. 5). Although strains HD21R-R7 and HD21-66 and strains HD26-67, HD26-43, HD26-R4, and HD26-R5 were isolated from two individual cattle on Farm A, they were represented in all three clusters (Fig. 5).
FIG. 5.
Dendrogram of cluster analysis based on PFGE banding patterns. We examined 26 strains isolated from PDD lesions and 2 type strains of Treponema species.
DISCUSSION
Culture methods appropriate for isolating treponemes effectively from PDD lesions have not previously been developed. A number of researchers have shown the difficulty of culturing and isolating Treponema species from PDD lesions, since the lesions are usually contaminated with feces and soil. Since other coinfecting bacteria (7, 22, 27, 34, 45) may have different growth characteristics and sensitivities to selective agents, as the growth rate of many of Treponema species is poor, and as the presence of other bacteria in PDD lesions often prevents the isolation of treponemes because of their overgrowth (43), varying the culture media, temperature, and time could improve isolation.
In the present study, we developed a simple two-step culture method that improved the rate of isolation. Initially, we used enrichment culture with selective antibiotics at 35°C, followed by direct plating on anaerobic agar plates. However, the enrichment culture allowed other bacteria to proliferate. Next, we inoculated biopsy specimens on PDDTp agar plates immediately after sampling, but we isolated only one spirochete from about 60 PDD lesions (data not shown). The rate of the isolation of treponemes from the samples in PDDTp broth, in which they were transported over 2 days, was improved over previous methods. After incubation at 4°C for 48 to 72 h in the PDDTp broth, nontreponeme bacteria inoculated on PDDTp agar plates grew slowly, and it was easier to separate the treponemes from them. Although why the other bacteria did not grow well remains unclear, many of them may have been killed by the antibiotics in the broth during incubation at low temperature, conditions which the isolated treponemes may be better able to survive. All treponemes isolated by this method were identified as T. phagedenis-like spirochetes.
Walker et al. isolated T. phagedenis-like, T. denticola-like, and T. vincentii/T. medium-like spirochetes in the United States (43). Recently, Evans et al. isolated these three groups from PDD lesions in United Kingdom cattle (16). Furthermore, Schrank et al. isolated and characterized the newly proposed species T. brennaborense (35). Since it is still unclear which Treponema is the most predominant species in PDD lesions in Japan, we cannot conclude that our culture method is suitable for the isolation of treponemes other than T. phagedenis-like spirochetes. However, T. denticola and T. phagedenis isolated from humans were able to grow on the PDDTp medium used in the present study (data not shown). Thus, the existence of treponemes other than T. phagedenis-like spirochetes in PDD lesions of cattle in Japan should be determined by molecular biological methods, such as PCR and in situ hybridization.
All 16S rRNA gene sequences of the isolates showed 99 to 100% identity with each other and with some isolates from the United States (43) and the United Kingdom (9, 16). Phylogenic analysis showed a close relationship between our PDD isolates and T. phagedenis ATCC 27087. Our isolates were catalase positive and oxidase negative and showed weak beta-hemolytic activity. Enzyme activities of all isolates examined were identical, and the enzyme profile was more closely related to that of T. phagedenis ATCC 27087 but not that of T. denticola JCM8225. However, the activities were not identical to those of strains reported previously (11, 16, 43), even though the 16S rRNA gene sequences were more than 99% identical. The 16S rRNA sequence might be similar, but the sequences that encode various enzymes might be more variable. These phenotypic characteristics may change with passage number or culture conditions.
In previous studies, the genetic diversity of T. phagedenis-like organisms isolated from PDD was assessed by molecular techniques such as PFGE and PCR-based restriction fragment length polymorphism (41), because epidemiological methods such as serotyping, biotyping, and outer membrane profiling have not been established for T. phagedenis-like spirochetes. Here, we used PFGE and RAPD to detect the heterogeneity of DNAs of T. phagedenis-like strains. The two typing methods revealed considerable diversity among the strains, not only from the same farm but also from the same animals. Trott et al. showed by PFGE analysis that 6 strains of T. phagedenis-like spirochetes were genetically heterogeneous (41). Walker et al. similarly showed that eight treponemes from various farms had different restriction patterns (43). Our result is consistent with these reports. Interestingly, we found genetic diversity among strains isolated from the same PDD lesion.
