Abstract
This study used a PCR-based approach targeting 16S rRNA gene fragments to determine the occurrence and association of the three bovine digital dermatitis (BDD) treponeme phylogroups within lesions found in cattle from the United Kingdom. Examination of 51 BDD lesions collected from infected cattle across the United Kingdom revealed that BDD treponeme group 1 (Treponema medium/Treponema vincentii-like), group 2 (Treponema phagedenis-like), and group 3 (Treponema putidum/Treponema denticola-like) were present in 96.1%, 98%, and 76.5% of BDD lesions, respectively. The three phylogroups were present together in 74.5% of lesions. The PCR assays enabled the isolation of further treponeme strains from previously mixed primary BDD lesion cultures. Here a representative from each of the three distinct treponeme phylogroups was isolated from a single BDD lesion for the first time. These data highlight the extent to which this disease is polytreponemal. Immunohistochemistry and electron microscopy were used to investigate lesional hoof tissues, resulting in treponemes being identified copiously in hair follicles and sebaceous glands, suggesting a potential route of exit and/or entry for these pathogens. This study gives further evidence for the importance of the three treponeme groups in BDD pathogenesis and reiterates the value of molecular genetic approaches for isolating and identifying fastidious anaerobes.
Bovine digital dermatitis (BDD) is a worldwide disease responsible for lameness in cattle. The disease is exceptionally painful for the animal involved and has economic implications such as reduction in milk yield and reproductive performance (1, 15, 16, 34). It has now been more than 30 years since the disease was first recognized, and while regular foot bathing has been shown to have some efficacy, an effective preventative measure or treatment that completely eliminates the disease has yet to be identified (19). The result is that the disease is endemic in cattle across the world, and one recent report suggested that the economic impact resulting from milk production in the United States alone is a loss of as much as $190 million per year (20).
The environmental conditions of housed dairy cows, whereby they stand in slurry continuously, are likely to result in the isolation of a plethora of different microorganisms from BDD lesions. Indeed, various types of bacteria have been identified within BDD manifestations including spirochetes, Bacteroides species, Campylobacter species, Fusobacterium species, and Peptococcus species (2, 3, 10, 18, 25, 28). The result of this complex bacteriological environment is that the exact etiology of BDD remains unresolved. However, identification of the bacteria involved in the initial infection would allow specific targeting treatment and, hopefully, eradication of this disease. Treponemes are the only bacteria for which there is considerable evidence for a consistent presence and involvement within BDD lesions. Cloning of bacterial 16S rRNA genes from BDD lesions in Germany indicated five phylogroups of treponemes present within lesions (5). We recently isolated a large number of treponemes from BDD lesions in the United Kingdom and demonstrated that the isolates belong to three distinct phylogenetic/taxonomic groups (13). We designated the BDD treponemes group 1, group 2, and group 3, which correspond to Treponema medium/Treponema vincentii-like, Treponema phagedenis-like, and Treponema putidum/Treponema denticola-like spirochetes, respectively. These isolated BDD treponemes are nearly identical to three phylotypes described by a smaller study in the United States (31) and are nearly identical to three of the five phylotypes identified in BDD lesions in Germany (5).
We previously identified a strong association of Treponema species with BDD using a PCR-based method (6). However, that study focused solely on group 1, T. medium/T. vincentii-like spirochetes. In this study, we used PCR assays specific for each of the three BDD-associated treponeme groups in order to determine the presence of each group in BDD lesions drawn from infected animals across the United Kingdom. Furthermore, the localization of BDD-associated treponemes within lesion material was investigated using immunohistochemistry and electron microscopy (EM).
MATERIALS AND METHODS
Bacterial strains and clinical samples.
Twenty-three strains isolated from BDD lesions and characterized previously were used to validate the PCR tests (13). A previously isolated group 2 strain, G356 (8); a group 3 strain, G179, isolated from a case of contagious ovine DD (CODD) (7); and several reference strains considered to be the closest relatives were also included (complete list in Table 1).
TABLE 1.
