Abstract
Commonly used internal controls (ICs) to monitor the efficiency of nucleic acid testing (NAT) assays do not allow verification of nucleic acid extraction efficiency. Since microbial cells are often difficult to lyse, it is important to ensure that nucleic acids are efficiently extracted from any target organism. For this purpose, we developed a cellular IC based on the use of nonpathogenic Bacillus spores. Purified Bacillus atrophaeus subsp. globigii (referred to hereafter as simply B. atrophaeus) spores were added to vaginal and anal samples, which were then subjected to rapid DNA extraction and subsequent PCR amplification. The proof of concept of this cellular IC was made through the use of both manual and automated DNA extraction methods, using vaginal or anal samples spiked with B. atrophaeus spores, combined with a multiplex real-time PCR assay for the specific detection of group B streptococci (GBS) and B. atrophaeus. The performance of the cellular IC was compared to that of a standard IC plasmid added to PCRs. Approximately 500 B. atrophaeus spores per PCR was found to be optimal since this did not interfere significantly with GBS detection for either DNA extraction method and yielded reproducible amplification and/or detection of B. atrophaeus genomic DNA serving as an IC template. Performance of the cellular IC was comparable to that of the standard IC. This novel IC system using nonpathogenic and hard-to-lyse B. atrophaeus spores allowed validation of both the DNA extraction procedure and the amplification and detection process. Use of a spore-based control also provides a universal control for microbial cell lysis.
Highly sensitive nucleic acid testing (NAT) technologies, such as the widely used PCR, represent increasingly popular molecular diagnostic tools for the detection of microbial pathogens (6, 9, 17, 19). Commercial and in-house NAT assays detecting target microbial nucleic acids often incorporate a nontarget nucleic acid serving as internal control (IC) template to verify the efficiency of each amplification and/or detection reaction (2, 6, 8, 10, 14, 17). A number of different strategies have been used for the construction of optimal IC nucleic acid templates for molecular diagnostic assays (3-5, 7, 11, 15, 20, 21). Concomitant amplification and/or detection of such IC nucleic acid templates allows verification of the NAT assay performance. In the absence of assay inhibition, the IC template should be detected efficiently from test samples negative for the target microbial nucleic acids. An absent or low IC detection signal observed with a test sample negative for the target microbial nucleic acids is indicative of complete or partial inhibition of the NAT assay by the test sample, which may contain inhibitors of nucleic acid amplification and/or detection. Technical failure(s) at the reagent or instrument level can also prevent normal amplification of the IC nucleic acid template. However, these ICs do not allow verification of the efficiency of the test sample preparation, cell lysis, and nucleic acid extraction procedures because they are only based on the detection of a target nucleic acid template added to each NAT assay reaction. Therefore, they can only reveal the failure of a NAT assay reaction.
Many microbial species have a thick and/or sturdy cell wall (e.g., gram-positive bacteria, mycobacteria, bacterial spores, and yeasts), which makes them difficult to lyse. Efficient lysis of any target microbial cell is required to release their nucleic acids and allow amplification and/or detection of target nucleic acids. Indeed, an optimal test sample preparation procedure for NAT must efficiently release nucleic acids from any target microbial cells or viruses and treat them to eliminate, neutralize, or inactivate nucleases and inhibitory substances that are potentially present in the test sample. Therefore, it is important to also validate the sample preparation and nucleic acid extraction procedure for NAT assays to ensure their adequate performance for target microbial cell lysis and nucleic acid recovery (5, 8, 15). Considering that efficient microbial cell lysis and target nucleic acid recovery are two key prerequisites for molecular methods detecting microbial pathogens (9, 17), there is a need to develop an IC system validating simultaneously both the nucleic acid extraction procedure and the amplification/detection process. Furthermore, such an IC would be in line with the current trend in molecular diagnostics, which is to integrate sample preparation and nucleic acid amplification and/or detection into a single device (1, 9).
In the present study, we developed an IC technology for NAT based on the use of hard-to-lyse Bacillus atrophaeus subsp. globigii (referred to hereafter as simply B. atrophaeus) spores that are added to each test sample. The proof of concept of this nonpathogenic cellular IC is made through the use of both manual and fully automated, rapid DNA extraction methods coupled with multiplex real-time PCR detection of group B streptococci (GBS) and B. atrophaeus in vaginal and anal samples collected from pregnant women at delivery.
MATERIALS AND METHODS
Bacterial spore preparation.
