Abstract
The first U.S. multicenter clinical trial to assess the performance of the Cepheid Xpert MRSA assay (Xpert MRSA) was conducted. The assay is a qualitative test designed for the rapid detection of methicillin-resistant Staphylococcus aureus (MRSA) directly from nares swabs. This novel test combines integrated nucleic acid extraction and automated real-time PCR for the detection of a MRSA-specific signature sequence. A total of 1,077 nares specimens were collected from seven geographically distinct health care sites across the United States with prevalence rates ranging from 5.2% to 44%. Nares specimens were tested by (i) the Xpert MRSA assay, (ii) direct culture on CHROMagar MRSA medium (direct CM culture), and (iii) broth-enriched culture (Trypticase soy broth with 6.5% sodium chloride) followed by plating onto CHROMagar MRSA medium (broth-enriched CM culture). When direct CM culture was designated the reference method, the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the Xpert MRSA assay were 94.3%, 93.2%, 73.0%, and 98.8%, respectively. When broth-enriched CM culture was used as the reference method, the clinical sensitivity, specificity, PPV, and NPV of the Xpert MRSA assay were 86.3%, 94.9%, 80.5%, and 96.6%, respectively. The BD GeneOhm MRSA (BDGO) assay was performed as a comparative molecular method. No statistical performance differences were observed between the Xpert MRSA and BDGO assays when they were compared to culture methods. From this large-scale, multicenter clinical comparison, we conclude that the Xpert MRSA assay is a simple, rapid, and accurate method for performing active surveillance for MRSA in a variety of health care populations.
The increasing prevalence and spread of methicillin-resistant Staphylococcus aureus (MRSA) is a worldwide pandemic (31). The National Healthcare Safety Network (NHSN) estimates that hospitalized patients in the United States acquire 2 million health care-associated infections (HAIs) each year, causing 90,000 deaths and $4.5 billion in excess health care costs; a large percentage of HAIs are known to be caused by MRSA (12). Infections are commonly associated with poor patient outcomes, elevated morbidity and mortality, and negative economic impacts; deaths occur both in hospitalized patients and in otherwise healthy individuals (6, 22, 25). For these reasons, tremendous amounts of effort and resources have recently been focused on the development of several new rapid screening tests for MRSA.
MRSA is harbored primarily in human nares, on skin, and on mucosal surfaces of the vagina and rectum. Colonized patients (carriers) have a high likelihood of developing and transmitting infections (10, 21, 25). Therefore, routine infection prevention practices, such as active surveillance programs that screen nares for MRSA, have been recommended and established by many accredited health care facilities in North America and Europe (5, 7, 20, 27, 28, 33). Most active surveillance programs rely on rapid detection methods, such as chromogenic agar (4, 9, 14, 32) or real-time PCR (2, 7, 11, 15, 19, 23, 29), to support their efforts, because delays in the identification of MRSA carriers offer an opportunity for MRSA to be transferred to other patients. Recently published results have shown that such MRSA surveillance programs can be combined with other infection prevention practices to significantly reduce the rates of HAIs caused by MRSA (28, 33), but questions remain about the comparative clinical performance and utility of the MRSA screening methods.
Most traditional MRSA screening methods focus on the phenotypic identification of the mecA gene, the structural gene responsible for methicillin resistance via production of an altered penicillin binding protein, PBP2a, which maintains staphylococcal cell wall integrity because of its low affinity to β-lactam antibiotics. The gene is present within a transposon-encoded genetic region known as the staphylococcal cassette chromosome mec (SCCmec).
The BD GeneOhm (BDGO) MRSA assay, first introduced to clinical laboratories as the IDI-MRSA assay, focuses on the detection of a unique junction fragment created by integration of the mecA-containing transposon into a single unique site (orfX), an open reading frame within the S. aureus genome (18), rather than on the detection of the mecA gene itself. The Xpert MRSA assay is a novel second-generation real-time PCR assay for the detection of the SCCmec-orfX junction (Xpert MRSA assay package insert; Cepheid) and is currently the most rapid of all the commercial methods. It is an easy-to-use test that, by virtue of its moderate-complexity CLIA (Clinical Laboratory Improvement Act) categorization, can be performed on-demand by virtually any laboratory technologist or technician. However, its clinical performance against culture-based methods, and against other FDA-cleared high-complexity molecular diagnostic tests, required assessment.
This study is the first comprehensive multicenter U.S. evaluation of the Xpert MRSA assay. It is the first study to assess assay performance stratified by prevalence and by health care setting. It is the first study to report assay performance by operators with limited molecular training and the first to report performance compared with two gold-standard methods, broth enrichment and another FDA-cleared MRSA PCR. The study closes gaps in the published literature by providing an extensive clinical comparison of commercial methods for MRSA detection for subjects and strains found in the United States.
MATERIALS AND METHODS
Analytical performance of the Xpert MRSA assay. (i) Sensitivity.