There are two possible main reasons for the DNA diversity among the strains: multiple infections with different strains and the acquisition of genetic diversity in vivo. Some evidence of genomic recombination or rearrangement in pathogens has been demonstrated (13, 40). Such mechanisms allow organisms to generate antigenic diversity, helping them to evade the immunological response of the host (17, 18). PDD is a persistent infection, and spirochetes are observed in most stages of infection (6). Although systemic antibodies against T. phagedenis-like spirochetes were detected (42), they may not be protective. Furthermore, recurrent infection has also been reported (26). These findings may provide further evidence for the role of these organisms in the pathogenesis of PDD. Long-term genotyping and phenotyping in experimental infection studies are needed to assess the genomic diversity among isolates over time in the same lesion.
The etiologic involvement of treponemes in PDD is incompletely known. However, the result that treponemes with very similar 16S rRNA gene sequences are common in PDD lesions in different countries implies that they may be one of the most predominant populations in the lesions and play a role in the development of PDD. Zuerner et al. showed that T. phagedenis-like spirochetes from PDD lesions suppress the innate immune response in cattle (47). Elliott et al. reported that T. phagedenis-like spirochetes induced abscesses in mice (15). These discoveries imply that T. phagedenis-like spirochetes have pathogenic potential. Further studies of the pathogenic features of PDD-related treponemes are required to clarify the etiology of PDD.
Acknowledgments
This study was supported by Japan Farriers Association, KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas “Applied Genomics” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and Project for Zoonoses Education and Research, University of Miyazaki.
Footnotes
Published ahead of print on 14 January 2009.
REFERENCES
- 1.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 205137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blowey, R. W., S. H. Done, and W. Cooley. 1994. Observations on the pathogenesis of digital dermatitis in cattle. Vet. Rec. 135115-117. [DOI] [PubMed] [Google Scholar]
- 3.Blowey, R. W., and M. W. Sharp. 1988. Digital dermatitis in dairy cattle. Vet. Rec. 122505-508. [DOI] [PubMed] [Google Scholar]
- 4.Cheli, R., and C. M. Mortellaro. 1974. La dermatite digitale del bovino, p. 208-213. In P. Gallarati (ed.), Proceedings of the 8th International Conference on Diseases of Cattle. International Conference on Diseases of Cattle, Piacenza, Italy.
- 5.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 313497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, B. K., H. Nattermann, S. Grund, W. Haider, and U. B. Gobel. 1997. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int. J. Syst. Bacteriol. 47175-181. [DOI] [PubMed] [Google Scholar]
- 7.Collighan, R. J., and M. J. Woodward. 1997. Spirochaetes and other bacterial species associated with bovine digital dermatitis. FEMS Microbiol. Lett. 15637-41. [DOI] [PubMed] [Google Scholar]
- 8.Cruz, C. E., C. A. Pescador, Y. Nakajima, and D. Driemeier. 2005. Immunopathological investigations on bovine digital epidermitis. Vet. Rec. 157834-840. [DOI] [PubMed] [Google Scholar]
- 9.Demirkan, I., S. D. Carter, C. A. Hart, and M. J. Woodward. 1999. Isolation and cultivation of a spirochaete from bovine digital dermatitis. Vet. Rec. 145497-498. [DOI] [PubMed] [Google Scholar]
- 10.Demirkan, I., S. D. Carter, R. D. Murray, R. W. Blowey, and M. J. Woodward. 1998. The frequent detection of a treponeme in bovine digital dermatitis by immunocytochemistry and polymerase chain reaction. Vet. Microbiol. 60285-292. [DOI] [PubMed] [Google Scholar]
- 11.Demirkan, I., H. F. Williams, A. Dhawi, S. D. Carter, C. Winstanley, K. D. Bruce, and C. A. Hart. 2006. Characterization of a spirochaete isolated from a case of bovine digital dermatitis. J. Appl. Microbiol. 101948-955. [DOI] [PubMed] [Google Scholar]
