Abstract
In the 2006-to-2007 influenza season, amantadine-sensitive strains were found among the N-lineage influenza A (H3N2) viruses, which were previously believed to be associated with amantadine resistance. Whole-genome sequencing results indicated that this was due to a further reassortment event.
Amantadine has been used for the treatment of influenza A virus infections. Recently, there has been a substantial increase in amantadine-resistant strains of the influenza A virus (1). The most common mutation linked to the resistance is a substitution of asparagine for serine at position 31 (S31N) in the M2 gene (3, 10).
Previous studies reported specific mutations in the hemagglutinin (HA) gene of the influenza A (H3N2) virus in the 2005-to-2006 season, including substitutions at position 193, from serine to phenylalanine (S193F), and at position 225, from aspartic acid to asparagine (D225N). Interestingly, all of the strains with these mutations in the HA gene had the amantadine-resistant S31N mutation in the M2 gene (4, 7, 8). Simonsen et al. demonstrated that these strains were generated by a reassortment event (9). It is still unclear whether the spread of the amantadine-resistant strains was related to drug selection pressure or whether it resulted from an interaction with advantageous mutations located elsewhere in the viral genome (2, 5, 6, 8, 9). Further clarification of whether the S193F and D225N double mutations in the HA gene, either alone or in combination, have a role in maintaining the S31N mutation in the M2 gene requires further study. Our objective was to determine whether these specific mutations in the HA gene were consistently associated with amantadine resistance in the influenza A (H3N2) virus strains.
Influenza A (H3N2) virus strains isolated from clinical specimens in the 2005-to-2006 and 2006-to-2007 seasons in Sendai (in the northern part of Japan) and Fukuoka (in the southern part of Japan) were used for the analysis. Nasopharyngeal swabs were inoculated on MDCK cells for virus isolation. Viral RNA was extracted from the isolated viruses, and PCR was performed to amplify parts of the HA1 and M2 genes. Sequences were analyzed using a 3130 genetic analyzer (Applied Biosystems, Foster City, CA).
For the 2005-to-2006 and 2006-to-2007 influenza seasons, 223 strains were isolated from Sendai and 117 were isolated from Fukuoka. We selected 66 isolates (44 isolates from Sendai and 22 from Fukuoka) for sequencing of the part of the HA1 gene (amino acids 17 to 317) and for the section of the M2 gene (amino acids 10 to 61) that included sites of mutations responsible for amantadine sensitivity. Fifty-one (77%) strains had an amantadine-resistant S31N mutation in the M2 gene. We did not find any other mutations linked to amantadine resistance except S31N.
A phylogenetic tree for the HA1 gene was inferred using the neighbor-joining method (see Fig. SA2 in the supplemental material). The strains were grouped into two lineages. (i) N-lineage strains (7) were characterized by S193F and D225N mutations in the HA gene and were found in the 2005-to-2006 and 2006-to-2007 seasons. All N-lineage strains from the 2005-to-2006 season were amantadine resistant, whereas for strains from the 2006-to-2007 season, we identified both amantadine-sensitive and amantadine-resistant strains. (ii) S-lineage strains (8) were found only in the 2005-to-2006 season, and all were amantadine sensitive (Table 1; also see Fig. SA2 in the supplemental material). The occurrence of amantadine-sensitive N-lineage strains was surprising, as previous studies indicated that almost all N-lineage strains are amantadine resistant (8, 9). We classified the strains into three groups: (i) N (Res) strains, or strains with both S193F and D225N mutations in the HA gene and the S31N mutation in the M2 gene; (ii) N (Sen) strains, or strains with both S193F and D225N mutations in the HA gene but without the S31N mutation in the M2 gene; and (iii) S (Sen) strains, or S-lineage strains.
TABLE 1.
Numbers of strains sensitive or resistant to amantadine, grouped by lineage and season
| Strain lineage | No. of strainsa
|
|||
|---|---|---|---|---|
| 2005-to-2006 season
|
2006-to-2007 season
|
|||
| Sensitive | Resistant | Sensitive | Resistant | |
| S lineage | 8 | 0 | 0 | 0 |
| N lineage | 0 | 26 | 7 | 25 |
Strains were defined as sensitive or resistant to amantadine by genetic analysis.
To explore the genomic events resulting in the genesis of N (Sen) strains, we sequenced all genes from 10 randomly selected strains (see Fig. SA2 in the supplemental material). We calculated pairwise distances between N (Sen) and S (Sen) strains and between N (Sen) and N (Res) strains for the full lengths of the HA and MP genes (data not shown). N (Sen) strains were close to N (Res) strains according to the HA genes, whereas N (Sen) strains were close to S (Sen) strains according to the MP genes. The results indicated the possibility of a reassortment event.
