Abstract
Influenza A virus surveillance studies of wild bird populations are essential to improving our understanding of the role of wild birds in the ecology of low-pathogenic avian influenza viruses and their potential contribution to the spread of H5N1 highly pathogenic avian influenza viruses. Whereas the primary results of such surveillance programs have been communicated extensively, practical considerations and technical implementation options generally receive little attention. In the present study, the data obtained from 39,490 samples were used to compare the impacts of variables such as the sampling procedure, storage and transport conditions, and the choice of molecular and classical diagnostic tests on the outcome of the results. Molecular diagnostic tests allowed estimation of the virus load in samples, which has implications for the ability to isolate virus. Virus isolation in embryonated eggs was more sensitive than virus isolation in cell cultures. Storage and transport conditions had less of an impact on diagnostics by the use of molecular tests than by the use of classical approaches. These findings indicate that molecular diagnostic tests are more sensitive and more reliable than classical tests. In addition, molecular diagnostic tests facilitated analyses in real time and allowed the discrimination of H5 influenza viruses with low and high pathogenicities without the need for virus isolation. Critical assessment of the methods used in large surveillance studies like this will facilitate comparison of the results between studies. Moreover, the lessons learned from current large-scale influenza A virus surveillance activities could be valuable for other pathogen surveillance programs in the future.
Highly pathogenic avian influenza (HPAI) viruses constitute a continuous concern from public health, veterinary, and wildlife perspectives. Whereas aquatic wild birds serve as the main reservoir for low-pathogenic avian influenza (LPAI) viruses, the emergence of HPAI viruses is primarily the result of large-scale poultry husbandry (1, 16, 23, 39). Outbreaks of HPAI predominantly occur in poultry and are restricted to influenza A viruses of the H5 and H7 subtypes. The last decade has seen a marked increase in outbreaks of HPAI in poultry around the world. While most HPAI outbreaks have been controlled relatively quickly, the H5N1 HPAI virus has continuously been circulating in poultry since 1997 (7, 10). The H5N1 HPAI virus is also unusual in the unprecedented scale and geographical spread of the outbreak that it has caused; its transmission to a wide variety of mammalian species, including humans; and the introductions of H5N1 HPAI virus in wild birds (5, 22, 28, 40). These recent introductions of H5N1 HPAI virus in wild birds and the subsequent spread of the virus throughout Asia, the Middle East, Africa, and Europe have put a focus on the role of wild birds in the geographical spread of the H5N1 HPAI virus (29). Large-scale surveillance programs have been implemented in several parts of the world to determine the role of wild birds in the spread of the H5N1 HPAI virus and to serve as a sentinel system for the introduction of the H5N1 HPAI virus into new geographical regions (4, 14, 21, 30, 36). Whereas the primary results of these surveillance programs have been communicated extensively (2, 6, 9, 14, 15, 17, 18, 27, 30, 31, 32), the practical considerations and technical implementation of large-scale influenza A virus surveillance techniques into various field and laboratory settings have received little attention. Here, the results of long-term avian influenza surveillance studies of wild birds were analyzed (24, 26) to determine the effects of sample collection procedures, sample storage conditions, and screening methods for the detection of influenza A viruses in samples obtained from wild bird samples on test results and virus isolation rates.
MATERIALS AND METHODS
Specimens.
Wild birds were trapped by expert ornithologists. Cloacal and/or oropharyngeal swab specimens were collected with sterile cotton swabs and stored in 1 ml transport medium consisting of Hanks balanced salt solution containing 0.5% lactalbumin, 10% glycerol, 200 U/ml penicillin, 200 μg/ml streptomycin, 100 U/ml polymyxin B sulfate, 250 μg/ml gentamicin, and 50 U/ml nystatin (ICN, The Netherlands).
Storage conditions.
The samples were stored at 4°C for less than 2 weeks, including the time required for molecular testing and virus isolation. The samples were stored at −80°C if freezers with such a capability were available near the sampling site and at −20°C if rapid transport or storage at −80°C was not possible. Frozen samples were stored at −80°C in the laboratory upon arrival and were thawed once for analysis.
RNA isolation and virus detection.
RNA was isolated by using a MagnaPure LC system with a MagnaPure LC total nucleic acid isolation kit (Roche Diagnostics, Almere, The Netherlands), and influenza A virus was detected by a generic real-time reverse transcriptase PCR (RRT-PCR) assay targeting the matrix (M) gene (M RRT-PCR). Amplification and detection were performed on an ABI 7700 machine with a TaqMan EZ RT-PCR core reagents kit (Applied Biosystems, Nieuwerkerk aan den IJssel, The Netherlands) and 20 μl of eluate in an end volume of 50 μl. M RRT-PCR-positive samples were subsequently used for the detection of H5 influenza A viruses by using a RRT-PCR targeting the H5 gene (H5 RRT-PCR). H5 gene-positive samples were subsequently characterized further by nucleotide sequencing of the hemagglutinin (HA) region spanning the cleavage site. RRT-PCR primers and probes were designed on the basis of sequence information obtained from publicly available nucleotide sequences (www.ncbi.nlm.nih.gov). RRT-PCR primer and probe combinations were designed with the software package Primer Express (version 2.0; Applied Biosystems).
Oligonucleotides RF1073 (5′-AAG-ACC-AAT-CCT-GTC-ACC-TCT-GA-3′) and RF1074 (5′-CAA-AGC-GTC-TAC-GCT-GCA-GTC-C-3′) and the double dye-labeled probe RF1293 (5′-6-carboxyfluorescein-TTT-GTG-TTC-ACG-CTC-ACC-GTG-CC-6-carboxytetramethylrhodamine-3′) were designed for the detection of the M gene segment of influenza A virus. The primers were taken from Ward et al. (38), but the probe sequence was adjusted for proper detection of avian influenza A viruses. Oligonucleotides RF1151 (5′-GGA-ACT-TAC-CAA-ATA-CTG-TCA-ATT-TAT-TCA-3′) and RF1152 (5′-CCA-TAA-AGA-TAG-ACC-AGC-TAC-CAT-GA-3′) and the double dye-labeled probe RF1153 (5′-6-carboxyfluorescein-TTG-CCA-GTG-CTA-GGG-AAC-TCG-CCA-C-6-carboxytetramethylrhodamine-3′) were designed for the detection of the H5 gene segment of the LPAI and the HPAI viruses.