B. atrophaeus strain CCRI-9827 was grown for 24 h at 35°C on sporulation agar medium containing 5 g of peptone, 3 g of beef extract, 5 mg of MnSO4, and 15 g of agar per liter. Subsequently, the B. atrophaeus spores were purified on a sodium bromide gradient as previously described (13). The purity of the spore preparation was assessed by using optical microscopy with a Petroff-Hausser counting chamber (Hausser Scientific) to determine the ratio of spores versus vegetative cells. This protocol yielded bacterial spore preparations having a purity of at least 99.9%. Quality control of spore preparations also included real-time PCR analysis for B. atrophaeus DNA contamination. B. atrophaeus-specific, real-time PCRs were prepared with 104 spores. The spores had either been subjected to the manual lysis and DNA preparation method, as described below, or were untreated. An acceptable purity was considered to be when the lysed spores gave a cycle threshold value at least 10 cycles lower than the untreated spores (i.e., at least 1,000 times more DNA). The purified B. atrophaeus spore preparations were stored as aliquots at −20°C at a concentration of 2.4 × 105 spores per μl.
Highly purified genomic DNA.
Highly purified bacterial genomic DNA was prepared from mid-log-phase cells of GBS strain ATCC 12973 and B. atrophaeus strain CCRI-9827 by using the MagneSil KF (Promega) genomic DNA isolation kit on a KingFisher ML instrument (Thermo Scientific).
Clinical specimens.
Vaginal and anal samples were collected from consenting women admitted for delivery at the Centre Mère-Enfant of the Centre Hospitalier Universitaire de Québec (Pavillion CHUL) using the swab transport system Copan Venturi Transystem (Copan Diagnostics) that contains liquid Stuart's medium. This ongoing clinical study was approved by both the hospital ethics committee and the U.S. National Institutes of Health (DMID protocol number 05-0107). Selected GBS-positive and GBS-negative vaginal and anal samples (freshly collected or stored at −80°C) collected through this clinical study were used.
Vaginal and anal swab sample preparation for DNA extraction.
The cotton tip portion of each collected vaginal or anal swab was broken and placed in a two-ml screw-cap tube containing 1.0 ml of TE buffer (10 mM Tris-Cl [pH 7.5], 1 mM EDTA). The vaginal or anal swab sample was resuspended by vigorous mixing on a vortex mixer for 15 s and then used directly for microbial DNA extraction.
Crude microbial DNA extraction procedures.
Crude microbial DNA extraction was performed from the resuspended vaginal or anal samples by using either a manual or an automated procedure. The manual method was performed with 60 μl of the vaginal or anal sample by using a BD GeneOhm lysis kit (BD Diagnostics), which is a simple 10-min method for universal microbial cell lysis. The automated procedure is based on the use of a prototype microfluidic system on a compact disc (CD), which is a rapid bead-beating microbial cell lysis platform (12). This cell lysis platform utilizes small metal disks placed inside each lysis chamber that oscillate during CD spinning through interaction with stationary magnets. A volume of approximately 60 μl of each vaginal or anal swab sample resuspension was added to a lysis chamber by using a micropipettor. Following a 12-min spinning profile of the CD device, to generate centrifugal forces allowing bead-beating and fluid movement into the microfluidic system, extracted DNA was removed via collection ports for subsequent testing by real-time PCR.
Purified spores of B. atrophaeus were incorporated directly either into the lysis tube of the BD GeneOhm lysis kit or into each lysis chamber of the microfluidic system to allow verification of the efficiency of the DNA extraction or detection process from the vaginal or anal swab resuspension fluids. The crude DNA extracts obtained with both procedures were heat treated for 2 min at 95°C prior to PCR.
Real-time PCR detection.