Collection of strains used to test sensitivity and specificity of BDD-associated Treponema-specific PCR assaysa
| Species and/or group and strain | Result
|
|||
|---|---|---|---|---|
| Specific PCR for group:
|
Treponema PCR | |||
| 1 | 2 | 3 | ||
| Group 1 (T. medium/T. vincentii-like) strains T184, T19, T54, T56, and T18A | + | − | − | + |
| Group 2 (T. phagedenis-like) strains T167, T52A, T136, T119A, T257, T296A, T354B, T2721A, T320A, T380A, T323C, W35, G169A, and G187 | − | + | − | + |
| Group 3 (T. denticola/T. putidum-like) strains T354A, T3552B, G819CB, and T18B | − | − | + | + |
| Bovine spirochete strain G356 | − | + | − | + |
| Ovine spirochete strain G179 | − | − | + | + |
| T. medium ATCC 700293T | + | − | − | + |
| T. phagedenis CIP 62.29 | − | + | − | + |
| T. phagedenis biotype Reiter | − | + | − | + |
| T. denticola ATCC 35405T | − | − | − | + |
| T. denticola GM-1 | − | − | − | + |
The assays include a specific PCR for groups 1, 2, and 3 and a general Treponema PCR.
Twenty-nine BDD lesion biopsies were collected from Holstein-Friesian cows at farms in Merseyside, Cheshire, Lancashire, Gloucestershire, and Shropshire, United Kingdom, from 2003 to 2007. All biopsies were used for DNA extraction, and 20 of the 29 biopsies were used in an attempt to cultivate treponemes. Biopsies for bacterial cultivation and DNA extraction were collected and transported back to the laboratory as described previously (13), placed immediately in an anaerobic cabinet, and divided into two samples for said procedures. Samples for DNA extraction only were transported on ice without transport medium. All samples for DNA extraction were stored at −80°C. The genomic DNAs from a further 22 BDD lesion biopsies collected during 1996 to 1997 and previously investigated by this laboratory using a different PCR method (6) were also included in this investigation. Single rear-hoof skin biopsies were collected postmortem from nine young bullocks and seven dairy cows (sent to slaughter) that did not have any evidence of BDD. For immunohistochemical and electron microscopical studies, biopsies from BDD lesions (three acute cases and one chronic case) and from healthy feet (nine sampled postmortem) were taken immediately into 10% formalin-saline and paraffin embedded.
Strain isolation.
Strain isolations from BDD lesional tissues, isolate 16S rRNA gene sequencing, and phylogenetic analyses were carried out as previously described (13).
DNA extraction.
DNA was extracted from treponeme cultures as previously described (13). Tissues from infected and uninfected hooves were thawed, and DNA was extracted using a DNeasy kit (Qiagen, United Kingdom) according to the manufacturer's instructions, and the resulting extracted genomic DNA was stored at −20°C.
PCR assays.
Nested PCR assays that were specific for the three BDD treponeme groups recently characterized and described by our laboratory were developed (13). The initial PCR step used a universal bacterial primer pair encompassing the majority of the 16S rRNA gene. PCR mixtures used Taq polymerase (Qiagen, United Kingdom) according to the manufacturers' instructions, with 1 μl of the DNA template per 25-μl reaction mixture volume and incubation at 95°C for 5 min, 25 cycles of 94°C for 1 min, 55°C for 3 min, and 72°C for 3 min, with a final extension step at 72°C for 7 min. The second/nested PCR step used primers encompassing smaller (300-to 500-bp) regions within the 16S rRNA gene. Primers were identified using a 16S rRNA gene CLUSTALW alignment of the 23 strains isolated in a previous study (13) with all known treponeme sequences present in GenBank. Stringent PCR conditions were identified using a Mastercycler gradient thermocycler (Eppendorf, Germany). The BDD treponeme-specific PCRs were applied to culture and tissue-derived DNA samples using 25-μl reaction mixes as described above with 1 μl PCR product template from the initial reaction. Temperature cycling entailed 95°C for 5 min followed by 40 cycles of 95°C for 1 min; annealing for either 2 min at 68°C for group 1 primers, 1 min at 64°C for group 2 primers, or 30 s at 68°C for group 3 primers; an extension step at 72°C for 2 min; and then a final elongation step at 72°C for 10 min. To ensure validity in each assay, water was used as a negative control, and positive controls included genomic DNA from each of the three treponeme groups. All biopsy extractions were further subjected to a Treponema genus-specific PCR assay as originally described (21). All PCR primers used are listed in Table 2.
TABLE 2.