Virus isolation and characterization.
All M RRT-PCR-positive samples (cycle threshold [CT] value, <40) were used for virus isolation. For isolation of influenza A virus from M RRT-PCR-positive samples, the original specimen was briefly centrifuged and 100 μl was inoculated into the allantoic cavity of 11-day-old embryonated hens' eggs. The allantoic fluid was harvested after 2 days, and influenza A virus was detected by a hemagglutination assay with turkey erythrocytes (41). When no influenza A virus was detected upon the initial virus isolation attempt, an aliquot of the allantoic fluid was inoculated blindly in the allantoic cavity of a second embryonated egg. The HA subtypes of the virus isolates were characterized by a hemagglutination inhibition assay with turkey erythrocytes and subtype-specific hyperimmune rabbit antisera raised against all 16 HA subtypes (12). The neuraminidase (NA) subtypes of the virus isolates were determined by RT-PCR and sequencing with primers specific for the noncoding regions of NA, as described previously (24).
Viruses and cells.
Influenza virus A/Common Teal/Netherlands/10/00 (H1N1), A/Mallard/Netherlands/3/99 (H5N2), and A/Mallard/Netherlands/12/00 (H7N3) were obtained from cloacal swab specimens from migratory ducks and were subsequently passaged twice in embryonated hens' eggs (24).
Madin-Darby canine kidney (MDCK) cells were cultured in Eagle minimal essential medium (Lonza, Heerhugowaard, The Netherlands) supplemented with 10% fetal calf serum (FCS), 100 IU/ml penicillin, 100 mg/ml streptomycin, 1.5 mg/ml sodium bicarbonate, 2 mM glutamine, 10 mM HEPES, and nonessential amino acids. CCL-141 (duck embryo) cells were cultured in minimal essential medium with Hank's salts supplemented with 10% FCS, 100 IU/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 2 mM glutamine, and nonessential amino acids. CCL-169 (goose embryonic kidney) cells were cultured in Ham's F-12 medium (Gibco) supplemented with 10% FCS, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine. Duck embryo fibroblast (DEF) and chicken embryo fibroblast (CEF) cells were cultured in Dulbecco minimal essential medium supplemented with 100 IU/ml penicillin, 100 mg/ml streptomycin, 2 mM glutamine, 1 mM sodium pyruvate, and nonessential amino acids.
Infections and titrations.
The sensitivity of detection and propagation of avian influenza A viruses in different cell lines was performed by inoculation of MDCK, CCL-141, CCL-169, DEF, and CEF cells with 100 μl of serially diluted virus stocks in infection medium. For each cell line used, the effect of trypsin on cell viability was tested by incubation with the infection media containing different concentrations of trypsin and observing the cultures for signs of toxicity. The highest concentration of trypsin at which cells showed a normal morphology was used in each infection medium. After incubation, the cells were washed once with phosphate-buffered saline and cultured in infection medium. For the infection media for CCL-141 and CCL-169, the 10% FCS in the initial cell culture medium was replaced by 3% FCS and 1 μg/ml trypsin (Cambrex) was added; for the infection media for DEF and CEF cells, the 10% FCS in the initial cell culture medium was replaced by 4% bovine serum albumin and 1 μg/ml trypsin (Cambrex) was added. Supernatants were harvested after 5 days, and influenza A virus was detected by the hemagglutination assay (41). To evaluate MDCK cells for use for primary virus isolation attempts, the MDCK cells were inoculated with 100 μl of the original material positive for the M gene by RRT-PCR in 1 ml MDCK infection medium (Eagle minimal essential medium supplemented with 4% bovine serum albumin [Gibco], 100 IU/ml penicillin, 100 μg/ml streptomycin, 1.5 mg/ml sodium bicarbonate, 2 mM glutamine, 10 mM HEPES, nonessential amino acids, and 20 μg/ml trypsin [Cambrex]) and incubated for 1 h. The cells were subsequently washed once with phosphate-buffered saline and cultured in infection medium. The cells were checked daily for cytopathic effects by microscopy. The supernatants were harvested after 5 days, and influenza A virus was detected by the hemagglutination assay. When no influenza A virus was detected upon the initial virus isolation attempt, the supernatant was passaged once more in MDCK cells.
Viruses were titrated by endpoint dilution in MDCK cells, as described previously (8). In short, 10-fold serial dilutions were used to inoculate MDCK cells. Three days after inoculation, the supernatants of the infected cell cultures were tested by hemagglutination assay as an indicator of infection. Infectious titers were calculated from five replicates by the Spearman method of Karber (19).
RESULTS
Analyses of wild bird surveillance.
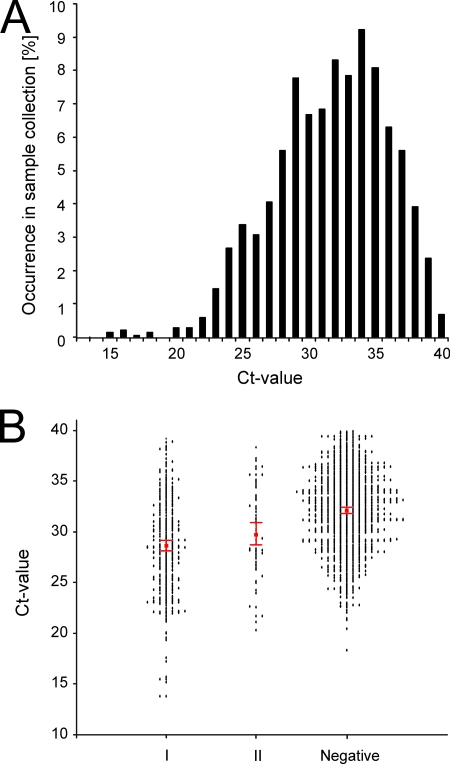
From a total of 39,490 samples collected from wild birds from 1998 to 2006, 1,483 influenza A virus-positive samples (3.8%) were detected by the M RRT-PCR. The use of the M RRT-PCR allowed the analysis of the distribution of the CT values among these samples (Fig. 1A). The CT value is the first real-time amplification cycle in which target gene amplification is detectable, and a small CT value indicates a high number of virus genome copies and thus virus particles in the sample, whereas a large CT value indicates a small amount of virus. The cutoff for negative samples was set at a CT value of 40 on the basis of the findings for multiple amplification curves. For the wild bird samples, the CT values ranged from 15 to 40, and a CT value of 34 had the highest rate of occurrence (9.4%).
