Abstract
We established a real-time PCR assay with melting curve analysis to rapidly genotype quinolone resistance-determining regions (QRDRs) of gyrase A and topoisomerase IV genes in Haemophilus influenzae. This assay is a useful tool for the detection of fluoroquinolone resistance and for the early detection of preexisting QRDR mutations.
Haemophilus influenzae is a major causative pathogen isolated from infections, including acute and chronic respiratory infections, acute otitis media, sinusitis, and meningitis in pediatric patients. Recent reports have noted the prevalence of fluoroquinolone (FQ)-resistant H. influenzae (1, 3, 4, 9, 11, 14, 20, 22, 26). FQ-resistant H. influenzae often carries mutations in the quinolone resistance-determining regions (QRDRs) of the gyrA and the parC genes, which encode subunits of DNA gyrase and topoisomerase IV, respectively (9, 11, 16, 18, 23). A combination of real-time PCR methods and melting curve analysis (PCR-MCA) is a useful tool for the rapid detection of key gene mutations associated with drug resistance in various microorganisms (13, 24, 25), but there are no reports about H. influenzae. The aim of this study was to develop a PCR-MCA method for detecting H. influenzae strains by targeting a total of four QRDR positions in the gyrA (codons 84 and 88) and the parC (codons 84 and 88) genes that are frequently associated with FQ resistance (9, 11, 16, 23). This current method could simultaneously identify the gyrA and the parC mutations by using only one PCR performance.
Seventeen H. influenzae clinical isolates were used. Ten of the strains were susceptible to FQ, and seven of the strains had low susceptibility or were resistant to FQ. The seven FQ-resistant/low-susceptibility strains consisted of one strain (NUH-1) from Nagasaki University Hospital, one strain (BY-1) from Bayer (Osaka, Japan), two strains (DR-1 and DS-2) from Daiichi-Sankyo (Tokyo, Japan), and three strains (MSC24060, MSC27995, and MSC11438) kindly provided by Meiji-Seika Kaisha (Tokyo, Japan) (21). The 10 FQ-susceptible strains were isolated from patients at Nagasaki University Hospital. Identification of H. influenzae was confirmed by colony morphology, Gram staining, growth on chocolate agar, and the X and V factor requirements. The MICs of ciprofloxacin (CPFX), sparfloxacin, levofloxacin (LVFX), gatifloxacin, moxifloxacin, garenoxacin, and sitafloxacin were determined by a broth dilution method using Haemophilus test medium according to the recommendations of the Clinical and Laboratory Standards Institute (5). H. influenzae ATCC 51907 was used for quality control.
DNA was extracted from each strain by using a QIAamp DNA mini-kit (Qiagen, Hilden, Germany). Sequences of the oligonucleotides and probes are shown in Tables 1 and 2. The sequences are from the known sequences of the parC and gyrA genes, which were derived from GenBank accession no. NP439678 and NP439419, respectively. To identify mutations in the QRDRs of gyrA and parC in these strains, we performed PCR and direct DNA sequencing according to the method described by Vila et al. (26).
TABLE 1.
Characteristics of specific primers used for sequencing gyrA and parC
| Target gene | Primer | Sequence | Nucleotide positions | Amplicon size (bp) |
|---|---|---|---|---|
| gyrA | Forward | 5′-TGGATCGCGAAGGCAATAC-3′ | 158-176 | 197 |
| Reverse | 5′-TGGCGCATCACCATCAAT-3′ | 354-337 | ||
| parC | Forward | 5′-GTATTGTATATGCGATGTCTGAAC-3′ | 143-166 | |
| Reverse | 5′-CCATCTACAAGTGGATAACGA-3′ | 317-297 | 175 |
TABLE 2.
Characteristics of specific probes used to detect amino acid mutations in QRDRs
| Target | Probe | Sequencea |
|---|---|---|
| Codon 84 of GyrA | Anchor | 5′AGCGCGTGTTGTGGGTGATGTAATCGGTAAATATCAC3′-FITC |
| Codon 84 of GyrA | Sensor | 5′-LCRed610-CGCATGGTGACTCCGC-3′-P |
| Codon 88 of GyrA | Anchor | 5′CCATCAACCAACATATAGCGAAGTGAGAAGGGTTGT3′-FITC |
| Codon 88 of GyrA | Sensor | 5′-Cy5-CCATACGAACGATGGTATCGTACA-3′-P |
| Codon 84 of ParC | Anchor | 5′TCGGTGATGTACTCGGTAAATTCCATCCACATGGT3′-FITC |
| Codon 88 of ParC | Sensor | 5′-LCRed610-ACAGTGCTTGTTATGAAGCTAT-3′-P |
FITC, fluorescein isothiocyanate; P, phosphorylated.