The analytical sensitivity of the Xpert MRSA assay was determined using six strains of MRSA representing the six SCCmec types and subtypes (I, II, III, IV, IVa, and V). Cultures of these strains were quantified by hemocytometer counting and then diluted to densities spanning a range of 10 to 1,000 CFU per swab. All dilutions were tested in replicates of four. The limit of detection (LoD) was determined for each type or subtype tested. The lowest density at which all four replicates were reported positive was designated the LoD-100%. In addition, studies using MRSA SCCmec type II cells were performed to determine the 95% confidence interval for the analytical LoD of this assay. The LoD was defined as the lowest number of MRSA CFU per swab that could be reproducibly distinguished from negative samples with 95% confidence.
(ii) Specificity.
A total of 95 Staphylococcus strains were tested using the Cepheid Xpert MRSA assay. All strains were tested in triplicate at densities of ≥1 × 106 CFU per swab. Cultures were obtained from the American Type Culture Collection and the Network on Antimicrobial Resistance in S. aureus. MRSA strains representing the six SCCmec types and subtypes (I, II, III, IV, IVa, and V) were included in testing. A total of 51 strains, representing species phylogenetically related to S. aureus, were tested. Other members of the nasal commensal flora, including 32 strains of methicillin-sensitive, coagulase-negative staphylococci and 12 strains of methicillin-resistant, coagulase-negative staphylococci, were also tested.
(iii) Reproducibility/intersite agreement.
Reproducibility (intersite agreement) was assessed using a panel of specimens with varying densities of MRSA and methicillin-sensitive Staphylococcus epidermidis (MSSE). Specimens were tested in triplicate on 10 different days at each of three different sites (4 specimens × 3 times/day × 10 days × 3 sites) by individuals with limited laboratory experience. One lot of the Xpert MRSA kit was used at each of the three testing sites.
Clinical experimental design.
The performance characteristics of the Xpert MRSA assay were determined in a multisite prospective study performed at seven health care organizations, with populations comprising patients at tertiary-care, extended-stay, and outpatient facilities. Nares swab specimens were tested by the Xpert MRSA assay, and the results were compared to those of direct culture on CHROMagar MRSA medium (direct CM culture), (Becton Dickinson, Sparks, MD) and a more sensitive reference method, broth-enriched culture followed by culture on CHROMagar MRSA medium (broth-enriched CM culture). In order to avoid the pitfalls of traditional discrepant analysis (17), samples were also tested by a third method, the FDA-cleared BDGO MRSA assay (Becton Dickinson, San Diego, CA).
Human subjects were enrolled in accordance to federal policy governing human subject protection and federal medical privacy standards (8). Nares specimens were collected from geographically distinct areas of the United States, including the East, Northeast, Midwest, Southwest, and West. Subjects included individuals from inpatient tertiary-care hospitals, nursing homes, and transplantation units, including health care staff. Each subject was enrolled in the study only once. Subjects who had received systemic or topical nasal antibiotics 48 h to 1 week prior to study enrollment, those under the age of 2 years, and those who had contraindications to nares swab collection were excluded from the study.
Four nares swabs, consisting of two double swabs (liquid Stuart Copan swabs, part 900-0370; Cepheid), were collected from persons in the populations listed in Table 2. After collection, the two double swabs were rubbed together to ensure the homogeneity of the samples. For one swab pair, one swab was tested by the Xpert MRSA assay within 24 h and the other swab was tested by the BDGO MRSA assay within 24 h. For the second swab pair, one swab was routed to a centralized laboratory for culture testing and the other was retained in case repeat testing was necessary.
TABLE 2.
Demographic description of Xpert MRSA assay results derived from different study populations
| Subject population tested | No. (%) of samples that were:
|
Total no. of individuals (% of total population)a | |
|---|---|---|---|
| Positive | Negative | ||
| Residents at nursing homes (long-term and extended-stay facilities) | 62 (25.5) | 181 (74.5) | 243 (22.6) |
| Patients hospitalized >3 days | 61 (23.0) | 204 (77.0) | 265 (24.7) |
| Patients hospitalized ≤3 days | 29 (13.1) | 193 (86.9) | 222 (20.7) |
| Patients at outpatient clinic | 46 (17.7) | 214 (82.3) | 260 (24.2) |
| Staff and others | 11 (12.9) | 74 (87.1) | 85 (7.9) |
| Total | 209 (19.4) | 866 (80.6) | 1,075a |
The total subject group (n = 1,077) includes two culture-positive hospitalized subjects whose admission dates were unknown.
Molecular methods.
Both the Xpert MRSA and BDGO MRSA molecular assays were performed according to the manufacturer's instructions at each participating institution. For the Xpert MRSA assay, a commercial lyophilized external control (Kwik-Stik MRSA, catalog no. 0158; MicroBioLogics, Inc., St. Cloud, MN) was used as the positive control; MSSE (catalog no. 0371; MicroBioLogics, Inc.) was used as the negative control. External controls were performed daily during the clinical study. The cutoff for a positive sample was established at a threshold cycle (CT) of 36.
Within-cartridge controls included reagents for the simultaneous detection of the target MRSA DNA and a sample-processing control (SPC), which serves both to monitor for appropriate sample processing and PCR conditions and to monitor for the presence of substances that can inhibit PCR. In this design, the SPC does not affect the sensitivity of the target sequence, because the concentration of the internal control is adjusted such that in the presence of inhibitors or MRSA DNA, the SPC will be repressed. A second control mechanism within the cartridge, the probe check control, verified reagent rehydration, PCR tube filling in the cartridge, hydrolysis probe integrity, and dye stability.
