Abstract
The utility of peptide nucleic acid fluorescence in situ hybridization (PNA FISH) for the detection of Acinetobacter spp. and Pseudomonas aeruginosa was evaluated on broth suspensions and spiked blood cultures of ATCC strains and clinical isolates with select gram-negative rods. After testing 60 clinical isolates, PNA FISH had a sensitivity and specificity of 100% and 100%, respectively, for Acinetobacter spp. and 100% and 95%, respectively, for P. aeruginosa. PNA FISH was able to detect both pathogens simultaneously and directly from spiked blood cultures.
Pseudomonas aeruginosa and Acinetobacter baumannii have emerged as two of the most troublesome pathogens for health care institutions globally, contributing significantly to the morbidity and mortality of hospitalized patients (8, 9). Novel, rapid diagnostics are urgently required to assist in the epidemiological control and early treatment of infection caused by these organisms (10).
Fluorescence in situ hybridization (FISH) using labeled peptide nucleic acid (PNA) probes is a methodology that has been applied for the rapid diagnosis of infectious diseases (7, 11). Due to their neutral charge, PNA probes have more-robust hybridization characteristics than those of DNA probes. They target naturally abundant rRNA molecules and, thus, allow the detection of individual microorganisms without the need for an amplification step (5). Finally, and adding to its clinical applicability, PNA FISH is less susceptible to inhibition by impurities in clinical samples and has been used effectively for the direct testing of blood, sputum, and wound cultures (2, 6, 7). Thus far, this technology has not been applied to A. baumannii and has only recently been described for P. aeruginosa (7). In this study, we assessed the effectiveness of a new, genus-specific Acinetobacter sp. PNA probe (AdvanDx, Inc., Woburn, MA) by testing a range of reference and clinical isolates. Given the similarity of infectious syndromes caused by A. baumannii and P. aeruginosa, we also assessed the ability of using the Acinetobacter sp. and P. aeruginosa probes (AdvanDx, Inc., Woburn, MA) simultaneously. Currently, both probes are for research use only.
The PNA FISH assay was performed according to published methods (11, 12). In brief, a 10-μl aliquot from an overnight culture (grown in brain heart infusion agar at 37°C) was mixed with 1 drop of fixation solution on a slide. After fixation, 1 drop of the probe solution was placed on the slide and hybridized at 55°C for 90 min (12). The slides were then washed for 30 min and were read using a fluorescence microscope with a dual fluorescein isothiocyanate/Texas Red filter. The final PNA FISH result was available in ∼2.5 h. Two independent investigators, who were blinded to the laboratory identification of the samples, examined all slides.
Initially, a group of ATCC reference strains was assessed using the genus-specific Acinetobacter sp. probe and included A. baumannii (ATCC 19606), Acinetobacter calcoaceticus (ATCC 14987), Acinetobacter haemolyticus (ATCC 19002), Acinetobacter lwoffii (ATCC 15309), Acinetobacter sp. (ATCC 49137, originally “Acinetobacter anitratus”), Escherichia coli (ATCC 35218), Klebsiella pneumoniae (ATCC 10031), P. aeruginosa (ATCC 10145), Pseudomonas fluorescens (ATCC 49838), Pseudomonas putida (ATCC 49128), and Pseudomonas stutzeri (ATCC 17588). PNA FISH testing with the Acinetobacter sp. probe correctly identified all Acinetobacter ATCC reference strains and no cross-hybridization with other ATCC reference gram-negative organisms were observed. We also assessed, in a separate experiment, the same group of ATCC strains with the previously designed P. aeruginosa probe (7). The P. aeruginosa probe correctly identified P. aeruginosa ATCC 10145 and showed no cross-hybridization with other Pseudomonas species or with other ATCC reference gram-negative organisms.