Real-time PCR-MCA was performed with a total volume of 10 μl containing 2 μl of DNA template, 5 μl of LightCycler 480 Probe Master mixture (Roche Diagnostics, Basel, Switzerland), a 0.2 μM concentration of each probe, and a 0.5 μM concentration of each primer. Thermal cycling was performed with an initial hold for 5 min at 95°C, followed by 30 cycles of 10 s at 95°C, 10 s at 58°C, and 12 s at 72°C. A melting curve was generated by cooling the reaction mixture to 35°C for 10 s, followed by heating it to 90°C at a rate of 0.2°C/s. The PCR-MCA was performed by using LightCycler 480 Basic software (Roche Diagnostics, Basel, Switzerland). The total assay time was approximately 1 h. The resulting QRDR DNA sequences were compared with the sequence of strain Rd (GenBank accession no. NC000907), which was used as the wild-type standard strain.
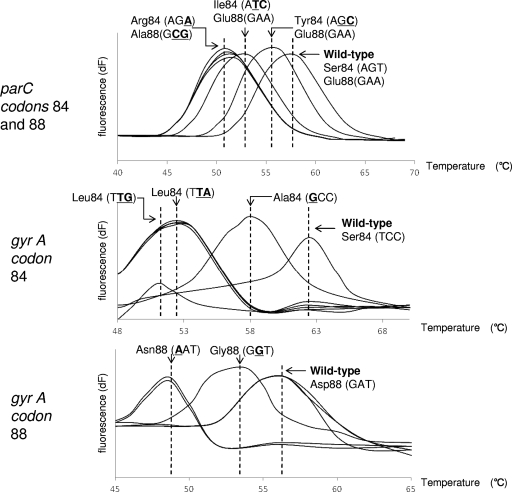
Using specific probes for the wild-type strain, we showed that all of the mutant strains had characteristic melting peaks with distinct melting temperature (Tm) values, as shown in Fig. 1. The minimum Tm shifts for mutant strains compared to that of the wild-type strain were 6.13°C for parC codons 84 and 88, 11.58°C for gyrA codon 84, and 7.9°C for gyrA codon 88. The PCR-MCA correctly detected seven LVFX low-susceptibility/resistant strains, as determined by a comparison with sequencing results (Table 3). From the sequencing results, all seven strains with low susceptibility/resistance to FQ had at least two single-amino-acid substitutions at four QRDR positions (Table 4). All LVFX-susceptible H. influenzae strains had the same Tm values as that of the wild-type strain, and sequencing results confirmed that these were in fact wild-type strains (data not shown).
FIG. 1.
Melting peak patterns for QRDR mutants with substitutions in parC and gyrA. MCA was performed with the 175-bp amplicon of the parC gene and the 197-bp amplicon of the gyrA gene. Top, middle, and bottom panels show melting peak patterns for codons 84 and 88 of the parC gene, codon 84 of the gyrA gene, and codon 88 of the gyrA gene, respectively. Each value on the y axis reveals the ratio of the first negative derivative of the change in fluorescence (dF) to the variation in temperature.
TABLE 3.
Seven LVFX low-susceptibility/resistant strains detected by PCR-MCA
| Strain | MIC (μg/ml) to LVFX | Susceptibility at QRDRsa
|
||||||
|---|---|---|---|---|---|---|---|---|
|
parC codon 84
|
parC codon 88
|
gyrA codon 84
|
gyrA codon 88
|
|||||
| Tm (°C) | Sequence/aa | Sequence/aa | Tm (°C) | Sequence/aa | Tm (°C) | Sequence/aa | ||
| Rd (wild type) | 0.003 | 56.86 | AGT/Ser | GAA/Glu | 62.44 | TCC/Ser | 56.49 | GAT/Asp |
| DS-1 | 8 | 55.05 | AGC/Thr | WT | 52.04 | TTA/Leu | 53.81 | GGT/Gly |
| NUH-1 | 16 | 50.73 | AGA/Arg | GCG/Ala | 52.04 | TTA/Leu | 53.81 | AAT/Asn |
| BY-1 | 8 | 50.73 | AGA/Arg | WT | 50.86 | TTG/Leu | 48.59 | AAT/Asn |
| MS24060 | 1 | 50.73 | AGA/Arg | GCG/Ala | 57.93 | GCC/Ala | 56.49 | WT |
| MS27995 | 1 | 52.11 | ATC/Ile | WT | 52.04 | TTA/Leu | 56.49 | WT |
| MS11438 | 4 | 50.73 | AGA/Arg | GCG/Ala | 52.04 | TTA/Leu | 48.59 | TAT/Tyr |
| DS-2 | 4 | 50.82 | AGA/Arg | GAT/Asp | 53.24 | TAC/Thr | 49.70 | TAT/Tyr |
aa, amino acid. WT, wild type.