Culture methods.
Direct cultures were performed on site at each of the seven study sites. In addition, a swab was sent to a centralized laboratory for a reference culture. The central laboratory was blind to the on-site results. Specimen swabs were transported to the centralized laboratory at ambient temperatures and were either tested upon receipt or stored at a refrigerated temperature of 2 to 8°C for as long as 72 h. Swabs were directly streaked on to CHROMagar MRSA medium, and in addition, the swab was then transferred to Trypticase soy broth with 6.5% sodium chloride (Becton Dickinson, Sparks, MD) and incubated for 18 to 24 h at 35 ± 2°C (broth-enriched CM culture). If the direct CM culture was negative at 24 h, the enriched Trypticase soy broth was streaked onto another CM plate and incubated for 24 to 48 h at 35 to 37°C. For all cultures, presumptively positive mauve colonies from either culture method were confirmed to be MRSA when the isolate was tube coagulase positive and exhibited typical staphylococcal Gram stain morphology.
Statistical analysis.
Descriptive statistical performance characteristics were calculated for the Xpert MRSA assay relative to direct-CM and broth-enriched-CM culture results, performed at the central laboratory (reference culture), and relative to the on-site CM culture. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated according to standard formulas. A priori power analysis was performed for a power of ≥90%, and the pooled sample size was calculated for analysis with 95% confidence intervals. The criterion for significance (alpha) was established at 0.05 (two-tailed).
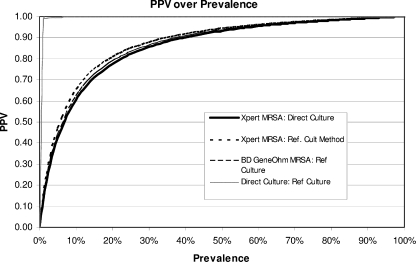
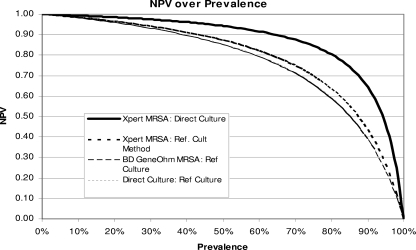
Since the PPV and NPV of each assay will change as the prevalence changes, comparisons of performance data by study site can be misleading if MRSA prevalence is not considered. Therefore, site-specific data for sensitivity and specificity were matched with corresponding prevalence data to create graphical representations of the predictive capacities of all test methods compared to that of the reference culture method.
For Fig. 1 and 2, the following equations and the “smooth-curve” function in Excel 2003 were used to create a “y = −1/x” type curve for each test method:
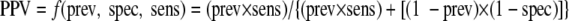 |
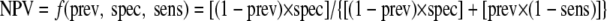 |
where prev is the prevalence at a particular site, sens is the sensitivity of each assay per site, spec is the specificity for each assay per site, and f is a constant (16).
FIG. 1.
Graphical representation of changes in the PPV with changes in prevalence. The PPVs for four MRSA detection methods at various prevalences are shown.
FIG. 2.
Graphical representation of changes in the NPV with changes in prevalence. The NPVs for four MRSA detection methods at various prevalences are shown.
RESULTS
Analytical performance.
Analytical performance was characterized with regard to sensitivity, specificity, and intersite agreement. For sensitivity, the Xpert MRSA assay yielded positive results for 100% of the six SCCmec types and subtypes (I, II, III, IV, IVa, and V). The calculated analytical specificity of the Xpert MRSA assay was 100%, since none of the 95 specimens with non-MRSA isolates tested were reported as positive for MRSA by the assay.
For MRSA SCCmec type II cells, the assay sporadically identified samples as positive at bacterial densities as low as 10 CFU/swab, while the LoD results indicate that the Xpert MRSA assay will produce a positive result with 95% confidence for a swab containing 80 CFU. Assay linearity was achieved throughout the range of 68 CFU/swab to 6.8 × 107 CFU/swab (4 replicates for each density), with corresponding CT values from 14.7 to 34.0.
Intersite agreement testing confirmed a high concordance of results across study sites on multiple testing days, as depicted in Table 1. For negative results, agreement between sites ranged from 96.8 to 100%; the only source of discordant results was clerical error. For weakly and moderately positive specimens, total agreement ranged from 97.8 to 100%, with clerical error accounting for discordant results. For strongly positive samples, total agreement was 100% across all sites. The overall total agreement across three sites was 99.2%.
TABLE 1.
Summary of intersite agreement for the Xpert MRSA assay
| Specimen | MRSA density (CFU/swab)a | Reproducibilityb at:
|
Total agreement across all sitesb | ||
|---|---|---|---|---|---|
| Site 1 | Site 2 | Site 3 | |||
| Negative | 0 | 30/30 | 30/30 | 30/31c | 90/91 (98.9) |
| Weakly positive | 117 | 30/30 | 30/30 | 27/29c | 87/89 (97.8) |
| Positive | 800 | 30/30 | 30/30 | 30/30 | 90/90 (100.0) |
| Strongly positive | 2.6 × 104 | 30/30 | 30/30 | 30/30 | 90/90 (100.0) |
| Total | 120/120 (100.0) | 120/120 (100.0) | 117/120 (97.5)d | 357/360 (99.2)d | |
In each specimen, the density of MSSE was 2.6 × 106 CFU/swab.