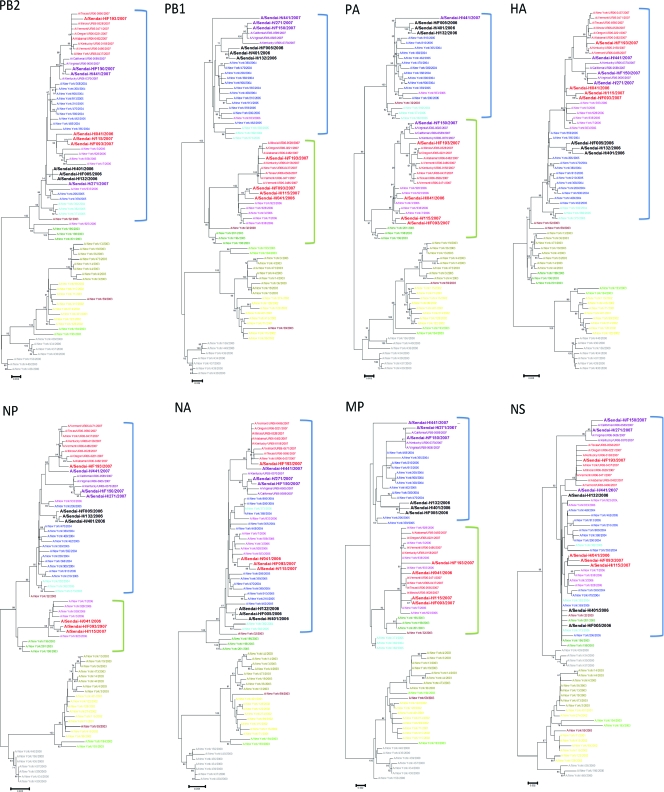
Next, we constructed phylogenetic trees for the 10 strains and for other representative strains available in the online Influenza Virus Resource database (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html). The representative strains had been isolated from the Northern Hemisphere since 2000 and were classified into 11 groups by a phylogenetic analysis of the HA gene (see Fig. SA3 in the supplemental material). Strains that could be classified as S (Sen), N (Res), or N (Sen) strains according to our definitions were found in other countries. All S (Sen) strain segments originated from strains found in the 2004-to-2005 season (Fig. 1); however, segments of the N (Res) and N (Sen) strains originated from strains found in the 2002-to-2003 and 2004-to-2005 seasons (Fig. 1).
FIG. 1.
Phylogenetic trees for various strain segments. The phylogenetic trees were inferred using the neighbor-joining method. The bars at the bottom are scales of branch lengths, which show the evolutionary distances computed using the maximum-composite-likelihood method. Strains used in this study are written in boldface type. S, N (Res), and N (Sen) strains are written in black, red, and purple types, respectively. Other colors for each taxon coincide with the colors in Fig. SA3 in the supplemental material. Blue brackets indicate strains whose segment originated from a strain found in the 2004-to-2005 season, and green brackets indicate strains whose segment originated from a strain found in the 2002-to-2003 season. (Also see a larger version of this figure in Fig. SA1 in the supplemental material.)
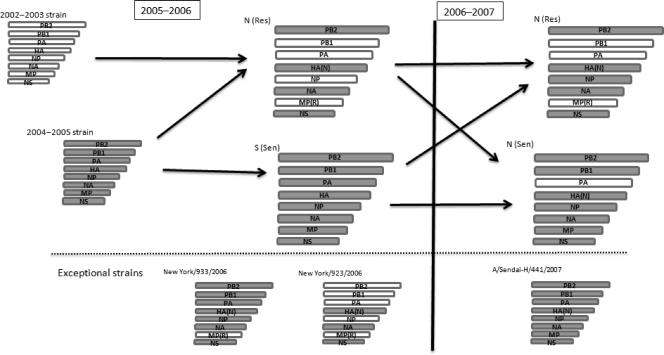
It is likely that four segments (the PB2, HA, NA, and NS genes) of N (Res) strains taken from the isolates found in the 2005-to-2006 season originated from strains found in the 2004-to-2005 season and that the other four segments (the PB1, PA, NP, and MP genes) originated from strains found in the 2002-to-2003 season (Fig. 2). These were likely to have been generated by a reassortment event. Both N-lineage amantadine-resistant and -sensitive [N (Res) and N (Sen)] strains were found in the 2006-to-2007 season. Although both types had specific HA gene mutations that characterized the N lineage (S193F and D225N), they had different combinations of segments from N (Res) strains found in the 2005-to-2006 season. In the 2006-to-2007 season, five segments (the PB2, HA, NA, NP, and NS genes) of the N (Res) strains originated from a strain found in the 2004-to-2005 season and three segments (the PB1, PA, and MP genes) originated from a strain found in the 2002-to-2003 season. In the 2006-to-2007 season, all segments, except for the PA gene, of N (Sen) strains originated from a strain from the 2004-to-2005 season; the PA gene originated from a strain found in the 2002-to-2003 season. In the 2006-to-2007 season, both N (Sen) and N (Res) strains were generated by further reassortment events. We also found exceptional strains that seemed to be products of other reassortment events (Fig. 2).