TABLE 4.
Relationship between FQ susceptibility, amino acid changes in gyrA and parC, and results of PCR-MCA
| Strain | MCAa | Amino acid change encoded by:
|
MIC (μg/ml) ofb:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| gyrA | parC | LVFX | SPFX | GFLX | MFLX | CPFX | GRNX | STFX | ||
| Rd | WT | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.0015 | 0.0006 | ||
| DS-1 | MT | Ser84→Leu | Gly82→Asp | 8 | 4 | 2 | 4 | 4 | 4 | 0.25 |
| Asp88→Gly | ||||||||||
| NUH-1 | MT | Ser84→Leu | Ser84→Arg | 16 | 16 | 8 | 16 | 16 | 8 | 1 |
| Asp88→Asn | Glu88→Ala | |||||||||
| BY-1 | MT | Ser84→Leu | Ser84→Arg | 8 | 8 | 8 | 32 | 16 | >32 | 2 |
| Asp88→Asn | Gly82→Asp | |||||||||
| MSC24060 | MT | Ser84→Ala | Ser84→Arg | 1 | 1 | 0.5 | 2 | 2 | 0.5 | 0.12 |
| Glu88→Ala | ||||||||||
| MSC27995 | MT | Ser84→Leu | Ser84→Ile | 1 | 1 | 0.5 | 0.5 | 1 | 1 | 0.03 |
| MSC11438 | MT | Ser84→Leu | Ser84→Arg | 4 | 4 | 4 | 8 | 4 | 8 | 0.25 |
| Asp88→Tyr | Glu88→Ala | |||||||||
| DS-2 | MT | Ser84→Leu | Ser84→Arg | 4 | 8 | 2 | 2 | 8 | 4 | 0.12 |
| Asp88→Tyr | Glu88→Ala | |||||||||
Mutants (MT) were defined as the strains with a Tm value shift compared to that of the wild type (WT), detected by using H. influenza Rd.
SPFX, sparfloxacin; GTFX, gatifloxacin; MXFX, moxifloxacin; GRNX, garenoxacin; STFX, sitafloxacin.
We compared the ability of the present PCR-MCA to detect FQ susceptibility in 17 H. influenzae strains with that of the conventional phenotypic method. All LVFX-susceptible strains that had no mutation in codons 84 and 88 of gyrA and parC were classified as susceptible according to Clinical and Laboratory Standards Institute criteria (6) (data not shown). As shown in Table 4, the mutation profiles for the QRDRs in the gyrA and parC genes revealed a close relationship between the MIC level and the number of QRDR mutations. Previous studies have found that the conventional phenotypic method failed to detect strains that have a single QRDR mutation; these strains have the potential to develop into a highly resistant pathogen (8). Several reports have noted that a significant number of Streptococcus pneumoniae isolates already have a single-step mutation and are prone to acquiring a second-step mutation (19). Unfortunately, our study lacks a collection of single mutant strains. It was reported that the selection window for CPFX with wild-type cells was below serum drug concentrations in human volunteers receiving twice-daily doses of 500 mg (10). However, Odoul et al. reported that the median area under the inhibitory curve was decreased by about half the proposed target value for CPFX in a cystic fibrosis patient receiving an oral regimen of 15 mg of CPFX/kg of body weight twice a day (15). Increasing the times that the drug concentrations fall in the mutant selection window causes the mutation (7). Actually, the previously reported strains that failed treatment had a double mutation in the gyrA and the parC genes (2). Furthermore, Pérez-Vázquez et al. reported that hypermutability is a risk condition for the development of FQ-resistant H. influenzae infection (17), and Li et al. reported that stepwise selection induced high-level resistance in H. influenzae (12). We emphasize the clinical importance of the detection of first-step QRDR mutations in either gyrA or parC for attempting to predict a strain's evolution into FQ resistance. In addition, we should consider the FQ dosage carefully to avoid the low FQ concentration when we treat patients who have chronic lung disease.