Expressed as the number of times the specimen was correctly identified as positive or negative/number of times the specimen was tested (percentage).
An Xpert MRSA assay was inadvertently performed on one additional negative specimen and one less weakly positive specimen at site 3.
Correction of clerical errors at site 3 would change the agreement to 100%.
Clinical performance.
A total of 1,077 unique specimens were collected from eligible subjects enrolled at the seven clinical study sites. The numbers and percentages of positive and negative cases, as determined by the reference culture method (broth-enriched CM culture), were calculated and are presented in Table 2 by the substrata of the study population. Among this population, 22.6% were residents at nursing homes (long-term and extended-stay facilities), 22.4% were patients hospitalized >3 days, 20.7% were patients hospitalized ≤3 days, 24.2% were outpatients, and 7.9% were staff and other subjects. Estimates of MRSA prevalence, derived from subject sampling at the seven study sites, ranged from 5.2 to 44.4%.
The performance characteristics of the Xpert MRSA assay (Table 3) were as follows. Compared to the direct CM culture method, the sensitivity (agreement with positive results), specificity (negative agreement), PPV, and NPV of the Xpert MRSA assay were 94.3%, 93.2%, 73.0%, and 98.8%, respectively. Relative to broth-enriched CM culture, sensitivity was 86.3%, specificity was 94.9%, the PPV was 80.5%, and the NPV was 96.6%. For 5 to 10% of the specimens tested, the Xpert MRSA assay results were positive despite a culture-negative status. Table 3 also compares Xpert MRSA assay results to the results of broth-enriched and direct CM cultures. In comparison to direct CM culture, sensitivity ranged from 81.3 to 100%, specificity from 81.8% to 100%, the PPV from 60 to 100%, and the NPV from 93.8 to 100%. Compared to broth-enriched CM culture, sensitivity ranged from 80 to 100%, specificity from 93.2 to 100%, the PPV from 75 to 100%, and the NPV from 94 to 100%. Three of 1,077 specimens did not give results by the Xpert MRSA assay after two attempts (0.3%); because their inclusion did not statistically alter the final result of the study, only samples for which results were obtained by all methods are used in Table 3.
TABLE 3.
Performance of the Xpert MRSA assay by site and prevalence compared to the performance of the combined reference culture method and of direct culture
| Site | MRSA prevalence (% [no. of positive samples/total samples])b | Result (% [95% CI])a for the Xpert MRSA assay by comparison to the:
|
No. of indeterminate results | |||
|---|---|---|---|---|---|---|
| Direct method
|
Reference method
|
|||||
| Sensitivity | Specificity | Sensitivity | Specificity | |||
| Site 1 | 20.2 (78/387) | 95.4 (87.1-99.0) | 92.2 (88.8-94.9) | 87.2 (77.7-93.7) | 93.9 (90.6-96.3) | 10 |
| Site 2 | 5.2 (3/58) | 100.0 (29.2-100.0) | 98.2 (90.3-100.0) | 100.0 (29.2-100.0) | 98.2 (90.3-100.0) | 3 |
| Site 3 | 44.4 (12/27) | 91.7 (61.5.-99.8) | 100.0 (78.2.-100.0) | 91.7 (61.5.-99.8) | 100.0 (78.2.-100.0) | 3 |
| Site 4 | 12.3 (20/162) | 81.3 (54.4-96.0) | 95.2 (90.4-98.1) | 80.0 (56.3-94.3) | 97.2 (92.9-99.2) | 10 |
| Site 5 | 20.5 (46/224) | 94.9 (82.7-99.4) | 93.0 (88.3-96.2) | 89.1 (76.4-96.4) | 94.9 (90.6-97.7) | 1 |
| Site 6 | 22.3 (42/188) | 97.1 (84.7-99.9) | 92.9 (87.6-96.4) | 81.0 (65.9-91.4) | 93.2 (87.8-96.7) | 6 |
| Site 7 | 35.7 (10/28) | 100.0 (54.1-100.0) | 81.8 (59.7-94.8) | 90.0 (55.5-99.8) | 94.4 (72.7-99.9) | 2 |
| Total | 19.6 (211/1,077)b | 94.3 (89.7-97.2) | 93.2 (91.4-94.8) | 86.3 (80.9-90.6) | 94.9 (93.2-96.3) | 35 |
CI, confidence interval; direct method, direct CM culture; reference method, broth-enriched CM culture.
The total row includes three samples for which Xpert MRSA results could not be determined after two attempts. For performance measures, only those samples with results for all methods could be calculated (n = 1,074). Removal of three samples did not statistically alter the final results.
The performance of the Xpert MRSA assay in comparison with other methods is reported in Table 4. The performances of the Xpert MRSA assay, the BDGO MRSA assay, and direct CM culture for each site, relative to broth-enriched CM culture, are presented. Although the Xpert MRSA assay appeared to be the most sensitive, the results were not statistically different from those of the BDGO MRSA assay or direct CM culture when summary data were compared by 95% confidence intervals. The overall site-specific clinical sensitivity of the Xpert MRSA assay ranged from 81% to 100%; site-specific specificity ranged from 93.2 to 100%.