Primers used to detect the three BDD treponeme phylogroups and all treponemes
| Primer specificity | Primer (sequence) | Predicted band size (bp) | Region of 16S targeted (positions)a | Reference or source |
|---|---|---|---|---|
| Universal | 16S F (5′-AGAGTTTGATCCTGG-3′) | 7-26 | 27 | |
| 16S R (5′-TACCTTGTTACGACTT-3′) | 1,526 | 1491-1506 | ||
| Group 1 (T. medium/T. vincentii-like) | TmF (5′-GAATGCTCATCTGATGACGGTAATCGACG-3′) | 472-500 | This study | |
| TmR (5′-CCGGCCTTATCTAAGACCTTCTACTAG-3′) | 475 | 1001-1029 | ||
| Group 2 (T. phagedenis-like) | TbF (5′-GAAATACTCAAGCTTAACTTGAGAATTGC-3′) | 612-640 | This study | |
| TbR (5′-CTACGCTACCATATCTCTATAATATTGC-3′) | 400 | 1006-1029 | ||
| Group 3 (T. denticola/T. putidum-like) | TpF (5′-GGAGATGAGGGAATGCGTCTTCGATG-3′) | 459-484 | This study | |
| TpR (5′-CAAGAGTCGTATTGCTACGCTGATATATC-3′) | 475 | 1017-1045 | ||
| Treponema sp. | TPF (5′-AARCATGCAAGTCGARCGGCAAG-3′) | 335 | 49-71 | 21 |
| TPR1 (5′-TCCATTGCGGAATATTCTTA-3′) | 365-384 |
Locations relative to those for the Escherichia coli 16S rRNA gene sequence reported under GenBank accession number M25588 (11) are given.
Production of antitreponemal antibodies.
Antigens were prepared from each of the three groups of treponemes by sonication and repeated freeze-thawing (9). These antigens were then pooled and supplied to a commercial concern for the generation of rabbit antisera. During this procedure, rabbits were immunized in a multisite regimen using Freund's complete and incomplete adjuvants over a period of 3 months. At the terminal bleed, the antisera were tested for reactivity by enzyme-linked immunosorbent assay and Western blotting against the purified antigens and were shown to cross-react with all three treponeme groups (data not shown).
Immunohistochemistry.
Paraffin-embedded tissues were sectioned by a microtome, deparaffinized with xylene, and blocked with 3% hydrogen peroxide and 2% bovine serum albumin. Each slide was incubated with antitreponeme antibodies (1:1,000 dilutions) overnight, washed with phosphate-buffered saline three times and then probed with a 1-in-500 solution of goat anti-rabbit antibody conjugated to peroxidase for 2 h. After a second washing step, bound peroxidase was localized with chromogen (diaminobenzidine) for 30 min and then counterstained with hematoxylin and eosin prior to light microscopy.
Electron microscopy.
For EM, paraffin-embedded tissues were prepared according to a previously described method (14). The resin-embedded tissues were sectioned with an ultramicrotome, stained with 2% uranyl acetate (pH 7), and subjected to EM.
Nucleotide sequence accession numbers.
The 16S rRNA gene GenBank accession numbers determined as part of this study for BDD Treponema strains T645C3, T116, T122, T100, T200, T52B, T136E, and T136P2 are FJ204236 to FJ204243, respectively.
RESULTS
PCR validation.
Within the original 23 spirochete strains used to characterize the three novel BDD treponeme groups, each PCR test detected only the group it was designed to identify (Table 1). When including the nearest designated relatives for each phylogroup, which were human oral and genital treponemes, the group 1 and group 2 PCR tests detected no other strains except the nearest designated relatives, T. medium and T. phagedenis, respectively, while the group 3 PCR test did not cross-react with its nearest designated relative, T. denticola. The detection limit for the group 1, 2, and 3 assays were stock culture DNA extraction dilutions of 1 × 10−3, 1 × 10−4, and 1 × 10−4, corresponding to 88, 11, and 33 treponemes per PCR, respectively.
Group-specific PCR survey of biopsied BDD lesions.
From the 29 BDD lesions biopsies collected during the current study (2002 to 2007), group 1, 2, and 3 treponemes were present in 96.6%, 100%, and 72.4% of lesions, respectively (Table 3). The only two acute lesions tested and a single case of interdigital dermatitis were positive for only two phylotypes: groups 1 and 2. All three groups were present in 72.4% of BDD lesions. From the 22 BDD lesions previously surveyed for group 1 isolates only (6), the group 1, 2, and 3 treponemes were present in 95.5%, 95.5%, and 81.8% of lesions (Table 4), with all three groups present in 77.3% of lesions.
TABLE 3.