Highly purified GBS and B. atrophaeus genomic DNAs were used to optimize a multiplex real-time PCR assay detecting both a GBS- and a B. atrophaeus-specific genomic targets (Table 1). This PCR assay was further validated by using crude DNA extracted from vaginal or anal samples spiked with various amounts of B. atrophaeus spores serving as the cellular IC. Each optimized 25-μl PCR contained 1.5 μl of either a purified genomic DNA preparation or a crude DNA extract prepared from a vaginal or anal sample with added B. atrophaeus spores, 0.4 μmol (each) of GBS-specific primers/liter, 0.1 μmol (each) of B. atrophaeus-specific primers/liter, 0.2 μmol of GBS-specific FAM-labeled TaqMan probe/liter, 0.1 μmol of B. atrophaeus-specific TET-labeled TaqMan probe/liter, 4.5 mmol of MgCl2/liter, 3.3 g of bovine serum albumin/liter, 200 μmol of deoxynucleoside triphosphate/liter, 50 mmol of KCl/liter, 10 mmol of Tris-HCl (pH 9.0)/liter, 0.1% Triton X-100, and 0.6 U of Taq polymerase (Promega) coupled with TaqStart antibody (Clontech). PCR amplifications (180 s at 94°C and then 45 cycles of three steps consisting of 5 s at 95°C, 15 s at 56°C, and 5 s at 72°C) were performed by using a Rotor-Gene thermocycler (Corbett Research). The performance of this assay, incorporating the B. atrophaeus-based cellular IC, was compared to that of another GBS-specific real-time PCR assay previously developed by our group (11). The latter assay incorporates a standard nucleic acid IC, which is based on the use of a linearized recombinant plasmid added to each GBS-specific real-time PCR. This IC nucleic acid template is also PCR amplified by the GBS-specific primers (Table 1).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Target gene | Sequence (5′-3′)b | Amplicon size (bp) |
|---|---|---|---|
| GBS-specific primersa | |||
| Sag59 | cfb | TTTCACCAGCTGTATTAGAAGTA | 154 |
| Sag190 | cfb | GTTCCCTGAACATTATCTTTGAT | |
| B. atrophaeus-specific primers | |||
| ABgl158 | atpD | CACTTCATTTAGGCGACGATACT | 212 |
| ABgl345a | atpD | TTGTCTGTGAATCGGATCTTTCTC | |
| TaqMan probes | |||
| GBS-specific (cfbSag-T1-A1) | cfb | FAM-CCCAGCAAATGGCTCAAAAGC-BHQ1 | |
| B. atrophaeus specific (ABgl-T1-B1) | atpD | TET-CGTCCCAATGTTACATTACCAACCGGCACTGAAATAGG-BHQ1 | |
| Plasmid IC specific (pStrep180-T2-B1) | TET-CTTCACATTGCTCCACCTTTCCT-BHQ1 |
The GBS-specific primers also amplified a 180-bp amplicon from the linearized recombinant plasmid serving as standard IC template (11).
FAM, 6-carboxylfluorescein; TET, tetrachloro-6-carboxylfluorescein; BHQ1, Black Hole Quencher 1 (Biosearch Technologies).
RESULTS
Optimization of the multiplex PCR assay.
Purified genomic DNA from GBS and B. atrophaeus was used to optimize the multiplex real-time PCR assay. Optimal and reproducible detection of the B. atrophaeus-specific genomic target, without any significant detrimental effect on the efficiency of specific GBS DNA detection, was achieved by using standard real-time PCR conditions with 0.4 μmol of the two GBS-specific primers/liter and only 0.1 μmol of the two B. atrophaeus-specific primers/liter. This optimized multiplex PCR assay was subsequently used for vaginal and anal swab sample testing for GBS detection in the presence of spiked B. atrophaeus spores.
Optimization of B. atrophaeus spore detection.
Crude DNA, extracted from vaginal and anal samples with added B. atrophaeus spores using the manual procedure, was subjected to analysis by real-time PCR. The optimized PCR assay for specific GBS and B. atrophaeus detection was initially used to determine the appropriate amount of bacterial spores to be incorporated directly into BD GeneOhm lysis tubes or lysis chambers of the prototype microfluidic system. Optimization of B. atrophaeus spore detection was performed by adding 2 × 103 to 2 × 105 spores (2 μl of spore preparation dilutions) per lysis tube. Subsequently, 58 μl of a GBS-negative vaginal or anal swab sample resuspension was added to each tube. After completion of the cell lysis process, a volume of 1.5 μl of the crude DNA extract representing the equivalent of approximately 50, 200, 500, 2,000, and 5,000 spores per PCR was used. The optimal number of spores per PCR was found to be approximately 500 since it allowed reproducible detection of the B. atrophaeus-specific genomic target from a vaginal or anal sample without significant detrimental effect on the performance of specific GBS DNA detection (data not shown). This amount of spores per PCR was obtained by initially introducing 2 × 104 spores per lysis tube. Fewer spores led to less reproducible B. atrophaeus DNA detection, while more spores reduced the efficiency of GBS detection, probably due to competition for PCR reagents (data not shown). Therefore, all subsequent experiments to evaluate the performance of the cellular IC with clinical samples were performed using the equivalent of 500 B. atrophaeus spores per PCR.
Evaluation of the cellular IC.