FIG. 1.
CT values and virus isolation. (A) Distribution of CT values for 1,483 M RRT-PCR-positive samples obtained from 39,490 wild migratory birds during wild bird surveillance studies. (B) Correlation between CT value and virus isolation results. Group I, all samples from which an influenza A virus was isolated during the first virus isolation attempt; group II, samples from which an influenza A virus was isolated only after blind passaging; negative, no influenza A virus was isolated after two isolation attempts. Isolation attempts were performed with embryonated hens' eggs. Each dot represents the CT value for an individual bird sample. The 95% CI is represented by the red error bars for each of the three groups.
Virus isolation.
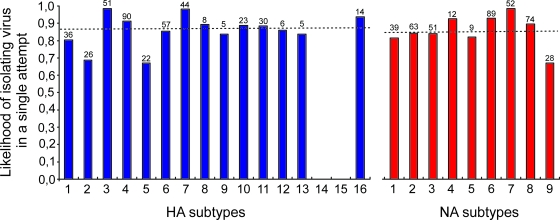
The correlation between the CT value of a positive sample and the ability to isolate virus in embryonated hens' eggs was determined. Three different outcomes of the virus isolation attempts were possible: virus isolation in the first attempt, virus isolation upon blind passage, and no virus isolation after two attempts. Statistically significant differences in CT values were observed between samples that were positive upon the first isolation attempt (mean CT value, 28.5; 95% confidence interval [CI], 28.0 to 29.0), samples positive only upon the second isolation attempt (mean CT value, 29.8; 95% CI, 28.7 to 30.9), and samples that remained negative after two isolation attempts (mean CT value, 32.1; 95% CI, 31.8 to 32.3), indicating that there was a correlation between the viral RNA copy numbers and the ability to isolate virus from samples (Fig. 1B). In total, 482 influenza A virus isolates were obtained from 1,483 M RRT-PCR-positive samples (32.5%). A total of 417 of the avian influenza A viruses were obtained after the first attempt to isolate virus (87%), whereas the remaining 65 (13%) were obtained only after blind passage. There were only marginal differences between the percentages of the HA and the NA subtypes of the virus isolates obtained after the first and the second virus isolation attempts. Of the HA and NA subtypes with the lowest likelihood of being isolated in the first attempt (H2, H5, N9), >65% were still obtained during the first isolation attempt (Fig. 2).
FIG. 2.
Likelihood of positive results during the first virus isolation attempt for 482 influenza A virus isolates obtained during wild bird surveillance studies. Blue bars, HA subtypes 1 to 16 (of note, HA subtypes 14 and 15 were not detected in this study); dotted line above the blue bars, the median (0.870) of the likelihood of positive results during the first virus isolation attempt for HA; red bars, NA subtypes 1 to 9; dotted line above the red bars, median (0.840) of the likelihood of positive results during the first virus isolation attempt for NA. The numbers above the bars indicate the numbers of viruses with a particular HA or NA subtype that were isolated during the first attempt.
Detection of H5 subtype of influenza A virus in original samples.
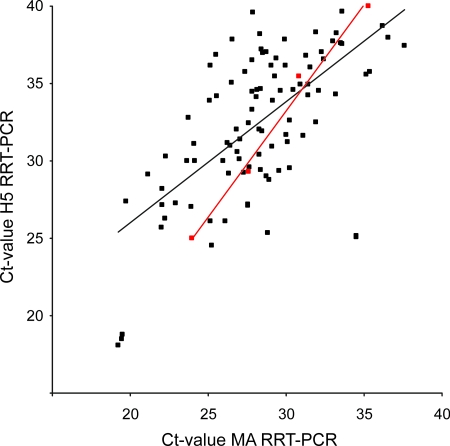
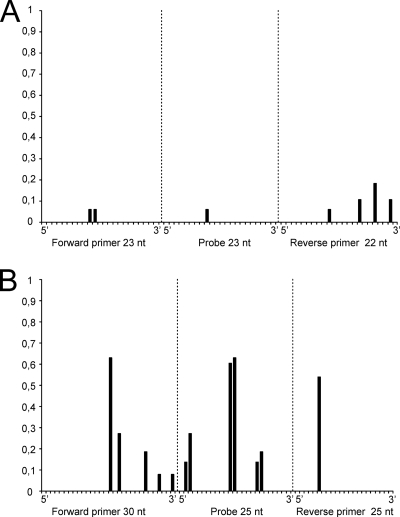
From 2005 onwards, the wild bird surveillance was performed in real time in order to serve as an early-warning system for the potential introduction of H5N1 HPAI virus in wild birds. M RRT-PCR analyses were performed within 1 week from the time that the samples were obtained from the birds, and positive samples were immediately analyzed by H5 RRT-PCR. H5 RRT-PCR-positive samples were further characterized by performing sequence analysis with the original sample without prior virus isolation. In 2005, 38 of 663 influenza A virus-positive samples (5.7%) were of the H5 subtype (determined by the H5 RRT-PCR); in 2006, 44 of 469 (9.4%) were of the H5 subtype; and in 2007, 3 of 503 (0.6%) were of the H5 subtype. Although virus could be isolated in embryonated hens' eggs from only 25% of the H5 subtype-positive samples, all 85 H5 influenza A virus-positive samples could be characterized by sequencing of the region of HA spanning the cleavage site. All samples positive for the H5 subtype were of the low-pathogenic genotype. Comparison of the H5 RRT-PCR CT values with the M RRT-PCR CT values revealed that the latter were about 4 CT value units lower (mean CT value for M RRT-PCR, 28.19 [95% CI, 27.39 to 29.00]; mean CT value for H5 RRT-PCR, 32.42 [95% CI, 31.48 to 33.37]). Comparison of the CT values obtained by the M RRT-PCR and the H5 RRT-PCR for serially diluted A/Mallard/Netherlands/3/99 (H5N2) virus also showed that the M RRT-PCR was more sensitive, but only by approximately 2.5 CT value units (Fig. 3). This suggests that the H5 RRT-PCR primers were better matched for influenza virus A/Mallard/Netherlands/3/99 than for some of the field strains. Therefore, the variability of the target sequences for each of the M RRT-PCR and H5 RRT-PCR primers and probes was analyzed by using sequences obtained from the influenza A virus isolates during the surveillance studies. Entropy plots generated from 87 M gene sequences revealed that the target sequences of the primers and probes were highly conserved, whereas for 64 H5 LPAI virus sequences, much more variation in the target sequences of the primers and probe used for the H5 RRT-PCR was observed (Fig. 4).