PCR detection of treponemes in BDD lesion biopsies, 2003 to 2007
| Sample | Biopsy date (day/mo/yr) | Description | Typea | Treponeme isolated (no. of isolates)d | Result
|
|||
|---|---|---|---|---|---|---|---|---|
| Specific PCR for group:
|
Treponema PCR | |||||||
| 1 | 2 | 3 | ||||||
| 1 | 1/12/03 | Cheshire, farm 1, cow 167, left hoof | Chronic | T167b (2) | + | + | + | + |
| 2 | 1/12/03 | Cheshire, farm 1, cow 167, right hoof | Acute | IF | + | + | − | + |
| 3 | 1/12/03 | Cheshire, farm 1, cow 13 | IF | + | + | − | + | |
| T136b (2) | + | + | + | + | ||||
| 4 | 26/1/04 | Shropshire, farm 1, cow 136 | T136Ec (1) | |||||
| T136P2c (3) | ||||||||
| 5 | 26/1/04 | Shropshire, farm 1, cow 52 | T52Ab (2) | + | + | + | + | |
| 26/1/04 | T52Bc (1) | |||||||
| 6 | 26/1/04 | Shropshire, farm 1, cow 200 | T200 (2)c | + | + | + | + | |
| 7 | 26/1/04 | Shropshire, farm 1, cow 119 | T119Ab (2) | + | + | + | + | |
| 8 | 13/2/04 | Merseyside, farm 1, cow 320 | T320Ab (2) | + | + | + | + | |
| 9 | 13/2/04 | Merseyside, farm 1, cow 380 | T380Ab (2) | + | + | + | + | |
| 10 | 13/2/04 | Merseyside, farm 1, cow 272 | T2721Ab (2) | + | + | + | + | |
| 11 | 13/2/04 | Merseyside, farm 1, cow 355 | T3552Bb (3) | + | + | + | + | |
| 12 | 26/4/04 | Gloucestershire, farm 1, cow 819 | G819CBb (3) | + | + | + | + | |
| 13 | 26/4/04 | Gloucestershire, farm 1, cow 317 | IF | + | + | + | + | |
| 14 | 23/4/04 | Gloucestershire, farm 2, cow 187 | IDD | G187b (2) | + | + | − | + |
| 15 | 16/5/04 | Gloucestershire, farm 3, cow 1 | IF | + | + | + | + | |
| 16 | 16/5/04 | Gloucestershire, farm 3, cow 169 | G169Ab (2) | + | + | + | + | |
| 17 | 9/7/04 | Merseyside, farm 1, cow 323 | T323Cb (2) | + | + | + | + | |
| 18 | 9/7/04 | Merseyside, farm 1, cow 645 | T645C3c (2) | + | + | + | + | |
| 19 | 28/4/05 | Merseyside, farm 2, cow 116 | T116Bc (2) | + | + | + | + | |
| 20 | 2/9/05 | Cheshire, farm 2, cow 100 | T100Ac (2) | + | + | + | + | |
| 21 | 2/9/05 | Cheshire, farm 2, cow 122 | T122Ac (2) | + | + | − | + | |
| 22 | 1/12/05 | Cheshire, farm 2, cow 5 | NIA | − | + | − | + | |
| 23 | 15/06/06 | Cheshire, farm 2, cow 67, left hoof | Chronic | NIA | + | + | − | + |
| 24 | 15/06/06 | Cheshire, farm 2, cow 67, right hoof | Chronic | NIA | + | + | + | + |
| 25 | 7/11/06 | Cheshire, farm 1, cow 577 | NIA | + | + | − | + | |
| 26 | 20/11/06 | Cheshire, farm 3, cow 87 | Chronic | NIA | + | + | + | + |
| 27 | 20/11/06 | Cheshire, farm 3, cow 574 | Chronic | NIA | + | + | + | + |
| 28 | 20/11/06 | Cheshire, farm 3, cow 265 | Acute | NIA | + | + | − | + |
| 29 | 11/01/07 | Lancashire, farm 1, cow 1 | NIA | + | + | + | + | |
All lesions were on rear hoofs with typical BDD-type manifestation unless otherwise stated.
Isolations reported previously.
Previously unreported isolations.
IF, isolation failed; NIA, no isolation attempted.
TABLE 4.