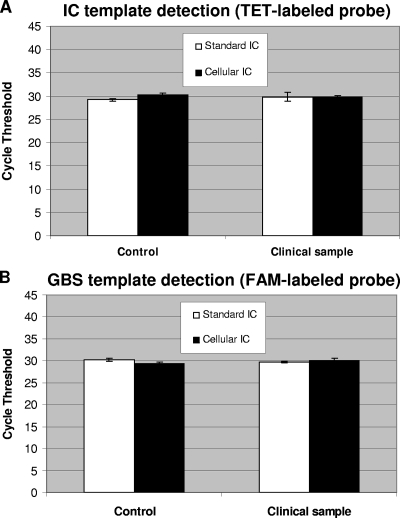
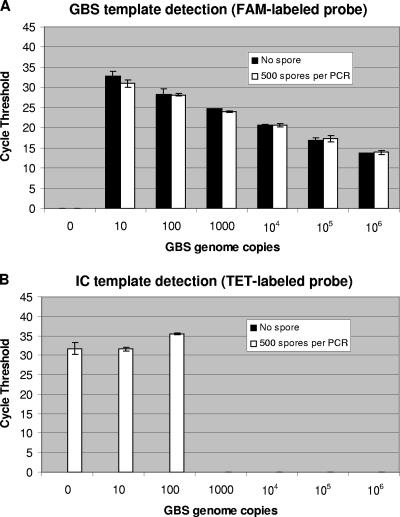
The performance of the cellular and standard IC systems was compared by using real-time PCR assays for GBS-specific DNA detection from vaginal and anal samples. Testing of the GBS-specific assay with the standard plasmid IC (11) was performed by using the same GBS-negative specimen spiked with B. atrophaeus spores. For testing of both IC systems with their respective PCR assays, PCRs containing the DNA extracted from the GBS-negative specimen plus B. atrophaeus spores was spiked with the equivalent of 100 genome copies of GBS DNA to also evaluate the performance of GBS detection. In terms of PCR cycle thresholds, the obtained values and reproducibility for the detection of both IC targets (i.e., DNA from lysed B. atrophaeus spores and the standard linearized recombinant plasmid) were similar (Fig. 1). The performance of both PCR assays to specifically detect GBS at a low number of genome copies (i.e., 100 copies, which is about 10 times the detection limit of the assay) was also similar (Fig. 1). Furthermore, the presence of B. atrophaeus spores in a GBS-negative specimen vaginal/anal sample with spiked GBS DNA did not influence significantly the performance of GBS detection with up to 106 GBS genome copies per PCR (Fig. 2A). As expected, increasing amounts of GBS DNA (i.e., 100 genome copies or more) caused a reduction (or complete inhibition) of the fluorescence signal for the cellular IC, which is associated with detrimental competition for PCR reagents by the GBS-specific amplification and detection (Fig. 2B).
FIG. 1.
Robustness of the cellular and standard IC systems. For both IC systems, a GBS-negative vaginal or anal sample was prepared by using the 10-min manual DNA extraction method. Control reactions without clinical sample were also performed. For the cellular IC, a portion of the clinical sample was spiked with 2 × 104 B. atrophaeus spores before cell lysis and DNA extraction (giving the equivalent of 500 spores per PCR). For the standard IC, 60 copies of linearized plasmid were added to each PCR. All PCR amplifications were carried out in the presence of approximately 100 GBS genome copies. Cycle threshold values obtained from both the TET-labeled IC-specific probes (A) and the FAM-labeled GBS-specific probes (B) are presented. Standard deviations are for three replicate tests.
FIG. 2.
Performance of GBS detection in the presence of lysed B. atrophaeus spores. Using the manual method, a DNA extract was prepared from a portion of a GBS-negative vaginal or anal sample spiked with 2 × 104 B. atrophaeus spores per lysis tube (giving the equivalent of 500 spores per PCR). PCR amplifications of 1.5 μl of DNA extract were performed in the presence of up to 106 GBS genome copies per PCR. Cycle threshold values obtained from both the FAM-labeled GBS-specific probes (A) and the TET-labeled IC-specific probes (B) are presented. The absence of cycle threshold value means that the nucleic acid target was undetectable after 45 cycles of PCR amplification. Standard deviations are for two replicate tests.