TABLE 4.
Performance of the Xpert MRSA assay, the BDGO MRSA assay, and the direct CM culture method compared to the broth-enriched CM culture reference method
| Site | % Agreement (95% CI)a with the reference method
|
|||||
|---|---|---|---|---|---|---|
| Positive
|
Negative
|
|||||
| Xpert MRSA assay | BDGO MRSA assay | Direct CM culture | Xpert MRSA | BDGO MRSA assay | Direct CM culture | |
| 1 | 87.2 (77.7-93.7) | 80.8 (70.3-88.8) | 83.3 (73.2-90.8) | 93.9 (90.6-96.3) | 92.2 (88.7-95.0) | 100.0 (98.8-100.0) |
| 2 | 100.0 (29.2-100.0) | 100.0 (29.2-100.0) | 100.0 (29.2-100.0) | 98.2 (90.3-100.0) | 98.2 (90.3-100.0) | 100.0 (93.6-100.0) |
| 3 | 91.7 (61.5.-99.8) | 83.3 (51.6-97.9) | 100.0 (73.5-100.0) | 100.0 (78.2.-100.0) | 100.0 (79.4-100.0) | 100.0 (79.4-100.0) |
| 4 | 80.0 (56.3-94.3) | 78.9 (54.4-93.9) | 80.0 (56.3-94.3) | 97.2 (92.9-99.2) | 97.9 (93.9-99.6) | 100.0 (97.5-100.0) |
| 5 | 89.1 (76.4-96.4) | 89.1 (76.4-96.4) | 84.8 (71.1-93.7) | 94.9 (90.6-97.7) | 93.8 (89.2-96.9) | 100.0 (97.9-100.0) |
| 6 | 81.0 (65.9-91.4) | 78.6 (63.2-89.7) | 81.0 (65.9-91.4) | 93.2 (87.8-96.7) | 94.5 (89.5-97.6) | 100.0 (97.5-100.0) |
| 7 | 90.0 (55.5-99.7) | 100.0 (69.2-100.0) | 60.0 (26.2-87.8) | 94.4 (72.7-99.9) | 94.4 (72.7-99.9) | 100.0 (81.5-100.0) |
| Total | 86.3 (80.9-90.6) | 83.3 (77.6-88.1) | 82.9 (77.2-87.8) | 94.9 (93.2-96.3) | 94.4 (92.7-95.9) | 100.0 (99.6-100.0) |
CI, confidence interval.
When site-specific data for sensitivity and specificity were combined with corresponding prevalence data to create a graphical representation of the predictive capacities of all test methods (16), a “y = −1/x” type curve was created for each test method. The relationship between prevalence and PPV is shown in Fig. 1, and that between prevalence and NPV is shown in Fig. 2, for all test methods, in comparison to the broth-enriched CM culture reference method. The graphs demonstrate how positive and negative predictive values will change based on assay-specific sensitivity, specificity, and site prevalence. As one would expect, the PPV increases with prevalence for all four methods. The Xpert MRSA assay is projected to have a higher NPV for populations with MRSA prevalences greater than 10%.
DISCUSSION
Rapid and accurate detection of MRSA carriers is increasingly important, because hospital-acquired infections, often due to antibiotic-resistant strains, have been associated with increased morbidity and mortality, as well as prolonged and more costly hospital stays (6, 21, 22, 24, 30). The challenges of implementing active surveillance programs to reduce infections are substantial. With those challenges in mind, a multisite clinical trial was conducted to assess the clinical performance of the Cepheid Xpert MRSA assay, the most rapid qualitative test available for the detection of MRSA directly from nares swabs.
Several studies have shown that PCR provides a sensitive method for identifying MRSA carrier status (15, 19, 26, 29) and have established PCR as an “improved gold standard” compared to direct routine culture (1). Data from the present study support this concept and demonstrate the performance of a new and totally integrated method for MRSA PCR, the Xpert MRSA assay. Of note, improved gold standard methods often have lower calculated PPVs than direct culture, due to their enhanced ability to identify microbes at low densities and produce PCR-positive but culture-negative results. In contrast, such methods exhibit very high NPVs, a performance characteristic that is suitable for laboratory “screening” methods. Consistent with the expectations of an improved gold standard method, Xpert MRSA assay results were MRSA positive, despite a culture-negative status, for 5 to 10% of specimens tested.
PCR-positive, culture-negative results could be consistent with a number of circumstances. For instance, false-negative culture results could arise due to opsonizing antibody responses to S. aureus, or due to antibiotic use. In addition, poor sampling or handling of the swabs may limit bacterial recovery in culture. Other explanations for “false”-positive results could include the presence of nonviable organisms, since the assay does not confirm the presence of live organisms. In addition, low bacterial densities in the nares can render the culture negative and the PCR positive, or vice versa, because low bacterial densities will cause both methods to produce sporadic positive or negative results under the parameters described by the statistical phenomenon known as the Poisson effect. While data describing the presence of MRSA from other body sites were not available for this study, there is a growing body of evidence to suggest that this condition occurs. For example, a recent study showed that among patients with apparently false positive PCR results in nares swabs, MRSA could be recovered by culture from other sites such as the inguinal region, which is most likely a surrogate for gastrointestinal tract colonization (29). As described by Paule et al., such results are often found in patients who have previously tested positive for MRSA or who harbor this organism at other body sites (26). In this example, the results would be considered positive from an infection control perspective.