FIG. 3.
Correlation between M RRT-PCR CT values and H5 RRT-PCR CT values. Black symbols, samples from the wild bird surveillance studies; red symbols, samples from a serially diluted virus stock of A/Mallard/Netherlands/3/99 (H5N2). Regression lines are shown for each series.
FIG. 4.
Entropy plots for oligonucleotide-annealing sites of the primers and probes of the M and H5 RRT-PCR assays. Nucleotide (nt) sequences from influenza A viruses isolated during the wild bird surveillance studies were aligned, and entropy was calculated for each nucleotide position of each oligonucleotide. Oligonucleotide positions are given in the 5′ to 3′ direction, with position 1 being the extreme 5′ nucleotide. (A) Analysis of 87 M gene sequences with primer RF1073, probe RF1293, and primer RF1074, shown from left to right, respectively. (B) Analysis of 64 H5 LPAI virus gene sequences with primer RF1151, probe RF1153, and primer RF1152, shown from left to right, respectively. The degree of heterogeneity (entropy) was defined as described elsewhere (33, 34).
Cloacal and pharyngeal excretion of avian influenza viruses.
Experimental studies with the H5N1 HPAI virus have shown that these viruses are excreted predominantly via the respiratory tract rather than via the intestinal tract (3, 20, 35). To increase the chance of detection of H5N1 HPAI viruses in wild birds, the sampling efforts in many surveillance studies have changed from cloacal sampling to cloacal plus oropharyngeal sampling. In the present study, we had 1,964 paired cloacal and oropharyngeal swab specimens obtained from migratory mallards (Anas platyrhynchos). The rate of influenza A virus detection in the cloacal samples was significantly higher than the rate of detection in the oropharyngeal samples: 8.0% (158 of 1,964) in cloacal samples and 3.7% (73 of 1964) in oropharyngeal samples (McNemar's test, P < 0.001). The majority of birds with positive oropharyngeal swabs also had positive cloacal swabs (56%). The rate of virus detection in the cloaca and the pharynx combined (9.6%) was higher than the rate of detection of virus in samples obtained from the cloaca only. The CT values for the samples obtained from the cloaca (mean CT value, 32.5; 95% CI, 31.8 to 33.1) were significantly lower than for those obtained from the respiratory tract (mean CT value, 35.5; 95% CI, 34.9 to 36.1; one-way analysis of variance, P < 0.001). Virus could be isolated from only 2 of 73 (2.7%) of the M RRT-PCR-positive samples obtained from the pharynx and from 19 of 158 (12%) of the M RRT-PCR-positive samples obtained from the cloaca in a single virus isolation attempt.
Virus stability under different storage conditions.
The effect of different cold-chain conditions on the virus isolation rate was determined for the samples obtained from the wild bird surveillance studies. M RRT-PCR-positive samples obtained from mallards that had CT values ranging from 25 to 35 and that were stored at either 4°C (207 samples), −20°C (262 samples), or −80°C (222 samples) until analysis were selected. Among the M RRT-PCR-positive samples, virus could be isolated from 39% of the samples stored at −80°C, 20% of the samples stored at −20°C, and 17% of the samples stored at 4°C. A significant difference was observed between the ability to isolate virus and the CT value for each of the three storage temperatures (Table 1).
TABLE 1.
Relation between different storage temperature conditions and the ability to isolate influenza A viruses from M RRT-PCR-positive samples
| Temp (°C) | No. of samples positive by RRT-PCR | No. (%) of samples from which virus was isolated | Mean CT value (95% CI)
|
P valuea | |
|---|---|---|---|---|---|
| Negative samples | Positive samples | ||||
| −80 | 222 | 87 (39.2) | 31.8 (31.4-32.3) | 29.6 (29.0-30.2) | <0.001 |
| −20 | 262 | 53 (20.2) | 29.8 (29.4-30.2) | 28.7 (28.0-29.4) | 0.019 |
| +4 | 207 | 36 (17.4) | 31.5 (31.1-31.9) | 29.9 (29.0-30.8) | 0.001 |
P value of the difference between the mean CT values for M RRT-PCR-positive samples that were positive or negative upon virus isolation attempts for three different storage temperatures (one-way analysis of variance).
The effect of different cold-chain conditions was also assessed experimentally. Tenfold serial dilutions of A/Mallard/Netherlands/10/00 (H1N1), A/Mallard/Netherlands/3/99 (H5N2), and A/Mallard/Netherlands/12/00 (H7N3) were made in transport medium containing feces and 10-fold serial dilutions of A/Mallard/Netherlands/3/99 (H5N2) were made in transport media without feces, and the samples were stored under different temperature conditions. The integrity of the RNA, as determined by the CT value, was maintained for up to 3 weeks in samples stored at 4°C and up to 3 months in samples stored at −20°C and −80°C (Fig. 5A). Analyses could not be completed for samples stored at 4°C from 4 weeks onwards due to the outgrowth of bacteria and fungi. No differences in RNA stability were observed between samples with and without duck feces, and no differences were observed between the three influenza A virus isolates tested (data not shown). Virus was isolated from all samples independently of the virus dilution, the storage temperature, and the duration of storage. However, a blind passage was required for virus isolation from the samples stored for 21 days at 4°C.