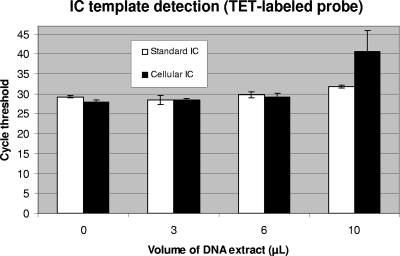
The ability of both IC systems to monitor PCR inhibition was evaluated by testing various amounts of a crude DNA extract prepared from a GBS-negative sample using the manual DNA extraction method. PCR amplifications were performed using 0, 3.0, 6.0, and 10 μl of the DNA extract in the presence of 500 B. atrophaeus spores (added before the lysis step) or 60 copies of linearized plasmid. Both IC systems were shown to detect partial PCR inhibition associated with the presence of at least 10 μl of the DNA extract, which is associated with a reduction in the efficiency of the B. atrophaeus and plasmid IC DNA target amplification (i.e., higher cycle threshold value) (Fig. 3). Control reactions with 0 to 10 μl of lysed spores without the vaginal or anal sample (i.e., in TE buffer) confirmed that there was no significant PCR inhibition in the absence of clinical sample (data not shown).
FIG. 3.
Performance of the cellular IC to monitor PCR inhibition compared to the standard IC. PCR amplifications were performed in the presence of various amounts of a crude DNA extract prepared from a GBS-negative vaginal/anal sample using the manual method. For the cellular IC, a portion of the clinical sample was spiked with 2 × 104 B. atrophaeus spores before cell lysis and DNA extraction (giving the equivalent of 500 spores per PCR). For the standard IC, 60 copies of linearized plasmid were added to each PCR. Cycle threshold values from the TET-labeled IC-specific probes obtained with the different volumes of DNA extract tested are presented. Standard deviations are for three replicate tests.
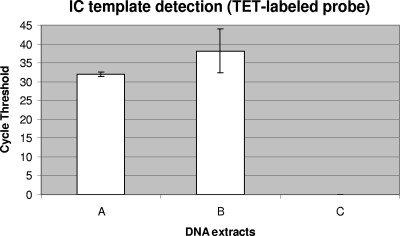
The ability of the cellular IC to detect inefficiency of the test sample preparation and/or DNA extraction level was also investigated. Truncated versions of the manual DNA extraction method, in which critical steps were voluntarily omitted, were tested with B. atrophaeus spores mixed in a GBS-negative vaginal/anal sample to compare their performance with the complete extraction method. It was demonstrated that omission of the glass bead agitation step for cell lysis, as well as removal of the final heating step at 95°C to inactivate PCR inhibitors, resulted in less efficient B. atrophaeus DNA detection (Fig. 4). The cycle threshold value for the incomplete method B was six cycles higher than that for the complete method, which represents 100-fold fewer target DNA molecules. This experiment demonstrates the effectiveness of the cellular IC to detect diminished efficiency of the DNA extraction process.
FIG. 4.
Ability of the cellular IC to detect inefficiency of DNA extraction. Truncated versions of the manual DNA extraction method were tested with 2 × 104 B. atrophaeus spores mixed in a GBS-negative vaginal/anal sample to compare their performance with the complete DNA extraction method. DNA extracts were prepared as follows: complete DNA extraction method (column A), DNA extraction without the glass bead agitation step for cell lysis (B), and DNA extraction without both the glass bead agitation step and the final heating step at 95°C to inactivate PCR inhibitors (column C). The absence of cycle threshold value means that the nucleic acid target was undetectable after 45 cycles of PCR amplification. Standard deviations are for three replicate tests.
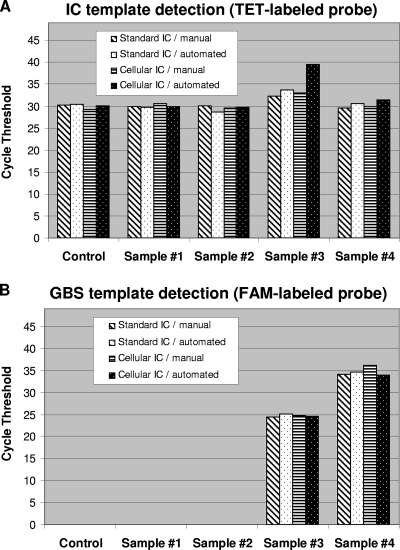
Finally, the performance of both IC systems was evaluated with selected GBS-positive and GBS-negative vaginal and anal samples collected from consenting women admitted for delivery through an ongoing clinical study at our university hospital. Crude microbial DNA extraction was performed from each selected clinical sample using both the manual BD GeneOhm lysis kit and the automated prototype microfluidic system on CD. To test the cellular IC, purified B. atrophaeus spores were incorporated directly either into each lysis tube for the manual method or into each lysis chamber of the microfluidic system prior to the addition of a vaginal/anal sample. Testing of the standard plasmid IC was performed by adding 60 copies of the linearized recombinant plasmid to each real-time PCR prior to the addition of the crude DNA extracted from the same vaginal or anal samples that were tested with the cellular IC. The PCR cycle threshold values obtained with all selected vaginal/anal specimens for the detection of both IC targets were similar (Fig. 5). Furthermore, the performance of the two IC systems was comparable with both DNA extraction methods, as confirmed by the obtainment of similar cycle threshold values. It should also be mentioned that both PCR assays were able to efficiently detect GBS target DNA in the two tested GBS-positive specimens (naturally infected) in the presence of either the cellular or the standard IC (Fig. 5). As expected, the presence of a high load of GBS caused a reduction of the fluorescence signal from the cellular IC probe associated with competition for PCR reagents.