Of particular interest, genetic excisions within the SCCmec region of MRSA strains may also yield positive PCR results in the absence of a functional mecA gene; therefore, these “empty cassette variants” (sometimes called MR-MSSA) cause PCR-positive, culture-negative results. These results occur because current commercial PCR assays target genetic regions, upstream of the mecA gene, which can remain even after excision of the mecA gene from within the cassette. A limitation of this study is that there was no assessment of the prevalence of these variants; however, the prevalence of empty cassette variants has been shown to differ by geographical region and currently appears to be more common outside the United States (10, 13, 15). Awareness and periodic monitoring for the presence of these variants is prudent. Monitoring of the mecA gene without targeting the SCCmec region is not recommended, because the mecA gene may be carried in bacterial isolates that are not S. aureus by origin.
The performance of the Xpert MRSA assay supports its utility for U.S. active surveillance programs using nares samples. The analytical sensitivity of the Xpert MRSA assay was slightly higher than that of the BDGO MRSA assay (80 CFU/swab reported here versus 325 CFU/swab, reported in the BDGO MRSA assay package insert). The results are comparable to those obtained in Europe by Rossney et al., 58 CFU/swab (29).
Under the conditions of the multicenter clinical trial, false-negative PCR results were rare (10 of 1,074 versus direct CM culture and 29 of 1,074 versus broth-enriched CM culture). Such discrepancies can occur if the microbe density is below the assay's 95% LoD or if unrecognized genotypic variation occurs. The Xpert MRSA assay achieved an exceptional NPV, 96.6%, across a broad range of MRSA prevalences. Improved NPVs are projected for the Xpert MRSA assay in populations with MRSA prevalences higher than 10%, as depicted in Fig. 2, confirming its utility as a screening method in a variety of U.S. health care settings.
Compared to broth-enriched CM culture, the clinical sensitivity, specificity, PPV, and NPV of the Xpert MRSA assay were 86.3%, 94.9%, 80.5%, and 96.6%, respectively, for nares samples from seven different performance sites with different subject populations ranging in prevalence from 12.9 to 25.5%. The performance of the assay in this study was somewhat lower than that achieved in Ireland by Rossney et al. (29), reported as 95%, 98%, 90%, and 99%, respectively, when amended for patients who were known to be positive for MRSA previously. The differences in performance can be attributed to the prevalence in the Irish population (58/204 patients [28.4%]), which could elevate the PPV, and to the sampling of multiple specimen sources in that study (nares, throat, and groin), which adds to the likelihood of acquiring a positive sample. In addition, the Rossney performance characteristics were adjusted, in their final analysis, to include patients previously known to be MRSA positive, a strategy that added to the overall agreement in their study. One could also speculate that in a tertiary-care population, bacterial colonization densities would be higher and would contribute to improved performance of the PCR assay, in contrast to this study, which included outpatients as well as inpatients; that single-site laboratory performance would be improved over that found in our multicenter performance study, which included performance by staff who were not trained in molecular methods; and that differences in detection could occur to due to the fact that the MRSA strains from the Irish population may differ from the U.S. strains.
The clinical performance of the Xpert MRSA assay was statistically equivalent to those of the BD GeneOhm MRSA assay and direct CHROMAgar MRSA culture compared to the reference method of 6.5% salt broth enrichment followed by CHROMAgar MRSA culture. Despite the fact that the targets of each assay are operationally distinct, the two PCR assays produced similar results, since both assays target SCCmec insertion sequences and regions within the orfX gene.
The Xpert MRSA assay has several operational advantages. With a PCR cycling time of ≤72 min, this assay offers a significant advantage in speed over culture methods and other PCR assays for the identification of MRSA colonization. Another advantage is the assay's ease of use. Since the Xpert MRSA method can be performed as a moderate-complexity method, a variety of health care staff can participate in testing efforts. The method is amenable to a variety of health care settings, ranging from the clinical laboratory to off-site point-of-care testing. The method is highly reproducible, even when performed by personnel with limited experience, as evidenced by a total intersite agreement of 99.2% and its rating as a “moderate-complexity” assay according to the CLIA categorization. For laboratories which are accredited by the College of American Pathologists, the format of the assay requires the testing of external controls for the verification of new lots and shipments of reagents, while daily/run control requirements are met by the assay's built-in controls, designed to ensure precision and reagent integrity.
Reagent costs are higher for the Xpert MRSA assay than for other rapid methods. However, despite the overall higher reagent cost per test, the Xpert MRSA assay offers clinical utility when rapid results are required for appropriate MRSA isolation and cohorting practices, designed to optimize the use of isolation beds. Costs can be partially offset by personnel options, which are available due to the “moderate-complexity” rating of the assay, eliminating the costs associated with highly trained laboratory staff, who are required for high-complexity assays. The limited hands-on-time for assay setup can also be factored into the cost-benefit equation. Ultimately, when selecting a laboratory screening method, each health care institution will have its own particular resources and speed requirements to consider.