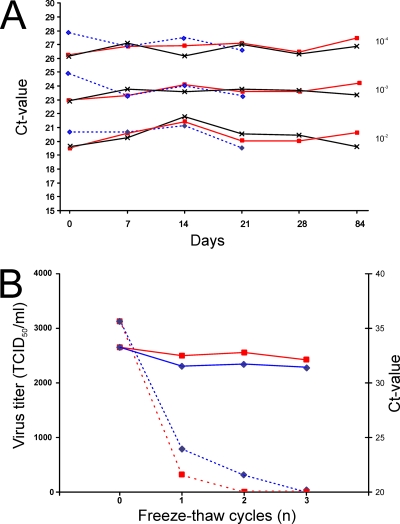
FIG. 5.
Virus stability under different storage conditions. (A) Stability of influenza virus A/Mallard/Netherlands/3/99 (H5N2) stored at different temperatures. Three 10-fold serial dilutions of virus were tested. Blue dotted lines (⋄), samples stored at 4°C; red lines (□), samples stored at −20°C; and black lines (×), samples stored at −80°C. (B) Impact of multiple freeze-thaw cycles on virus viability and integrity of the RNA of influenza virus A/Mallard/Netherlands/3/99 (H5N2). Blue and red lines, samples stored at −80°C and −20°C, respectively; solid and dotted lines, CT values (right axis) and virus titers (left axis), respectively. TCID50, 50% tissue culture infective dose.
In addition, the effect of freeze-thaw cycles on virus viability and RNA integrity was determined. Whereas the CT value of all samples remained relatively stable regardless of the number of freeze-thaw cycles, a decrease in the virus titers was observed with each freeze-thaw cycle (Fig. 5B). The decrease in the virus titer was more pronounced in samples with low virus titers (data not shown). A single freeze-thaw cycle of samples stored at −80°C resulted in an ∼4-fold decrease in virus titers, and a freeze-thaw cycle performed at −20°C resulted in an ∼10-fold decrease in virus titers.
Comparison of virus isolation techniques.
In order to compare virus isolation in cell lines with virus isolation in embryonated hens' eggs, the ability of avian influenza A viruses to replicate in a variety of cell lines, CCL-169, CCL-141, MDCK, DEF, and CEF cells, was tested. Three serially diluted avian influenza A viruses of different subtypes were used for inoculation of the different cell lines, and the highest dilution that resulted in influenza A virus detection was recorded as an indicator of the relative sensitivities of these cells for the detection of avian influenza A viruses. MDCK cells were the most sensitive cell line for the isolation of avian influenza A viruses; for each of the three virus subtypes, the highest dilution that yielded positive results was 10- to 100-fold higher in MDCK cells than in the other four cell types (data not shown).
Next, virus isolation in embryonated hens' eggs was compared to virus isolation in MDCK cells by the use of wild bird specimens. Twenty-five samples positive by the M RRT-PCR (CT values, 23 to 32; HA subtypes 1 to 7) and 80 negative by the M RRT-PCR samples were used. Influenza A viruses were isolated in embryonated hens' eggs for 23 of the 25 M RRT-PCR-positive samples, whereas with the MDCK cells, only 5 of 25 influenza A viruses were isolated. No influenza A viruses were obtained from the samples negative by the M RRT-PCR (data not shown). Virus isolation by inoculation of embryonated hens' eggs was thus more sensitive than virus isolation in MDCK cells and in cells of the other cell lines.
DISCUSSION
Previously, we reported the results of large-scale influenza A virus surveillance studies of wild birds (24). An evaluation of the methods used for these high-throughput influenza A surveillance studies is described here. The use of RRT-PCR allowed the quantification of the viral loads in the original samples. The CT values for influenza A virus-infected wild birds in the surveillance studies ranged from 15 to 40. There was a direct relation between the viral load in a sample and the ability to isolate virus from that sample. In general, virus isolates were obtained more frequently from samples with low CT values. A single cutoff CT value, above which virus isolation attempts were generally negative, could not be obtained. However, when a CT value of 37 was used as the cutoff, only 3.5% of the virus isolates would have been missed in these studies. For practical reasons, it may desirable to set such a cutoff CT value for virus isolation, but this would need to be done by each laboratory and for each RRT-PCR method individually.
The collection of cloacal and oropharyngeal samples from the same birds allowed the assessment of the role of the respiratory tract as the site of replication of avian influenza A viruses and as targets for surveillance studies. Numerous M RRT-PCR-positive oropharyngeal samples were obtained, indicating that LPAI viruses replicate in the respiratory tracts of wild birds, in addition to their intestinal tracts. However, the low frequency of detection of LPAI virus in oropharyngeal samples, the fact that most birds with positive oropharyngeal swabs also had positive cloacal swabs, and the significantly lower virus load in the oropharyngeal samples may suggest that the respiratory tract plays a limited role in the replication, transmission, and ecology of avian influenza A viruses. Our results are in agreement with recently published data from a wild bird surveillance study performed in Sweden (11). However, the exact contribution of LPAI virus replication in the respiratory tracts of wild birds to the transmission and ecology of avian influenza A viruses in this natural reservoir remains to be elucidated. The implementation of oropharyngeal sample collection from mallards in influenza A virus surveillance studies seems to be useful primarily for the surveillance of H5N1 HPAI viruses (3, 13, 20), while the added value for the surveillance of LPAI viruses appears to be limited.