PCR screen of BDD biopsies from a previous study from 1996 to 1997
| Sample | Biopsy date (yr) | Description | Result
|
|||
|---|---|---|---|---|---|---|
| Specific PCR for group:
|
Treponema PCR | |||||
| 1 | 2 | 3 | ||||
| 1 | 1996 | Cheshire, farm 1, cow 30 | − | + | + | + |
| 2 | 1996 | Cheshire, farm 1, cow 16 | + | + | − | + |
| 3 | 1996 | Cheshire, farm 1, cow 197 | + | + | + | + |
| 4 | 1997 | Cheshire, farm 1, cow 31 | + | + | + | + |
| 5 | 1997 | Cheshire, farm 1, cow 39 | + | + | + | + |
| 6 | 1997 | Cheshire, farm 1, cow 50 | + | + | + | + |
| 7 | 1997 | Cheshire, farm 1, cow 66 | + | + | + | + |
| 8 | 1996 | Cheshire, farm 2, cow 17 | + | + | − | + |
| 9 | 1997 | Cheshire, farm 2, cow 25 | + | + | + | + |
| 10 | 1997 | Cheshire, farm 2, cow 310 | + | + | + | + |
| 11 | 1997 | Cheshire, farm 2, cow 502 | + | + | + | + |
| 12 | 1996 | Cheshire, farm 3, cow 84 | + | + | + | + |
| 13 | 1997 | Cheshire, farm 3, cow 328 | + | − | − | + |
| 14 | 1997 | Cheshire, farm 3, cow 464 | + | + | + | + |
| 15 | 1996 | Cheshire, farm 3, cow 579 | + | + | + | + |
| 16 | 1997 | Cheshire, farm 3, cow 612 | + | + | + | + |
| 17 | 1997 | Cheshire, farm 3, cow 623 | + | + | + | + |
| 18 | 1997 | Cheshire, farm 3, cow 625 | + | + | + | + |
| 19 | 1997 | Cheshire, farm 3, cow 593 | + | + | + | + |
| 20 | 1996 | Cheshire, farm 4, cow 15 | + | + | + | + |
| 21 | 1997 | Cheshire, farm 4, cow 54 | + | + | + | + |
| 22 | 1997 | Cheshire, farm 4, cow 227 | + | + | − | + |
Including all 51 lesions tested, the group 1, 2, and 3 treponemes were present in 96.1%, 98%, and 76.5% of lesions, with all three groups present in 74.5% of lesions. All BDD lesion samples were positive for the general treponeme PCR.
PCR survey of healthy foot tissues.
Healthy (non-BDD) foot tissues were obtained from nine young bullocks, none of which had been reported to have suffered or had any symptoms of BDD. None of the biopsies from the young bullocks tested positive for any of the BDD-associated treponeme groups (Table 5). In order to test animals that had been present in the farmyard environment but did not have current BDD lesions, seven non-BDD cows that had been on farms that had endemic BDD were tested for the presence of the BDD treponemes in hoof tissues. For six of the seven samples tested, the PCRs did not reveal BDD treponemes in healthy hoof skin (Table 5). The single exception was one sample that tested positive for all three of the treponeme groups. Interestingly, all healthy hoof tissues, including that of the bullocks, tested positive by general treponeme PCR.
TABLE 5.
PCR detection of treponemes in hoof tissues from cows without BDD
| Sample (s) | Biopsy date (day/mo/yr) | Description | Result
|
|||
|---|---|---|---|---|---|---|
| Specific PCR for group:
|
Treponema PCR | |||||
| 1 | 2 | 3 | ||||
| 1-9 | 15/12/05 | Cheshire, farm 1, single rear foot from 8 young bullocks | − | − | − | + |
| 10 | 17/1/07 | Lancashire, farm 1, cow 182265 | − | − | − | + |
| 11 | 17/1/07 | Lancashire, farm 1, cow 00645 | + | + | + | + |
| 12 | 17/1/07 | Lancashire, farm 1, cow 600309 | − | − | − | + |
| 13 | 17/1/07 | Lancashire, farm 1, cow 391 | − | − | − | + |
| 14 | 17/1/07 | Lancashire, farm 1, cow 100233 | − | − | − | + |
| 15 | 17/1/07 | Lancashire, farm 1, cow 20560 | − | − | − | + |
| 16 | 17/1/07 | Lancashire, farm 1, cow 111B | − | − | − | + |
Isolation of further treponeme isolates.
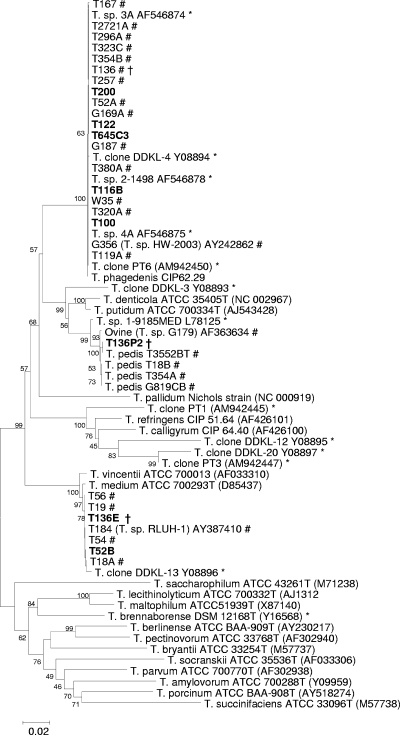
As part of this study, we isolated several more spirochetes, which are listed along with previous isolations from biopsies in Table 3. From the isolations listed, 12 of the 20 were reported previously (13). Strains T100, T122, and T116B were isolated as described previously and were confirmed as group 2 isolates, the most commonly isolated strains from BDD lesions (Fig. 1).