In conclusion, the PCR-MCA was easily and quickly performed and had an accuracy that was at least as satisfactory as that of the conventional phenotypic method. FQ-resistant H. influenzae is expected to become a more important pathogen in the future, because FQ is the most effective antibiotic against H. influenzae infection, and the number of FQ-resistant strains may rise further, along with the recent increase in FQ prescription. Although additional studies are needed, we anticipate that the PCR-MCA used in this study may be a useful tool for surveillance studies in the screening of FQ resistance as an alternative to DNA nucleotide sequencing, because this PCR-MCA can recognize the gyrA and the parC mutations more clearly, easily, and rapidly than sequencing.
Acknowledgments
No conflicts of interest and no funding sources are reported for this study.
Footnotes
Published ahead of print on 7 January 2009.
REFERENCES
- 1.Barriere, S. L., and J. Hindler. 1993. Ciprofloxacin resistant Haemophilus influenzae in a patient with chronic lung disease. Ann. Pharmacother. 27309-310. [DOI] [PubMed] [Google Scholar]
- 2.Bastida, T., M. Perez-Vazquez, J. Campos, M. C. Cortés-Lletget, F. Román, F. Tubau, A. G. de la Campa, and C. Alonso-Tarrés. 2003. Levofloxacin treatment failure in Haemophilus influenzae pneumonia. Emerg. Infect. Dis. 91475-1478. [DOI] [PubMed] [Google Scholar]
- 3.Biedenbach, D. J., and R. N. Jones. 2003. Five-year analysis of Haemophilus influenzae isolates with reduced susceptibility to fluoroquinolones: prevalence results from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 4655-61. [DOI] [PubMed] [Google Scholar]
- 4.Campos, J., F. Román, M. Georgiou, C. García, R. Gómez-Lus, R. Cantón, H. Escobar, and F. Baquero. 1996. Long-term persistence of ciprofloxacin-resistant Haemophilus influenzae in patients with cystic fibrosis. J. Infect. Dis. 1741345-1347. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M07-A7, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing, 17th ed. Approved standard M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Drlica, K. 2003. The mutant selection window and antimicrobial resistance. J. Antimicrob. Chemother. 5211-17. [DOI] [PubMed] [Google Scholar]
- 8.Fukushima, K. Y., Y. Hirakata, K. Sugahara, K. Yanagihara, A. Kondo, S. Kohno, and S. Kamihira. 2006. Rapid screening of topoisomerase gene mutations by a novel melting curve analysis method for early warning of fluoroquinolone-resistant Streptococcus pneumoniae emergence. J. Clin. Microbiol. 444553-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgiou, M., R. Munoz, F. Roman, R. Canton, R. Gomez-Lus, J. Campos, and A. G. De La Campa. 1996. Ciprofloxacin-resistant Haemophilus influenzae strains possess mutations in analogous positions of GyrA and ParC. Antimicrob. Agents Chemother. 401741-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez, M. A., A. H. Moranchel, S. Duran, A. Pichardo, J. L. Magana, B. Painter, A. Forrest, and G. L. Drusano. 1985. Multiple-dose pharmacokinetics of ciprofloxacin administered intravenously to normal volunteers. Antimicrob. Agents Chemother. 28235-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, X., N. Mariano, J. J. Rahal, C. M. Urban, and K. Drlica. 2004. Quinolone-resistant Haemophilus influenzae in a long-term-care facility: nucleotide sequence characterization of alterations in the genes encoding DNA gyrase and DNA topoisomerase IV. Antimicrob. Agents Chemother. 483570-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, X., N. Mariano, J. J. Rahal, C. M. Urban, and K. Drlica.2004. Quinolone-resistant Haemophilus influenzae: determination of mutant selection window for ciprofloxacin, garenoxacin, levofloxacin, and moxifloxacin. Antimicrob. Agents Chemother. 2004;484460-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motoshima, M., K. Yanagihara, K. Fukushima, J. Matsuda, K. Sugahara, Y. Hirakata, Y. Yamada, S. Kohno, and S. Kamihira. 2007. Rapid and accurate detection of Pseudomonas aeruginosa by real-time polymerase chain reaction with melting curve analysis targeting gyrB gene. Diagn. Microbiol. Infect. Dis. 5853-58. [DOI] [PubMed] [Google Scholar]