- 12.Döpfer, D., A. Koopmans, F. A. Meijer, I. Szakall, Y. H. Schukken, W. Klee, R. B. Bosma, J. L. Cornelisse, A. J. van Asten, and A. A. ter Huurne. 1997. Histological and bacteriological evaluation of digital dermatitis in cattle, with special reference to spirochaetes and Campylobacter faecalis. Vet. Rec. 140620-623. [DOI] [PubMed] [Google Scholar]
- 13.Dutta, C., and A. Pan. 2002. Horizontal gene transfer and bacterial diversity. J. Biosci. 2727-33. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes: characterization of a gene coding for 16S rRNA. Nucleic Acids Res. 177843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, M. K., D. P. Alt, and R. L. Zuerner. 2007. Lesion formation and antibody response induced by papillomatous digital dermatitis-associated spirochetes in a murine abscess model. Infect. Immun. 754400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans, N. J., J. M. Brown, I. Demirkan, R. D. Murray, W. D. Vink, R. W. Blowey, C. A. Hart, and S. D. Carter. 2008. Three unique groups of spirochetes isolated from digital dermatitis lesions in UK cattle. Vet. Microbiol. 131141-150. [DOI] [PubMed] [Google Scholar]
- 17.Futse, J. E., K. A. Brayton, M. J. Dark, D. P. Knowles, Jr., and G. H. Palmer. 2008. Superinfection as a driver of genomic diversification in antigenically variant pathogens. Proc. Natl. Acad. Sci. USA 1052123-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grogono-Thomas, R., M. J. Blaser, M. Ahmadi, and D. G. Newell. 2003. Role of S-layer protein antigenic diversity in the immune responses of sheep experimentally challenged with Campylobacter fetus subsp. fetus. Infect. Immun. 71147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez, J., J. K. Shearer, and J. B. Elliott. 1999. Comparison of topical application of oxytetracycline and four nonantibiotic solutions for treatment of papillomatous digital dermatitis in dairy cows. J. Am. Vet. Med. Assoc. 214688-690. [PubMed] [Google Scholar]
- 20.Kimura, Y., M. Takahashi, H. Matsumoto, H. Tsukuda, M. Sato, K. Ohgawara, M. Kanoe, N. Goto, O. Aoki, and M. Hataya. 1992. Verrucose dermatitis and digital papillomatosis in dairy cows. Hoof 16037-38. (In Japanese.) [Google Scholar]
- 21.Klitgaard, K., M. Boye, N. Capion, and T. K. Jensen. 2008. Evidence of multiple Treponema phylotypes involved in bovine digital dermatitis as shown by 16S rDNA analysis and fluorescent in situ hybridization. J. Clin. Microbiol. 463012-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koniarová, I., A. Orsag, and V. Ledeckŷ. 1993. The role anaerobes in dermatitis digitalis et interdigitalis in cattle. Vet. Med. 38589-596. [PubMed] [Google Scholar]
- 23.Moter, A., G. Leist, R. Rudolph, K. Schrank, B. K. Choi, M. Wagner, and U. B. Gobel. 1998. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology 144(Pt. 9)2459-2467. [DOI] [PubMed] [Google Scholar]
- 24.Nagai, F., H. Ota, K. Fujimoto, H. Abe, T. Munakata, T. Abe, M. Tanaka, A. Ito, Y. Yamamoto, T. Ando, M. Koiwa, H. Taniyama, N. Kikuchi, and N. Kusaba. 2000. Mass outbreak of verrucous dermatitis and digital papillomatous caused by Treponema-like spirochetes in daily cattle. J. Jpn. Vet. Med. Assoc. 53577-581. (In Japanese.) [Google Scholar]
- 25.Nordhoff, M., A. Moter, K. Schrank, and L. H. Wieler. 2008. High prevalence of treponemes in bovine digital dermatitis-A molecular epidemiology. Vet. Microbiol. 131293-300. [DOI] [PubMed] [Google Scholar]
- 26.Ohtake, O. 2003. Present status of leg and hoof diseases are frequently reported from various parts in Japan. Yamaguchi J. Vet. Med. 3041-50. (In Japanese.) [Google Scholar]
- 27.Ohya, T., H. Yamaguchi, Y. Nii, and H. Ito. 1999. Isolation of Campylobacter sputorum from lesions of papillomatous digital dermatitis in dairy cattle. Vet. Rec. 145316-318. [DOI] [PubMed] [Google Scholar]
- 28.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12357-358. [DOI] [PubMed] [Google Scholar]
- 29.Read, D. H., and R. L. Walker. 1998. Papillomatous digital dermatitis (footwarts) in California dairy cattle: clinical and gross pathologic findings. J. Vet. Diagn. Investig. 1067-76. [DOI] [PubMed] [Google Scholar]