FIG. 1.
Phylogenetic tree based on 16S rRNA gene sequence comparison over ∼1,330 aligned bases showing relationships between strains isolated here (boldface type) and related 16S rRNA gene sequences. Bootstrap confidence levels are shown as percentages of nodes, and only values above 40% are shown. GenBank accession numbers are shown in parentheses next to each strain or 16S rRNA gene fragment clone. *, previously reported 16S rRNA gene sequences from BDD lesions; #, previously reported isolated BDD treponemes from this laboratory; †, strains from three different groups isolated from the same lesion.
It was hypothesized that the successful development of group-specific PCRs would enable further isolations from primary BDD biopsy cultures, which we had considered to contain mixed cultures of two or more treponeme groups. After reviewing the data suggesting that many lesions contained all three phylogroups, we used the PCR assays to test original initial primary cultures of strains for the three phylogroups. We then used group-specific serum and growth conditions described previously (13), with several rounds of plating and subculture to isolate further strains. In this approach, the group-specific PCRs were a powerful tool for the identification of isolate purity. A group 2 isolate, T200, was isolated from a culture that had previously failed. A group 1 isolate, T52B, was isolated from a lesional culture which had previously yielded only a group 2 treponeme (T52A). In one instance, we managed to isolate a further two treponemes, one group 1 strain (T136E) and one group 3 strain (T136p1), from a culture from which we had already isolated one group 2 isolate (T136). All the newly found isolates had their 16S rRNA genes sequenced and clustered with their respective groups upon phylogenetic analysis (Fig. 1). In Table 3, where isolation attempts failed, this was typically due to heavy contamination, which we could not remove, as opposed to no treponemal presence (as determined by phase-contrast microscopy).
Immunohistochemistry of healthy and infected bovine foot tissues.
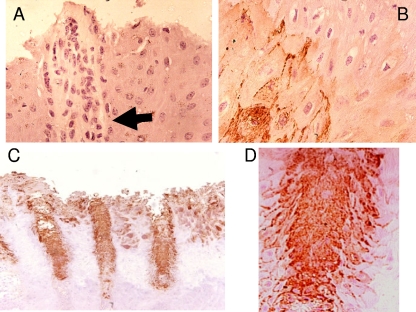
Healthy foot tissues showed no treponemal presence by immunohistochemistry using antisera raised against the BDD treponemes (Fig. 2A). In comparison, lesional tissues from BDD cases displayed very strong staining with the antitreponemal antisera. This was apparent particularly in the deep layers of the lesion (Fig. 2B) and, unexpectedly, in the hair follicles and sebaceous glands (Fig. 2C and D). This staining pattern was seen in all the cases tested. Differences were seen in the sublocalization of the treponemes: in the hair follicles, the treponemes appeared to be both intra- and extracellular, and in the surrounding tissues, they were almost entirely extracellular in location.
FIG. 2.
Immunohistochemical staining of treponemes in digital dermatitis lesions. (A) Staining of healthy hoof skin (hair follicle is identified by the arrow). (B) Staining of deep dermal layers showing treponemes tracking between cells. (C) Staining of hoof skin showing strong staining of hair follicles. (D) Staining of hair follicle showing tracking of infection from hair follicles into surrounding tissues.
EM of infected tissues.
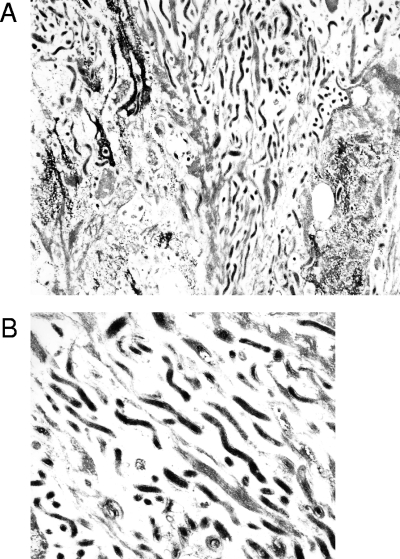
Confirmation that the antitreponeme antisera were detecting treponemes in these tissues was provided by EM studies, which provided characteristic treponemal morphology both in deep lesional tissues and in hair follicles (Fig. 3). No treponemes were observed by EM in healthy foot tissues (data not shown).