FIG. 5.
Performance comparison of both IC systems using selected GBS-positive and GBS-negative vaginal/anal samples. For both IC systems, crude DNA extracts were prepared from the selected vaginal and anal samples by using the manual or the CD-based automated DNA extraction methods. Control reactions without clinical sample were performed. For the cellular IC, a portion of each selected vaginal or anal sample was spiked with 2 × 104 B. atrophaeus spores before cell lysis and DNA extraction (giving the equivalent of 500 spores per PCR). For the standard IC, 60 copies of linearized plasmid were added to each PCR. Cycle threshold values obtained from both the TET-labeled IC-specific probes (A) and the FAM-labeled GBS-specific probes (B) are presented. Selected clinical samples are as follows: samples 1 and 2, GBS-negative vaginal/anal samples; samples 3 and 4, GBS-positive vaginal/anal samples collected from heavily and lightly colonized women, respectively. The absence of cycle threshold value means that the nucleic acid target was undetectable after 45 cycles of PCR amplification.
DISCUSSION
Detection of an IC nucleic acid template is often integrated into commercial and in-house NAT assays (2, 6, 10, 14, 17). This allows verification of their efficiency to detect target nucleic acids but does not permit verification of the efficiency of sample preparation, cell lysis, and nucleic acid extraction procedures. Considering that efficient microbial cell lysis and target nucleic acid recovery are two key prerequisites to detect microbial pathogens reliably using nucleic acid-based detection methods (9, 17), there is a need to develop IC systems validating the nucleic acid extraction procedure, as well as the amplification and/or detection processes.
In the present study, we have developed an IC technology for NAT assays that is based on the use of hard-to-lyse and nonpathogenic B. atrophaeus spores, which are added to each test sample prior to cell lysis and DNA extraction. Purified spores of B. atrophaeus can be incorporated directly into a cell lysis tube or chamber prior to the addition of the test sample, as described here. This allows simplification of the procedure and reduction of the potential risk of sample-to-sample contamination that would be associated with the pipetting of spores directly into the test sample. Another advantage of this spore-based IC is that it can serve as a universal control for microbial cell lysis and DNA extraction in molecular diagnostic assays for the detection of infectious disease agents and/or associated virulence factors such as antibiotic resistance or toxin genes. In fact, the use of IC templates inside bacterial spores (e.g., B. atrophaeus spores, as described here) provides a universal cell lysis control since bacterial spores are among the most difficult cells to lyse. Indeed, bacterial spores are highly resistant living structures (16). Spores have evolved as a survival mechanism able to resist harsh conditions, including heat and UV light. However, to serve as a universal cell lysis control, it is critical that the spore preparation contains very low amounts of vegetative cells, which are much easier to lyse than spores. We have found that B. atrophaeus spore preparations with a purity of at least 99.9% are suitable for this purpose.
Spore preparations should also be free of genomic DNA that is released from vegetative cells during their preparation. However, it should be noted that spore walls naturally incorporate copies of the organism's DNA, which can be detected by highly sensitive PCR without spore lysis. As previously stated, our quality control of spore preparations involves real-time PCR quantification of B. atrophaeus DNA from an aliquot of each preparation containing 104 spores. Spore preparations that have been treated with the manual lysis-DNA extraction procedure must give a cycle threshold at least 10 cycles lower than that of the untreated preparation to pass the quality control test. This equates to at least a 3-log difference in the amount of DNA detected by real-time PCR. In fact, our results routinely show a 12-cycle difference that approaches a 4-log difference in the amount of DNA. The fact that some DNA is detected in untreated samples could indicate the presence of free DNA in the preparation or that a small amount of DNA is released from spores during the aggressive, repeated thermal cycling of PCR. Nevertheless, this amount of DNA is insignificant compared to that released by our sample preparation methods.