- 30.Read, D. H., R. L. Walker, A. E. Castro, J. P. Sundberg, and M. C. Thurmond. 1992. An invasive spirochaete associated with interdigital papillomatosis of dairy cattle. Vet. Rec. 13059-60. [DOI] [PubMed] [Google Scholar]
- 31.Rebhun, W. C., R. M. Payne, J. M. King, M. Wolfe, and S. N. Begg. 1980. Interdigital papillomatosis in dairy cattle. J. Am. Vet. Med. Assoc. 177437-440. [PubMed] [Google Scholar]
- 32.Rijpkema, S. G., G. P. David, S. L. Hughes, and M. J. Woodward. 1997. Partial identification of spirochaetes from two dairy cows with digital dermatitis by polymerase chain reaction analysis of the 16S rRNA gene. Vet. Rec. 140257-259. [DOI] [PubMed] [Google Scholar]
- 33.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 34.Schlafer, S., M. Nordhoff, C. Wyss, S. Strub, J. Hubner, D. M. Gescher, A. Petrich, U. B. Gobel, and A. Moter. 2008. Involvement of Guggenheimella bovis in digital dermatitis lesions of dairy cows. Vet. Microbiol. 128118-125. [DOI] [PubMed] [Google Scholar]
- 35.Schrank, K., B. K. Choi, S. Grund, A. Moter, K. Heuner, H. Nattermann, and U. B. Gobel. 1999. Treponema brennaborense sp. nov., a novel spirochaete isolated from a dairy cow suffering from digital dermatitis. Int. J. Syst. Bacteriol. 49(Pt. 1)43-50. [DOI] [PubMed] [Google Scholar]
- 36.Schroeder, C. M., K. W. Parlor, T. L. Marsh, N. K. Ames, A. K. Goeman, and R. D. Walker. 2003. Characterization of the predominant anaerobic bacterium recovered from digital dermatitis lesions in three Michigan dairy cows. Anaerobe 9151-155. [DOI] [PubMed] [Google Scholar]
- 37.Shibahara, T., T. Ohya, R. Ishii, Y. Ogihara, T. Maeda, Y. Ishikawa, and K. Kadota. 2002. Concurrent spirochaetal infections of the feet and colon of cattle in Japan. Aust. Vet. J. 80497-502. [DOI] [PubMed] [Google Scholar]
- 38.Sneath, P. H., and R. R. Sokal. 1973. Numerical taxonomy: the principles and practice of numerical classification. W. H. Freeman & Co., San Francisco, CA.
- 39.Stamm, L. V., H. L. Bergen, and R. L. Walker. 2002. Molecular typing of papillomatous digital dermatitis-associated Treponema isolates based on analysis of 16S-23S ribosomal DNA intergenic spacer regions. J. Clin. Microbiol. 403463-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sung, S. Y., J. V. McDowell, J. A. Carlyon, and R. T. Marconi. 2000. Mutation and recombination in the upstream homology box-flanked ospE-related genes of the Lyme disease spirochetes result in the development of new antigenic variants during infection. Infect. Immun. 681319-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trott, D. J., M. R. Moeller, R. L. Zuerner, J. P. Goff, W. R. Waters, D. P. Alt, R. L. Walker, and M. J. Wannemuehler. 2003. Characterization of Treponema phagedenis-like spirochetes isolated from papillomatous digital dermatitis lesions in dairy cattle. J. Clin. Microbiol. 412522-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker, R. L., D. H. Read, K. J. Loretz, D. W. Hird, and S. L. Berry. 1997. Humoral response of dairy cattle to spirochetes isolated from papillomatous digital dermatitis lesions. Am. J. Vet. Res. 58744-748. [PubMed] [Google Scholar]
- 43.Walker, R. L., D. H. Read, K. J. Loretz, and R. W. Nordhausen. 1995. Spirochetes isolated from dairy cattle with papillomatous digital dermatitis and interdigital dermatitis. Vet. Microbiol. 47343-355. [DOI] [PubMed] [Google Scholar]
- 44.Wilson, K. 1987. Preparation of genomic DNA from bacteria, p. 293-332. In F. M. Ausubel, R. Brent, R. E., Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, unit 2.4. Wiley, New York, NY. [DOI] [PubMed]
- 45.Wyss, C., F. E. Dewhirst, B. J. Paster, T. Thurnheer, and A. Luginbuhl. 2005. Guggenheimella bovis gen. nov., sp. nov., isolated from lesions of bovine dermatitis digitalis. Int. J. Syst. Evol. Microbiol. 55667-671. [DOI] [PubMed] [Google Scholar]
- 46.Yeruham, I., and S. Perl. 1998. Clinical aspects of an outbreak of papillomatous digital dermatitis in a dairy cattle herd. J. S. Afr. Vet. Assoc. 69112-115. [DOI] [PubMed] [Google Scholar]
- 47.Zuerner, R. L., M. Heidari, M. K. Elliott, D. P. Alt, and J. D. Neill. 2007. Papillomatous digital dermatitis spirochetes suppress the bovine macrophage innate immune response. Vet. Microbiol. 125256-264. [DOI] [PubMed] [Google Scholar]