FIG. 3.
EM of hair follicle from hoof skin of a cow with BDD. (A) Treponemes tracking in parallel inside follicle at a 1:9,000 magnification. (B) Characteristic treponeme morphology in follicle at a 1:22,500 magnification.
DISCUSSION
One of the main aims of this study was to design and implement PCR tests that were capable of detecting treponemes, which we have previously identified as being associated with BDD (13). This would provide a powerful molecular epidemiology tool to help understand the involvement of individual organisms in the disease process. Immunohistochemistry and EM were also used to further delineate some relationships between the treponemes and foot infection in cows with BDD.
The development of the PCR tests was successful, and phylogroup-specific primers and PCR conditions were readily determined. The first use of these assays was to identify the distribution of the three phylogroups within lesions from cattle from different regions of the United Kingdom. This indicated the presence of two (or, most commonly, three) phylogroups in BDD lesions. Isolation data from previous studies suggested that the group 2 isolates (T. phagedenis-like) might be the only phylogroup specifically associated with BDD, as they have been the most commonly isolated ones (13, 32, 33). The current data clearly refute this premise, as the new PCR tests indicated the presence of group 2 treponemes in nearly all BDD lesions (98%) and detected group 1 treponemes (T. medium/T. vincentii-like) in the majority (96.1%) of BDD lesions. In comparison, the group 3 treponemes (T. putidum/T. denticola-like) were present in 74.5% of lesion biopsies. Previously, group 3 treponemes were identified only at the exterior of lesions, suggesting that they may prefer outer skin layers, while group 1 and group 2 isolates have been identified deeper within lesions (22). Therefore, we are unsure whether the comparatively lower association of group 3 isolates with BDD represents variations in sampling location and/or lesion age or whether the data reduce the importance of the group 3 isolates in the etiology of the disease. Interestingly, the healthy foot tissues were universally negative for any of the BDD-associated treponemes, although other treponemes were readily detected in these samples.
The high level of association of the group 1 and group 2 isolates with BDD is very interesting, especially as the association of oral spirochetes with periodontal infections is typically reported at a considerably lower percentage. For example, using a nested PCR method similar to that described here, T. denticola was found to be present in ∼79% of human endodontic abscesses, while Treponema socranskii was the next most prevalent at 26%, and T. medium was present in only 5% of samples (30). Similar percent associations were also previously reported for human primary root canal infections (26). Hence, the results presented here argue that the infection etiology of this disease may not be as polymicrobial in origin as that encountered with oral infections and implicate group 1 and 2 treponemes as being highly associated with infection. It would be interesting to postulate that these two treponemes are required together as a pathogenic complex to cause BDD, which might explain why the disease has been transmitted using lesion material (24) but why no single isolate has yet been reported to fulfill Koch's postulates.
Despite attempts to locate primers at unique sites on the 16S rRNA gene locus, the group 1 and group 2 treponeme primers still cross-reacted with their nearest relatives (T. medium and T. phagedenis, respectively). We expected cross-reactions with the nearest relatives of group 1 and 2 treponemes, as the respective 16S rRNA gene sequences are nearly identical. In fact, the taxonomic relationship with the closest relatives has still not been answered, although we have recently proposed the group 3 isolates as a novel species, “Treponema pedis” (12). The nearly identical 16S rRNA genes of these bovine and human treponemes could be considered a limitation, and future PCR methods might warrant the use of alternative genetic loci. It should be noted that the different Treponema pallidum subspecies that cause very different diseases in humans (e.g., syphilis and yaws) are distinguished thus far only by genes encoding putative surface-exposed proteins (4).
Retrospective isolations were carried out on initial cultures when the PCR assays identified more than one phylogroup present. Using the PCR protocol and further isolations, we have obtained several new strains from initial cultures, and when detected, we have been able to isolate these strains. To verify that the strains isolated belong to their expected PCR groups, these new isolates had their 16S rRNA genes sequenced, and it was confirmed that they belong to their respective groups. This is further validation of this PCR-based approach.