- 14.Nazir, J., C. Urban, N. Mariano, J. Burns, B. Tommasulo, C. Rosenberg, S. Segal-Maurer, and J. J. Rahal. 2004. Quinolone-resistant Haemophilus influenzae in a long-term care facility: clinical and molecular epidemiology. Clin. Infect. Dis. 381564-1569. [DOI] [PubMed] [Google Scholar]
- 15.Odoul, F., L. C. Guellec, C. Giraut, D. C. Gialluly, S. Marchand, G. Paintaud, M. C. Saux, J. C. Rolland, and E. Autret-Leca. 2001. Ciprofloxacin pharmacokinetics in young cystic fibrosis patients after repeated oral doses. Therapie 56519-524. [PubMed] [Google Scholar]
- 16.Pérez-Vázquez, M., F. Román, B. Aracil, R. Cantón, and J. Campos. 2004. Laboratory detection of Haemophilus influenzae with decreased susceptibility to nalidixic acid, ciprofloxacin, levofloxacin, and moxifloxacin due to gyrA and parC mutations. J. Clin. Microbiol. 421185-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pérez-Vázquez, M., F. Román, S. García-Cobos, and J. Campos. 2007. Fluoroquinolone resistance in Haemophilus influenzae is associated with hypermutability. Antimicrob. Agents Chemother. 511566-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pérez-Vázquez, M., F. Román, M. C. Varela, R. Cantón, and J. Campos. 2003. Activities of thirteen quinolones by three susceptibility testing methods against a collection of Haemophilus influenzae isolates with different levels of susceptibility to ciprofloxacin: evidence for cross-resistance. J. Antimicrob. Chemother. 51147-151. [DOI] [PubMed] [Google Scholar]
- 19.Richter, S. S., K. P. Heilmann, S. E. Beekman, N. J. Miller, C. L. Rice, and G. V. Doern. 2005. The molecular epidemiology of Streptococcus pneumoniae with quinolone resistance mutations. Clin. Infect. Dis. 40225-235. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez-Martínez, J. M., L. López, I. García, and A. Pascual. 2006. Characterization of a clinical isolate of Haemophilus influenzae with a high level of fluoroquinolone resistance. J. Antimicrob. Chemother. 57577-578. [DOI] [PubMed] [Google Scholar]
- 21.Sanbongi, Y., T. Suzuki, Y. Osaki, N. Senju, T. Ida, and K. Ubukata. 2006. Molecular evolution of β-lactam-resistant Haemophilus influenzae: 9-year surveillance of penicillin-binding protein 3 mutations in isolates from Japan. Antimicrob. Agents Chemother. 502487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokota, S., Y. Ohkoshi, K. Sato, and N. Fujii. 2008. Emergence of fluoroquinolone-resistant Haemophilus influenzae strains among elderly patients but not among children. J. Clin. Microbiol. 46361-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshizumi, S., Y. Takahashi, Y. Watanabe, E. Okezaki, Y. Ishii, and K. Tateda. 2004. In vitro antibacterial activities of new fluoroquinolones against clinical isolates of Haemophilus influenzae with ciprofloxacin-resistance-associated alterations in GyrA and ParC. Chemotherapy 50265-275. [DOI] [PubMed] [Google Scholar]
- 24.Vacher, S., A. Menard, E. Bernard, A. Santos, and F. Megraud. 2005. Detection of mutations associated with macrolide resistance in thermophilic Campylobacter spp. by real-time PCR. Microb. Drug Resist. 1140-47. [DOI] [PubMed] [Google Scholar]
- 25.Vernel-Pauillac, F., V. Falcot, D. Whiley, and F. Merien. 2006. Rapid detection of a chromosomally mediated penicillin resistance-associated ponA mutation in Neisseria gonorrhoeae using a real-time PCR assay. FEMS Microbiol. Lett. 25566-74. [DOI] [PubMed] [Google Scholar]
- 26.Vila, J., J. Ruiz, F. Sánchez, F. Navarro, B. Mirelis, M. T. Jiménez de Anta, and G. Prats. 1999. Increase in quinolone resistance in Haemophilus influenzae strain isolated from a patient with recurrent respiratory infections treated with ofloxacin. Antimicrob. Agents Chemother. 43161-162. [DOI] [PMC free article] [PubMed] [Google Scholar]