Comparison of the classical method of virus isolation in embryonated hens' eggs with virus detection by molecular diagnostic approaches indicated that the actual prevalence of influenza A virus is underestimated by the use of virus isolation methods. Even with samples stored and transported under optimal conditions (−80°C) for a minimal period of time, virus could be isolated from only 40% of the influenza A virus-positive samples. No differences in the ability to isolate influenza A viruses of different HA and NA subtypes in embryonated hens' eggs were observed. Surveillance studies that rely on virus isolation as the method of choice therefore do not appear to be biased toward the detection of particular avian influenza A virus subtypes that are more easily isolated in eggs. Virus isolation by inoculation of embryonated hens' eggs was more sensitive than virus isolation in five different cell lines and thus remains the best choice for the collection of as many virus isolates as possible. When the CT values for influenza A viruses diluted in transport medium and those diluted in transport medium containing duck feces were compared, no differences were observed. This indicates that RRT-PCR inhibitors in feces did not appear to affect the ability to detect influenza A virus-positive samples by RRT-PCR.
The detection of influenza A virus-positive samples by RRT-PCR was insensitive to differences in storage conditions and freeze-thaw cycles. This is likely explained by the fact that the detection of influenza A viruses by RRT-PCR is based on relatively small regions of the virus genome (∼100 nucleotides for both the M RRT-PCR and the H5 RRT-PCR), limiting the impact of the partial degradation of viral RNA on detection by RRT-PCR. Whereas we did not observe large differences in the rates of detection of influenza A virus by RRT-PCR in samples stored under different cold-chain conditions, there was an effect of different storage conditions on the ability to isolate virus from the RRT-PCR-positive samples. In general, the rate of success of virus isolation from RRT-PCR-positive samples stored at −80°C was twice as high that from samples stored either at −20°C or at 4°C.
Comparison of the CT values obtained by the M RRT-PCR and the H5 RRT-PCR revealed that the M RRT-PCR detected influenza viruses three to four PCR amplification cycles earlier. Under ideal PCR conditions, 3 CT value units corresponds to an ∼10-fold differences in genome copy numbers. We suspect that this difference in sensitivity between the two RRT-PCR assays was due to the heterogeneity of the H5 gene target sequences. The H5 RRT-PCR was specifically designed for the detection of the various LPAI virus and H5 HPAI virus lineages that are currently circulating, but the variability within the different H5 strains is quite high and is much higher than the variation in the M gene. Reinspection of publicly available LPAI virus and H5 HPAI virus sequences indicated that it is virtually impossible to obtain a higher degree of conservation across all H5 virus HA sequences. Therefore, for more critical studies, the use of two or more H5 RRT-PCR tests may be desirable to limit the chance of missing positive samples.
The initial detection of H5 influenza A viruses in these studies was based solely on the analyses of the original sample, without prior culturing. By using the M RRT-PCR, a subsequent H5 RRT-PCR, and the final characterization of H5-positive samples by sequencing the region spanning the cleavage site of the H5 gene segment, all 87 H5-positive samples were successfully characterized as LPAI virus. The major advantages of the implementation of such molecular tests were that high-level biocontainment conditions were not required for the majority of the work and that the analysis times were reduced. The speed of sample processing is crucial if surveillance networks are used as early-warning and risk assessment systems for the threat posed by H5N1 HPAI viruses from migratory birds (25). On average, H5 RRT-PCR-positive samples were fully characterized within 3 days, and this processing time could be reduced further if needed. The characterization of viruses by the use of classical virus isolation methods would certainly take considerably more time. In addition, if the analyses for the H5 subtype had been performed by virus isolation methods, 75% of the H5 viruses would not have been detected, due to an inability to isolate the virus in the first place.
A variety of cold-chain solutions suit to fit particular regions or purposes could be implemented without a loss of sensitivity for the detection of avian influenza A viruses by RRT-PCR. Expedition-like surveillance programs currently performed in remote locations in Africa, Alaska, or Siberia are hampered by the limited availability of dry shippers, liquid nitrogen, dry ice, and cryopacks. By using the more easily maintainable cold chains at 4°C or −20°C, the samples would still be sufficiently preserved to characterize H5 viruses without the need for prior culturing. Such a change to a more easily maintainable cold chain would be recommendable over methods such as preservation of the samples in ethanol (37), due to the ability to isolate viruses from at least a proportion of the samples. Although the focus of some of the surveillance efforts is solely to detect H5N1 HPAI viruses in wild birds, it would be a missed opportunity if the complete characterization of influenza A virus-positive samples were not attempted. The availability of virus isolates is essential for the further characterization of both H5N1 HPAI and LPAI viruses, for the assessment of the biological properties of these viruses, and for the development of vaccines and diagnostics. Whereas most surveillance studies focus on communicating the results of the studies, limited attention has been given to analyses of the methodologies used. Without critical assessment of the methodologies used within these surveillance studies, detailed comparison of the results will be difficult. The lessons learned from the current large-scale influenza A virus surveillance activities could also be valuable for application to other pathogen surveillance programs in the future.
Acknowledgments
We thank the following ornithologists and their organizations for their continuous support of the wild bird influenza A surveillance program: J. Berkhouwer, T. Boudewijn, L. Bruinzeel, D. Buehler, F. Cottaar, J. de Boer, T. de Vaal, C. Dove, B. Ebbinge, T. Eggenhuizen, C. Eising, G. Escudero, T. Groothuis, B. Herrman, J. Hooijmeijer, H. Horn, B. Hoye, M. Inia, D. Jonkers, J. Jukema, N. Kajiwara, G. Keijl, M. Klaassen, H. Koffeiberg, D. Kuiken, F. Leighton, J. Leyer, M. Loonen, E. Maassen, F. Majoor, L. Mendes, G. Muskens, J. Nienhuis, Björn Olsen, K. Oosterbeek, U. Ottosson, B. Pellegrom, T. Piersma, K. Polderdijk, J. Slijkerman, J. Smit, M. Stervander, T. Talsma, P. Tavernier, H. Vader, M. van Eerden, A. van Loon, E. van Oort, T. Veen, F. Velkers, C. Verbeek, S. Verhulst, B. Voslamber, J. Waldenström, and H. Zantinge and the Alterra Wageningen, the Eendenkooi Stichting, NIOZ, the Ottenby Bird Observatory, Sovon, the University of Groningen, Wetlands International, vogelringstation Amsterdamse Waterleiding duinen, ringstation Castricum, ringstation Schiermonnikoog, and Zuid-Hollands Landschap. We thank Monique Spronken, Ger van der Water, Robin Huisman, and Chantal Burghoorn-Maas for their excellent technical assistance.