Following this initial validation, the Acinetobacter sp. and P. aeruginosa PNA probes were combined and used simultaneously to test 60 recent clinical isolates. Each probe had a different fluorescent marker as follows: green (fluorescein isothiocyanate) for Acinetobacter spp. and red (Texas Red) for P. aeruginosa. The clinical isolates included 20 Acinetobacter sp. strains, 20 P. aeruginosa strains, and 20 other strains with gram-negative rods (Table 1). All isolates were obtained from different patients and were resistant to at least three antimicrobial groups. The Acinetobacter sp. and P. aeruginosa isolates were confirmed to be of different genetic types, as determined by pulsed-field gel electrophoresis, which was performed according to previously described methods (4). Organism identification and susceptibility testing had been performed in a clinical microbiology laboratory according to CLSI standards (3). Using the multiprobe PNA FISH assay on the 60 clinical isolates individually, the sensitivity and specificity for Acinetobacter spp. were 100% and 100%, respectively, and for P. aeruginosa, they were 100% and 95%, respectively (Table 1). Routine laboratory organism identification was used as the gold standard. The two false-positive results were from Stenotrophomonas maltophilia and Escherichia coli, which both produced a weak red signal identified as P. aeruginosa. After repeat testing, these results were negative. The positive and negative predictive values were 91% and 100%, respectively, for P. aeruginosa detection. There was no interobserver variability.
TABLE 1.
Characteristics of the 60 clinical isolates tested using the Acinetobacter sp. and P. aeruginosa PNA probes simultaneously
| Isolates (no. of organisms)a | PNA FISH results
|
|
|---|---|---|
| Positive for Acinetobacter spp. (no. of organisms) | Positive for Pseudomonas aeruginosa (no. of organisms) | |
| Acinetobacter spp. (20) | 20 | 0 |
| A. baumannii-calcoaceticus complex (18) | ||
| A. lwoffii (1) | ||
| Other Acinetobacter sp. (1) | ||
| Pseudomonas aeruginosa (20) | 0 | 20 |
| Other isolates with gram-negative rods (20) | ||
| Escherichia coli (5) | 0 | 1 |
| 2 ESBL producers | ||
| Enterobacter cloacae (2) | 0 | 0 |
| Klebsiella oxytoca (2) | 0 | 0 |
| 1 ESBL producer | ||
| Klebsiella pneumoniae (5) | 0 | 0 |
| 3 ESBL producers | ||
| Morganella morganii (1) | 0 | 0 |
| Proteus mirabilis (2) | 0 | 0 |
| Stenotrophomonas maltophilia (3) | 0 | 1 |
ESBL, extended-spectrum β-lactamase.
To determine the threshold of detection for the multiprobe PNA FISH assay, serial dilutions of an overnight culture of A. baumannii (ATCC 19606) and P. aeruginosa (ATCC 10145) were tested. The bacterial densities (CFU/ml) of the dilutions were confirmed by colony counts on brain heart infusion agar. We observed that the lower limit of detection for the multiprobe PNA FISH assay was 104 CFU/ml for both Acinetobacter spp. and P. aeruginosa. Given that hospital-acquired and ventilator-associated pneumonia is the most common infectious syndrome caused by these organisms, such a threshold is clinically applicable (1).
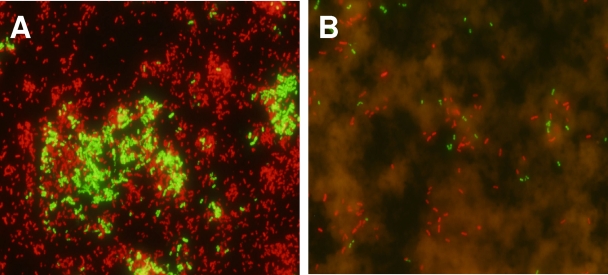
To assess whether Acinetobacter spp. and P. aeruginosa could be identified simultaneously, 10 mixed bacterial broth cultures were prepared. Each mixed culture consisted of a random selection of 3 isolates from the 60 clinical isolates described above. Each mixed culture was prepared by combining 1 ml of an overnight culture of each isolate. All mixed cultures, which were tested in a blinded fashion, were correctly identified by using the multiprobe PNA FISH assay. A total of 6 of these 10 mixed cultures contained both Acinetobacter spp. and P. aeruginosa (Fig. 1A).
FIG. 1.
FISH of a mixed culture of Acinetobacter baumannii (ATCC 19606) and Pseudomonas aeruginosa (ATCC 10145) using PNA probes specific for these organisms. Green fluorescence signifies hybridization with Acinetobacter spp., and red signifies hybridization with P. aeruginosa. A sample from an overnight broth culture (A) and a sample from a spiked blood culture bottle taken 3 h after inoculation (B) are shown, presenting evidence of both microbes and overlying red cells.