We conclude that the Xpert MRSA assay is a novel, rapid, and accurate method for the determination of MRSA carrier status to support active surveillance programs. The results of this study show that the Xpert MRSA assay is reliable and reproducible for on-demand testing in health care settings where such testing is desirable.
Acknowledgments
This study was funded by Cepheid, Inc., as part of the FDA submission for IVD approval of the Xpert MRSA assay.
We thank Linda Hilbert and Patricia Richards for their support, proofreading, and editing.
Footnotes
Published ahead of print on 7 January 2009.
REFERENCES
- 1.Alonzo, T. A., and M. S. Pepe. 1999. Using a combination of reference tests to assess the accuracy of a new diagnostic test. Stat. Med. 182987-3003. [DOI] [PubMed] [Google Scholar]
- 2.Boyce, J. M., and N. L. Havill. 2008. Comparison of BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR versus the CHROMagar MRSA assay for screening patients for the presence of MRSA strains. J. Clin. Microbiol. 46350-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll, K. C. 2008. Rapid diagnostics for methicillin-resistant Staphylococcus aureus: current status. Mol. Diagn. Ther. 1215-24. [DOI] [PubMed] [Google Scholar]
- 4.Cherkaoui, A., G. Renzi, P. Francois, and J. Schrenzel. 2007. Comparison of four chromogenic media for culture-based screening of meticillin-resistant Staphylococcus aureus. J. Med. Microbiol. 56500-503. [DOI] [PubMed] [Google Scholar]
- 5.Conterno, L. O., J. Shymanski, K. Ramotar, B. Toye, C. van Walraven, D. Coyle, and V. R. Roth. 2007. Real-time polymerase chain reaction detection of methicillin-resistant Staphylococcus aureus: impact on nosocomial transmission and costs. Infect. Control Hosp. Epidemiol. 281134-1141. [DOI] [PubMed] [Google Scholar]
- 6.Cosgrove, S. E., Y. Qi, K. S. Kaye, S. Harbarth, A. W. Karchmer, and Y. Carmeli. 2005. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect. Control Hosp. Epidemiol. 26166-174. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, R., P. Jenks, J. Northwood, M. Wallis, S. Ferguson, and S. Hunt. 2007. Effect on MRSA transmission of rapid PCR testing of patients admitted to critical care. J. Hosp. Infect. 6524-28. [DOI] [PubMed] [Google Scholar]
- 8.Department of Health and Human Services. 14 August 2002. Standards for privacy of individually identifiable health information; final rule. 45 CFR Parts 160 and 164. U.S. Government Printing Office, Washington, DC.
- 9.Diederen, B., I. van Duijn, A. van Belkum, P. Willemse, P. van Keulen, and J. Kluytmans. 2005. Performance of CHROMagar MRSA medium for detection of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 431925-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnio, P. Y., F. Février, P. Bifani, M. Dehem, C. Kervégant, N. Wilhelm, A. L. Gautier-Lerestif, N. Lafforgue, M. Cormier, the MR-MSSA Study Group of the Collège de Bactériologie-Virologie-Hygiène des Hôpitaux de France, and A. Le Coustumier. 2007. Molecular and epidemiological evidence for spread of multiresistant methicillin-susceptible Staphylococcus aureus strains in hospitals. Antimicrob. Agents Chemother. 514342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drews, S. J., B. M. Willey, N. Kreiswirth, M. Wang, T. Ianes, J. Mitchell, M. Latchford, A. J. McGeer, and K. C. Katz. 2006. Verification of the IDI-MRSA assay for detecting methicillin-resistant Staphylococcus aureus in diverse specimen types in a core clinical laboratory setting. J. Clin. Microbiol. 443794-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, J. R., K. D. Peterson, M. L. Andrus, J. S. Tolson, J. S. Goulding, M. A. Dudeck, R. B. Mincey, D. A. Pollock, T. C. Horan, and the NHSN Facilities. 2007. National Healthcare Safety Network (NHSN) Report, data summary for 2006, issued June 2007. Am. J. Infect. Control 35290-301. [DOI] [PubMed] [Google Scholar]
- 13.Farley, J. E., P. D. Stamper, T. Ross, M. Cai, S. Speser, and K. C. Carroll. 2008. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR assay to culture by use of BBL CHROMagar MRSA for detection of MRSA in nasal surveillance cultures from an at-risk community population. J. Clin. Microbiol. 46743-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flayhart, D., J. F. Hindler, D. A. Bruckner, G. Hall, R. K. Shrestha, S. A. Vogel, S. S. Richter, W. Howard, R. Walther, and K. C. Carroll. 2005. Multicenter evaluation of BBL CHROMagar MRSA medium for direct detection of methicillin-resistant Staphylococcus aureus from surveillance cultures of the anterior nares. J. Clin. Microbiol. 435536-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francois, P., M. Bento, G. Renzi, S. Harbarth, D. Pittet, and J. Schrenzel. 2007. Evaluation of three molecular assays for rapid identification of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 452011-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galen, R. S., and T. Peters. 1986. Analytical goals and clinical relevance of laboratory procedures, p. 398. In N. W. Tietz (ed.), Textbook of clinical chemistry. W. B. Saunders, Philadelphia, PA.