This work was sponsored by grants from the Dutch Ministry of Agriculture, the EU DG Sanco monitoring program, EU Framework six program NewFluBird (044490), the NWO Birdhealth program, and contract NIAIDNIH HHSN266200700010C.
Footnotes
Published ahead of print on 24 December 2008.
REFERENCES
- 1.Alexander, D. J. 2000. A review of avian influenza in different bird species. Vet. Microbiol. 743-13. [DOI] [PubMed] [Google Scholar]
- 2.Bragstad, K., P. H. Jorgensen, K. Handberg, A. S. Hammer, S. Kabell, and A. Fomsgaard. 2007. First introduction of highly pathogenic H5N1 avian influenza A viruses in wild and domestic birds in Denmark, Northern Europe. Virol. J. 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, J. D., D. E. Stallknecht, J. R. Beck, D. L. Suarez, and D. E. Swayne. 2006. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg. Infect. Dis. 121663-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, H., Y. Li, Z. Li, J. Shi, K. Shinya, G. Deng, Q. Qi, G. Tian, S. Fan, H. Zhao, Y. Sun, and Y. Kawaoka. 2006. Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J. Virol. 805976-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, H., G. J. Smith, S. Y. Zhang, K. Qin, J. Wang, K. S. Li, R. G. Webster, J. S. Peiris, and Y. Guan. 2005. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436191-192. [DOI] [PubMed] [Google Scholar]
- 6.Chen, H. X., H. G. Shen, X. L. Li, J. Y. Zhou, Y. Q. Hou, J. Q. Guo, and J. Q. Hu. 2006. Seroprevalance and identification of influenza A virus infection from migratory wild waterfowl in China (2004-2005). J. Vet. Med. B Infect. Dis. Vet. Public Health 53166-170. [DOI] [PubMed] [Google Scholar]
- 7.de Jong, J. C., E. C. Claas, A. D. Osterhaus, R. G. Webster, and W. L. Lim. 1997. A pandemic warning? Nature 389554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Wit, E., M. I. Spronken, T. M. Bestebroer, G. F. Rimmelzwaan, A. D. Osterhaus, and R. A. Fouchier. 2004. Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments. Virus Res. 103155-161. [DOI] [PubMed] [Google Scholar]
- 9.Ducatez, M. F., Z. Tarnagda, M. C. Tahita, A. Sow, S. de Landtsheer, B. Z. Londt, I. H. Brown, D. M. Osterhaus, R. A. Fouchier, J. B. Ouedraogo, and C. P. Muller. 2007. Genetic characterization of HPAI (H5N1) viruses from poultry and wild vultures, Burkina Faso. Emerg. Infect. Dis. 13611-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis, T. M., R. B. Bousfield, L. A. Bissett, K. C. Dyrting, G. S. Luk, S. T. Tsim, K. Sturm-Ramirez, R. G. Webster, Y. Guan, and J. S. Malik Peiris. 2004. Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wild birds in Hong Kong in late 2002. Avian Pathol. 33492-505. [DOI] [PubMed] [Google Scholar]
- 11.Ellström, P., N. Latorre-Margalef, P. Griekspoor, J. Waldenström, J. Olofsson, J. Wahlgren, and B. Olsen. 2008. Sampling for low-pathogenic avian influenza A virus in wild Mallard ducks: oropharyngeal versus cloacal swabbing. Vaccine 264414-4416. [DOI] [PubMed] [Google Scholar]
- 12.Fouchier, R. A., V. Munster, A. Wallensten, T. M. Bestebroer, S. Herfst, D. Smith, G. F. Rimmelzwaan, B. Olsen, and A. D. Osterhaus. 2005. Characterization of a novel influenza a virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 792814-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier, R. A. M., V. J. Munster, J. Keawcharoen, A. D. M. E. Osterhaus, and T. Kuiken. 2007. Virology of avian influenza in relation to wild birds. J. Wildl. Dis. 43S7-S14. [Google Scholar]
- 14.Gaidet, N., T. Dodman, A. Caron, G. Balanca, S. Desvaux, F. Goutard, G. Cattoli, F. Lamarque, W. Hagemeijer, and F. Monicat. 2007. Avian influenza viruses in water birds, Africa. Emerg. Infect. Dis. 13626-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gall-Recule, G. L., F. X. Briand, A. Schmitz, O. Guionie, P. Massin, and V. Jestin. 2008. Double introduction of highly pathogenic H5N1 avian influenza virus into France in early 2006. Avian Pathol. 3715-23. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert, M., P. Chaitaweesub, T. Parakamawongsa, S. Premashthira, T. Tiensin, W. Kalpravidh, H. Wagner, and J. Slingenbergh. 2006. Free-grazing ducks and highly pathogenic avian influenza, Thailand. Emerg. Infect. Dis. 12227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ip, H. S., P. L. Flint, J. C. Franson, R. J. Dusek, D. V. Derksen, R. E. Gill, Jr., C. R. Ely, J. M. Pearce, R. B. Lanctot, S. M. Matsuoka, D. B. Irons, J. B. Fischer, R. M. Oates, M. R. Petersen, T. F. Fondell, D. A. Rocque, J. C. Pedersen, and T. C. Rothe. 2008. Prevalence of influenza A viruses in wild migratory birds in Alaska: patterns of variation in detection at a crossroads of intercontinental flyways. Virol. J. 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jonassen, C. M., and K. Handeland. 2007. Avian influenza virus screening in wild waterfowl in Norway, 2005. Avian Dis. 51425-428. [DOI] [PubMed] [Google Scholar]
- 19.Karber, G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Exp. Pathol. Pharmakol. 162480-483. [Google Scholar]