Finally, to determine whether this multiprobe PNA FISH assay could be used directly on positive blood cultures, we prepared artificially spiked BacT/Alert SA blood culture bottles (bioMerieux, Inc., Durham, NC), which contained 10 ml of seeded human blood (Research Blood Components, Brighton, MA). One colony of A. baumannii (ATCC 19606) or P. aeruginosa (ATCC 10145) from a fresh agar plate was used to inoculate two bottles each, and they were incubated aerobically at 37°C for 3 to 6 h before testing. Two further bottles were also inoculated with the two organisms combined. We observed that the multiprobe PNA FISH assay was able to detect A. baumannii and P. aeruginosa from blood culture bottles either alone or when inoculated simultaneously (Fig. 1B).
PNA FISH testing is a rapid and highly sensitive and specific method for the detection of troublesome gram-negative pathogens such as P. aeruginosa and Acinetobacter spp. Importantly, the methodology has the potential to be used on direct clinical samples, and we have demonstrated its potential for pathogen detection from blood cultures. The commonality of infection types caused by these organisms, particularly pneumonia and bloodstream infection, makes simultaneous detection clinically useful. Such rapid diagnostics have the potential to not only improve therapeutic decision making but may also help optimize infection control interventions by more rapidly identifying patient or environmental reservoirs. Before the full utility of this new technology can be determined, it is important for further clinical studies to be performed.
Acknowledgments
AdvanDx, Inc., Woburn, MA, provided supplies for this project.
Potential conflicts of interest are that Y.T. and M.J.F. were employees of AdvanDx, Inc. at the time of the study.
Footnotes
Published ahead of print on 30 December 2008.
REFERENCES
- 1.American Thoracic Society, and Infectious Diseases Society of America. 2005. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 171388-416. [DOI] [PubMed] [Google Scholar]
- 2.Brown, A. R., and J. R. Govan. 2007. Assessment of fluorescent in situ hybridization and PCR-based methods for rapid identification of Burkholderia cepacia complex organisms directly from sputum samples. J. Clin. Microbiol. 451920-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2007. M7-A7. Dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.D'Agata, E., L. Venkataraman, P. DeGirolami, and M. Samore. 1997. Molecular epidemiology of acquisition of ceftazidime-resistant gram-negative bacilli in a nonoutbreak setting. J. Clin. Microbiol. 352602-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 2431360-1363. [DOI] [PubMed] [Google Scholar]
- 6.Forrest, G. N., M.-C. Roghmann, L. S. Toombs, J. K. Johnson, E. Weekes, D. P. Lincalis, and R. A. Venezia. 2008. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob. Agents Chemother. 523558-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirketerp-Moller, K., P. O. Jensen, M. Fazli, K. G. Madsen, J. Pedersen, C. Moser, T. Tolker-Nielsen, N. Hoiby, M. Givskov, and T. Bjarnsholt. 2008. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 462717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paterson, D. L. 2006. The epidemiological profile of infections with multidrug-resistant Pseudomonas aeruginosa and Acinetobacter species. Clin. Infect. Dis. 43(Suppl. 2)S43-S48. [DOI] [PubMed] [Google Scholar]
- 9.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel, J. D., E. Rhinehart, M. Jackson, L. Chiarello, and the Healthcare Infection Control Practices Advisory Committee. 2006. Management of multidrug-resistant organisms in healthcare settings. Centers for Disease Control and Prevention, Atlanta, GA. [DOI] [PubMed]
- 11.Stender, H. 2003. PNA FISH: an intelligent stain for rapid diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 3649-655. [DOI] [PubMed] [Google Scholar]
- 12.Wilson, D. A., M. J. Joyce, L. S. Hall, L. B. Reller, G. D. Roberts, G. S. Hall, B. D. Alexander, and G. W. Procop. 2005. Multicenter evaluation of a Candida albicans peptide nucleic acid fluorescent in situ hybridization probe for characterization of yeast isolates from blood cultures. J. Clin. Microbiol. 432909-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]