In rare cases, as shown in Fig. 4, an IC signal can be seen when only 500 spores are used in a clinical sample when the sample preparation is incomplete. However, in this example, this signal is six cycle thresholds higher than the completely treated sample, equating to a 2-log difference in the amount of DNA. Also, through many repeated experiments we have only rarely seen this phenomenon, and we normally only get a signal when the complete procedure is used. Moreover, during the development of this method, we found that 500 spores per real-time PCR were necessary to give a reproducible signal. As such, we conclude that the use of B. atrophaeus spores is a robust tool for verification of cell lysis/nucleic acid extraction and target amplification efficiencies.
Such a cellular IC may be applicable to any test sample, including clinical, veterinary, or environmental specimens, and may be used with any nucleic acid extraction and nucleic acid amplification/detection technologies. Furthermore, microbial cells other than bacterial spores may be used as a source of IC template, but the use of spores provides a particularly stable cellular IC. We have found that the purified B. atrophaeus spore preparations provide a reliable and efficient cellular IC that is stable for a period of at least 18 months when stored in aliquots at −20°C (data not shown).
GBS is a leading cause of sepsis and meningitis in newborns (18). Rapid methods for specific detection of GBS from clinical samples collected from pregnant women (preferentially at delivery) are important in the prevention of neonatal infections, as well as in the rationalization of antibiotic use. The proof of concept of our new cellular IC system was made through the use of a NAT assay for GBS detection using vaginal and anal samples collected from pregnant women at delivery. DNA was extracted from these samples in the presence of B. atrophaeus spores by using two different methods. Target genomic DNAs were detected by using a multiplex real-time PCR assay for GBS and B. atrophaeus. For this assay, the GBS-specific cfb gene and B. atrophaeus-specific sequences selected from the atpD gene were chosen as targets. We have demonstrated that the performance of this cellular IC was comparable to that of a standard IC plasmid added to each PCR, and this, for the two different DNA extraction methods. Clearly, other target genes or target sequences in B. atrophaeus could have also been used as long as the selected IC target sequences are sufficiently distinct from the other target sequences of the assay to allow their specific detection. The main advantage of using bacterial cells as the cellular IC over the use of standard plasmid ICs is that it allows a validation of both the sample preparation method and the amplification and/or detection processes. Indeed, amplification of the cellular IC template is dependent on the efficiency of the sample preparation protocol to (i) lyse the bacterial cells containing the IC template; (ii) release nucleic acids from the cells to allow their amplification and/or detection; and (iii) eliminate, inactivate, and/or neutralize amplification and/or detection inhibitors found in test samples. The use of a cellular IC is particularly relevant for detection of targeted nucleic acids found in low numbers in a test sample to ensure that each step of the NAT assay from sample to answer is efficient.
Cellular ICs may also be adapted for use with non-PCR nucleic acid amplification technologies or signal amplification methods for detection of any target nucleic acid. Indeed, utilization of cellular ICs with NAT assays is desirable considering that efficient target cell lysis and recovery of their nucleic acids are two key prerequisites for molecular diagnostic methods. Furthermore, such cellular ICs would be ideal to validate molecular diagnostic tests that integrate sample preparation and nucleic acid extraction with nucleic acid amplification and/or detection in a single device (1, 9).
In conclusion, a cellular IC provided by using B. atrophaeus spores added to a test sample prior to its preparation for PCR is as efficient and as reproducible as the commonly used IC plasmids incorporated directly into amplification/detection reaction mixtures. Nonpathogenic microbial cells other than bacterial spores could also be used to provide a cellular IC. However, spores offer the added advantages of being highly stable and serving as a universal cell lysis control. Such a cellular IC may be applicable to any NAT technology.
Acknowledgments
This research project was supported by grant 1 U01 AI060594-01 from the U.S. National Institutes of Health and by grant PA-15586 from the Canadian Institutes of Health Research.
Footnotes
Published ahead of print on 14 January 2009.