FIG. 2.
Scheme for reassortment events. The origination of each segment of each group is shown. No shading, segment originated from a strain found in the 2002-to-2003 season; gray shading, segment originated from a strain found in the 2004-to-2005 season. HA(N) indicates the HA gene with mutations characteristic of the N lineage. MP(R) indicates the MP gene with an amantadine-resistant mutation. As for A/Sendai-H/441/2007, although all of its segments originated from strains found in the 2004-to-2005 season, it has mutations in the HA gene characteristic of the N lineage. The mutations were not found in an original strain from the 2004-to-2005 season that was an S (Sen) strain. Therefore, we considered A/Sendai-H/441/2007 exceptional.
In the N (Res) strains, the MP gene coding for amantadine resistance was replaced, by a reassortment event, by a gene that did not code for resistance. N (Sen) strains did not have a combination of the S193F and D225N substitutions in the HA gene and the S31N substitution in the M2 gene. This suggested that the combination was not necessarily associated with acquired fitness. Acquiring or losing amantadine-resistant mutations seems to have occurred through a reassortment event. The advent of the N lineage was one of these events. We must continue to observe the genomic evolution and prevalence of amantadine-resistant strains, which should provide clues about the spread of these strains.
Nucleotide sequence accession numbers.
All accession numbers used in the study, including our newly determined gene sequences, are given in list SA1 in the supplemental material.
Supplementary Material
Acknowledgments
We thank Hidekazu Nishimura and all other staff members of the virus center of Sendai Medical Center. We are also indebted to Makoto Shoji and other medical practitioners for collecting clinical samples.
Financial support for this study was provided by Health Labor Sciences research grant 20-005-OH from the Ministry of Health, Labor, and Welfare, Japan.
Footnotes
Published ahead of print on 24 December 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Bright, R. A., M.-J. Medina, X. Xu, G. Perez-Oronoz, T. R. Wallis, X. M. Davis, L. Povinelli, N. J. Cox, and A. I. Klimov. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 3661175-1181. [DOI] [PubMed] [Google Scholar]
- 2.Bush, R. M., W. M. Fitch, C. A. Bender, and N. J. Cox. 1999. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 161457-1465. [DOI] [PubMed] [Google Scholar]
- 3.Hay, A. J., A. J. Wolstenholme, J. J. Skehel, and M. H. Smith. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 43021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li, D., R. Saito, M. T. Q. Le, H. L. K. Nguyen, Y. Suzuki, Y. Shobugawa, D. T. Dinh, P. V. M. Hoang, H. T. T. Tran, H. K. Nghiem, L. T. Hoang, L. P. Huynh, H. T. Nguyen, M. Nishikawa, and H. Suzuki. 2008. Genetic analysis of influenza A/H3N2 and A/H1N1 viruses circulating in Vietnam from 2001 to 2006. J. Clin. Microbiol. 46399-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medeiros, R., N. Naffakh, J. C. Manuguerra, and S. van der Werf. 2004. Binding of the hemagglutinin from human or equine influenza H3 viruses to the receptor is altered by substitutions at residue 193. Arch. Virol. 1491663-1671. [DOI] [PubMed] [Google Scholar]
- 6.Regoes, R. R., and S. Bonhoeffer. 2006. Emergence of drug-resistant influenza virus: population dynamical considerations. Science 312389-391. [DOI] [PubMed] [Google Scholar]
- 7.Saito, R., D. Li, C. Shimomura, H. Masaki, M. Q. Le, H. L. K. Nguyen, H. T. Nguyen, T. V. Phan, T. T. K. Nguyen, M. Sato, Y. Suzuki, and H. Suzuki. 2006. An off-seasonal amantadine-resistant H3N2 influenza outbreak in Japan. Tohoku J. Exp. Med. 21021-27. [DOI] [PubMed] [Google Scholar]
- 8.Saito, R., D. Li, Y. Suzuki, I. Sato, H. Masaki, H. Nishimura, T. Kawashima, Y. Shirahige, C. Shimomura, N. Asoh, S. Degawa, H. Ishikawa, M. Sato, Y. Shobugawa, and H. Suzuki. 2007. High prevalence of amantadine-resistance influenza A (H3N2) in six prefectures, Japan, in the 2005-2006 season. J. Med. Virol. 791569-1576. [DOI] [PubMed] [Google Scholar]
- 9.Simonsen, L., C. Viboud, B. T. Grenfell, J. Dushoff, L. Jennings, M. Smit, C. Macken, M. Hata, J. Gog, M. A. Miller, and E. C. Holmes. 2007. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol. Biol. Evol. 241811-1820. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki, H., R. Saito, H. Masuda, H. Oshitani, M. Sato, and I. Sato. 2003. Emergence of amantadine-resistant influenza A viruses: epidemiological study. J. Infect. Chemother. 9195-200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.