- 17.Hadgu, A. 2000. Discrepant analysis is an inappropriate and unscientific method. J. Clin. Microbiol. 384301-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huletsky, A., R. Giroux, V. Rossbach, M. Gagnon, M. Vaillancourt, M. Bernier, F. Gagnon, K. Truchon, M. Bastien, F. J. Picard, A. van Belkum, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 421875-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huletsky, A., P. Lebel, F. J. Picard, M. Bernier, M. Gagnon, N. Boucher, and M. G. Bergeron. 2005. Identification of methicillin-resistant Staphylococcus aureus carriage in less than 1 hour during a hospital surveillance program. Clin. Infect. Dis. 40976-981. [DOI] [PubMed] [Google Scholar]
- 20.Khoury, J., M. Jones, A. Grim, W. M. Dunne, Jr., and V. Fraser. 2005. Eradication of methicillin-resistant Staphylococcus aureus from a neonatal intensive care unit by active surveillance and aggressive infection control measures. Infect. Control Hosp. Epidemiol. 26616-621. [DOI] [PubMed] [Google Scholar]
- 21.Klein, E., D. L. Smith, and R. Laxminarayan. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg. Infect. Dis. 131840-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2981763-1771. [DOI] [PubMed] [Google Scholar]
- 23.Liassine, N., F. Decosterd, and J. Etienne. 2007. Evaluation of IDI-MRSA assay on a collection of community-acquired methicillin-resistant Staphylococcus aureus isolates and on carriage specimens. Pathol. Biol. (Paris) 55378-381. (In French.) [DOI] [PubMed] [Google Scholar]
- 24.Mauldin, P. D., C. D. Salgado, V. L. Durkalski, and J. A. Bosso. 2008. Nosocomial infections due to methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococcus: relationships with antibiotic use and cost drivers. Ann. Pharmacother. 42317-326. [DOI] [PubMed] [Google Scholar]
- 25.Muto, C. A., J. A. Jernigan, B. E. Ostrowsky, H. M. Richet, W. R. Jarvis, J. M. Boyce, and B. M. Farr. 2003. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus. Infect. Control Hosp. Epidemiol. 24362-386. [DOI] [PubMed] [Google Scholar]
- 26.Paule, S. M., D. M. Hacek, B. Kufner, K. Truchon, R. B. Thomson, Jr., K. L. Kaul, A. Robicsek, and L. R. Peterson. 2007. Performance of the BD GeneOhm methicillin-resistant Staphylococcus aureus test before and during high-volume clinical use. J. Clin. Microbiol. 452993-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson, L. R., D. M. Hacek, and A. Robicsek. 2007. Case study: an MRSA intervention at Evanston Northwestern Healthcare. Jt. Comm. J. Qual. Patient Saf. 33732-738. [DOI] [PubMed] [Google Scholar]
- 28.Robicsek, A., J. L. Beaumont, S. M. Paule, D. M. Hacek, R. B. Thomson, Jr., K. L. Kaul, P. King, and L. R. Peterson. 2008. Universal surveillance for methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Ann. Intern. Med. 148409-418. [DOI] [PubMed] [Google Scholar]
- 29.Rossney, A. S., C. M. Herra, G. I. Brennan, P. M. Morgan, and B. O'Connell. 2008. Evaluation of the Xpert methicillin-resistant Staphylococcus aureus (MRSA) assay using the GeneXpert real-time PCR platform for rapid detection of MRSA from screening specimens. J. Clin. Microbiol. 463285-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shurland, S., M. Zhan, D. D. Bradham, and M. C. Roghmann. 2007. Comparison of mortality risk associated with bacteremia due to methicillin-resistant and methicillin-susceptible Staphylococcus aureus. Infect. Control Hosp. Epidemiol. 28273-279. [DOI] [PubMed] [Google Scholar]
- 31.Spellberg, B., R. Guidos, D. Gilbert, J. Bradley, H. W. Boucher, W. M. Scheld, J. G. Bartlett, and J. Edwards, Jr. 2008. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 46155-164. [DOI] [PubMed] [Google Scholar]
- 32.van Loo, I. H., S. van Dijk, I. Verbakel-Schelle, and A. G. Buiting. 2007. Evaluation of a chromogenic agar (MRSASelect) for the detection of meticillin-resistant Staphylococcus aureus with clinical samples in The Netherlands. J. Med. Microbiol. 56491-494. [DOI] [PubMed] [Google Scholar]
- 33.van Trijp, M. J., D. C. Melles, W. D. Hendriks, G. A. Parlevliet, M. Gommans, and A. Ott. 2007. Successful control of widespread methicillin-resistant Staphylococcus aureus colonization and infection in a large teaching hospital in the Netherlands. Infect. Control Hosp. Epidemiol. 28970-975. [DOI] [PubMed] [Google Scholar]
- 34.Wagenvoort, J. H., W. Sluijsmans, and R. J. Penders. 2000. Better environmental survival of outbreak vs. sporadic MRSA isolates. J. Hosp. Infect. 45231-234. [DOI] [PubMed] [Google Scholar]