REFERENCES
- 1.Chen, L., A. Manz, and P. J. R. Day. 2007. Total nucleic acid analysis integrated on microfluidic devices. Lab. Chip 71413-1423. [DOI] [PubMed] [Google Scholar]
- 2.Chudy, M., I. Hewlett, J. Saldanha, C. Bianco, A. J. Conrad, T. Gierman, C. Heldebrant, G. G. Rautmann, W. K. Roth, S. Stramer, T. Weimer, B. Whitaker, and G. Zerlauth. 2003. Technical considerations for the performance of nucleic acid amplification technology. Biologicals 31153-159. [DOI] [PubMed] [Google Scholar]
- 3.Courtney, B. C., M. M. Smith, and E. A. Henchal. 1999. Development of internal controls for probe-based nucleic acid diagnostic assays. Anal. Biochem. 270249-256. [DOI] [PubMed] [Google Scholar]
- 4.Dingle, K. E., D. Crook, and K. Jeffery. 2004. Stable and noncompetitive RNA internal control for routine clinical diagnostic reverse transcription-PCR. J. Clin. Microbiol. 421003-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotsch, A., A. Schubert, A. Bombis, M. Wiedmann, M. Zauke, and S. Schorling. 2007. Nuclease-resistant single-stranded DNA controls for nucleic acid amplification assays. J. Clin. Microbiol. 452570-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoorfar, J., N. Cook, B. Malorny, M. Wagner, D. De Medici, A. Abdulmawjood, and P. Fach. 2003. Making internal amplification control mandatory for diagnostic PCR. J. Clin. Microbiol. 415835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 421863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoorfar, J., P. Wolffs, and P. Rådström. 2004. Diagnostic PCR: validation and sample preparation are two sides of the same coin. APMIS 112808-814. [DOI] [PubMed] [Google Scholar]
- 9.Huang, Y., E. L. Mather, J. L. Bell, and M. Madou. 2002. MEMS-based sample preparation for molecular diagnostics. Anal. Bioanal. Chem. 37249-65. [DOI] [PubMed] [Google Scholar]
- 10.Kaltenboeck, B., and C. Wang. 2005. Advances in real-time PCR: application to clinical laboratory diagnostics. Adv. Clin. Chem. 40219-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke, D., C. Ménard, F. J. Picard, M. Boissinot, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin. Chem. 46324-331. [PubMed] [Google Scholar]
- 12.Kido, H., M. Micic, D. Smith, J. Zoval, J. Norton, and M. Madou. 2007. A novel, compact disk-like centrifugal microfluidics system for cell lysis and sample homogenization. Colloids Surf. B Biointerfaces 5844-51. [DOI] [PubMed] [Google Scholar]
- 13.Laflamme, C., S. Lavigne, J. Ho, and C. Duchaine. 2004. Assessment of bacterial endospore viability with fluorescent dyes. J. Appl. Microbiol. 96684-692. [DOI] [PubMed] [Google Scholar]
- 14.Morré, S. A., P. Sillekens, M. V. Jacobs, P. van Aarle, S. de Blok, B. van Gemen, J. M. M. Walboomers, C. J. L. M. Meijer, and A. J. C. van den Brule. 1996. RNA amplification by nucleic acid sequence-based amplification with an internal standard enables reliable detection of Chlamydia trachomatis in cervical scrapings and urine samples. J. Clin. Microbiol. 343108-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasloske, B. L., C. R. Walkerpeach, R. D. Obermoeller, M. Winkler, and D. B. DuBois. 1998. Armored RNA technology for production of ribonuclease-resistant viral RNA controls and standards. J. Clin. Microbiol. 363590-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearce, S., and P. Fitz-James. 1971. Spore refractility in variants of Bacillus cereus treated with actinomycin D. J. Bacteriol. 107337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Picard, F. J., and M. G. Bergeron. 2002. Rapid molecular theranostics in infectious diseases. Drug Discovery Today 71092-1101. [DOI] [PubMed] [Google Scholar]
- 18.Picard, F. J., and M. G. Bergeron. 2004. Laboratory detection of group B Streptococcus for prevention of perinatal disease. Eur. J. Clin. Microbiol. Infect. Dis. 23665-671. [DOI] [PubMed] [Google Scholar]
- 19.Procop, G. W. 2007. Molecular diagnostics for the detection and characterization of microbial pathogens. Clin. Infect. Dis. 45S99-S111. [DOI] [PubMed] [Google Scholar]
- 20.Rosenstraus, M., Z. Wang, S. Chang, D. DeBonville, and J. P. Spadoro. 1998. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J. Clin. Microbiol. 36191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stöcher, M., V. Leb, and J. Berg. 2003. A convenient approach to the generation of multiple internal control DNA for a panel of real-time PCR assays. J. Virol. Methods 1081-8. [DOI] [PubMed] [Google Scholar]