- 20.Keawcharoen, J., D. van Riel, G. van Amerongen, T. Bestebroer, W. E. Beyer, R. van Lavieren, A. D. Osterhaus, R. A. Fouchier, and T. Kuiken. 2008. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 14600-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krauss, S., C. A. Obert, J. Franks, D. Walker, K. Jones, P. Seiler, L. Niles, S. P. Pryor, J. C. Obenauer, C. W. Naeve, L. Widjaja, R. J. Webby, and R. G. Webster. 2007. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog. 3e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, J., H. Xiao, F. Lei, Q. Zhu, K. Qin, X. W. Zhang, X. L. Zhang, D. Zhao, G. Wang, Y. Feng, J. Ma, W. Liu, J. Wang, and G. F. Gao. 2005. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 3091206. [DOI] [PubMed] [Google Scholar]
- 23.Martin, V., L. Sims, J. Lubroth, D. Pfeiffer, J. Slingenbergh, and J. Domenech. 2006. Epidemiology and ecology of highly pathogenic avian influenza with particular emphasis on South East Asia. Dev. Biol. (Basel) 12423-36. [PubMed] [Google Scholar]
- 24.Munster, V. J., C. Baas, P. Lexmond, J. Waldenstrom, A. Wallensten, T. Fransson, G. F. Rimmelzwaan, W. E. Beyer, M. Schutten, B. Olsen, A. D. Osterhaus, and R. A. Fouchier. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munster, V. J., J. Veen, B. Olsen, R. Vogel, A. D. Osterhaus, and R. A. Fouchier. 2006. Towards improved influenza A virus surveillance in migrating birds. Vaccine 246729-6733. [DOI] [PubMed] [Google Scholar]
- 26.Munster, V. J., A. Wallensten, C. Baas, G. F. Rimmelzwaan, M. Schutten, B. Olsen, A. D. Osterhaus, and R. A. Fouchier. 2005. Mallards and highly pathogenic avian influenza ancestral viruses, northern Europe. Emerg. Infect. Dis. 111545-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy, A., J. Machova, J. Hornickova, M. Tomci, I. Nagl, B. Horyna, and I. Holko. 2007. Highly pathogenic avian influenza virus subtype H5N1 in mute swans in the Czech Republic. Vet. Microbiol. 1209-16. [DOI] [PubMed] [Google Scholar]
- 28.OIE. 2006, posting date. Update on avian influenza in animals (type H5). OIE, Paris, France. http://www.oie.int/downld/AVIAN%20INFLUENZA/A_AI-Asia.htm.
- 29.Olsen, B., V. J. Munster, A. Wallensten, J. Waldenstrom, A. D. Osterhaus, and R. A. Fouchier. 2006. Global patterns of influenza A virus in wild birds. Science 312384-388. [DOI] [PubMed] [Google Scholar]
- 30.Parmley, E. J., N. Bastien, T. F. Booth, V. Bowes, P. A. Buck, A. Breault, D. Caswell, P. Y. Daoust, J. C. Davies, S. M. Elahi, M. Fortin, F. Kibenge, R. King, Y. Li, N. North, D. Ojkic, J. Pasick, S. P. Pryor, J. Robinson, J. Rodrigue, H. Whitney, P. Zimmer, and F. A. Leighton. 2008. Wild bird influenza survey, Canada, 2005. Emerg. Infect. Dis. 1484-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Runstadler, J. A., G. M. Happ, R. D. Slemons, Z. M. Sheng, N. Gundlach, M. Petrula, D. Senne, J. Nolting, D. L. Evers, A. Modrell, H. Huson, S. Hills, T. Rothe, T. Marr, and J. K. Taubenberger. 2007. Using RRT-PCR analysis and virus isolation to determine the prevalence of avian influenza virus infections in ducks at Minto Flats State Game Refuge, Alaska, during August 2005. Arch. Virol. 1521901-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saad, M. D., L. S. Ahmed, M. A. Gamal-Eldein, M. K. Fouda, F. Khalil, S. L. Yingst, M. A. Parker, and M. R. Montevillel. 2007. Possible avian influenza (H5N1) from migratory bird, Egypt. Emerg. Infect. Dis. 131120-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 186097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon, C. E. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27379-423. [Google Scholar]
- 35.Sturm-Ramirez, K. M., D. J. Hulse-Post, E. A. Govorkova, J. Humberd, P. Seiler, P. Puthavathana, C. Buranathai, T. D. Nguyen, A. Chaisingh, H. T. Long, T. S. Naipospos, H. Chen, T. M. Ellis, Y. Guan, J. S. Peiris, and R. G. Webster. 2005. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J. Virol. 7911269-11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallensten, A., V. J. Munster, N. Latorre-Margalef, M. Brytting, J. Elmberg, R. A. Fouchier, T. Fransson, P. D. Haemig, M. Karlsson, A. Lundkvist, A. D. Osterhaus, M. Stervander, J. Waldenstrom, and O. Bjorn. 2007. Surveillance of influenza A virus in migratory waterfowl in northern Europe. Emerg. Infect. Dis. 13404-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, R., L. Soll, V. Dugan, J. Runstadler, G. Happ, R. D. Slemons, and J. K. Taubenberger. 2008. Examining the hemagglutinin subtype diversity among wild duck-origin influenza A viruses using ethanol-fixed cloacal swabs and a novel RT-PCR method. Virology 375182-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward, C. L., M. H. Dempsey, C. J. Ring, R. E. Kempson, L. Zhang, D. Gor, B. W. Snowden, and M. Tisdale. 2004. Design and performance testing of quantitative real time PCR assays for influenza A and B viral load measurement. J. Clin. Virol. 29179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.WHO. 2008. Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO. WHO, Geneva, Switzerland. http://www.who.int/csr/disease/avian_influenza/country/cases_table_2008_03_05/en/index.html.
- 41.WHO. 2004, posting date. WHO manual on animal influenza diagnosis and surveillance. WHO, Geneva, Switzerland. http://www.who.int/csr/resources/publications/influenza/WHO_CDS_CSR_NCS_2002_5/en/index.html.