The PCR assays that we have developed can now be used to investigate environmental samples for potential transmission routes as well as whole-animal surveys for possible infection reservoir identifications. Furthermore, preliminary evidence presented here showed that acute lesions may be the result of only two phylotypes of treponeme, T. medium-like and T. phagedenis-like. Evidence of T. medium-like and T. phagedenis-like spirochetes being located deep within lesions has been identified before using fluorescent in situ hybridization (FISH) (22). Further studies of acute versus chronic lesions would give more evidence of whether this is a significant association and might allow the identification of the primary invader(s) so as to specifically target and eradicate BDD once and for all. Further PCR tests for other treponemes isolated from BDD lesions, such as Treponema brennaborense (29), and a global survey of BDD lesions would also be very valuable. These PCR assays could also be used to further investigate the ovine manifestation of DD called CO DD. One of our isolates (G179) is from a CO DD lesion, and the current work confirms it as a group 3 treponeme, as previously suggested (13). This result adds strength to the suggestion that the sheep form of the disease has been transmitted from the cow or a shared intermediary host (9).
In terms of the etiopathogenesis and molecular epidemiology of BDD, the most striking finding is that 75% of BDD lesions contain representatives of all three BDD treponeme groups. This is not readily explainable, unless a symbiotic relationship is optimal for disease induction and maintenance. Symbiosis of treponemes in BDD lesions has been used to explain recent results from FISH studies (17, 23), and further research into such a symbiosis might be appropriate. As a result of this study, we may be one step closer to dissecting such a symbiotic pathogenesis given that we have been able to isolate members of all three groups from the same lesion. However, it is important that a recent German BDD FISH study identified five groups as being the most important in BDD etiology (23), with three of the groups resembling the treponemes that we have isolated. Furthermore, a larger variety of treponemes have been identified in Danish BDD lesions, although only three groups were identified as being the most prevalent (17) and only one of the groups resembles the treponemes which we isolated, suggesting regional differences in BDD etiology.
Both PCR and immunohistochemistry suggest that healthy tissues on bovine feet do not (apart from one cow tested) harbor the BDD treponemes, although other treponemes are readily detected. Hence, the BDD treponemes do not appear to be commensals and/or opportunistic invaders. Indeed, it is likely that they are highly developed to exist in the environmental niche of the bovine foot, although they may also be present in large numbers in slurry.
If the treponemes are the primary infectious agent in BDD, they will need to be transmitted from a reservoir to foot tissues. While the reservoir for these organisms has not yet been identified, the immunohistochemistry shown here clearly indicates that the treponemes may be entering (and may be exiting) via hair follicles and/or sebaceous glands. This route of transmission would explain how they are able to breach such a hard physical barrier to establish infection in deep-lying tissues. This finding is also possibly relevant to the polytreponemal nature of BDD in that in most foot tissues, the organisms appear to be largely extracellular, and in hair follicles, they are extracellular and intracellular. The data presented in this paper justifies further investigation to attempt to identify routes of transmission of each of the three treponeme groups and how that may lead to the initiation of the BDD disease process.
Acknowledgments
This work was funded by a Department for Environment, Food, and Rural Affairs animal welfare grant (grant AW1010) and a Biotechnology and Biological Sciences Research Council CEDFAS research grant (grant BBE0189201).
Footnotes
Published ahead of print on 14 January 2009.
REFERENCES
- 1.Argaez-Rodriguez, F. J., D. W. Hird, J. Hernandez de Anda, D. H. Read, and A. Rodriguez-Lainz. 1997. Papillomatous digital dermatitis on a commercial dairy farm in Mexicali, Mexico: incidence and effect on reproduction and milk production. Prev. Vet. Med. 32275-286. [DOI] [PubMed] [Google Scholar]
- 2.Blowey, R. W., S. H. Done, and W. Cooley. 1994. Observations on the pathogenesis of digital dermatitis in cattle. Vet. Rec. 135115-117. [DOI] [PubMed] [Google Scholar]
- 3.Blowey, R. W., and M. W. Sharp. 1988. Digital dermatitis in dairy cattle. Vet. Rec. 122505-508. [DOI] [PubMed] [Google Scholar]
- 4.Centurion-Lara, A., B. J. Molini, C. Godornes, E. Sun, K. Hevner, W. C. Van Voorhis, and S. A. Lukehart. 2006. Molecular differentiation of Treponema pallidum subspecies. J. Clin. Microbiol. 443377-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, B. K., H. Nattermann, S. Grund, W. Haider, and U. B. Gobel. 1997. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int. J. Syst. Bacteriol. 47175-181. [DOI] [PubMed] [Google Scholar]
- 6.Demirkan, I., S. D. Carter, R. D. Murray, R. W. Blowey, and M. J. Woodward. 1998. The frequent detection of a treponeme in bovine digital dermatitis by immunocytochemistry and polymerase chain reaction. Vet. Microbiol. 60285-292. [DOI] [PubMed] [Google Scholar]
- 7.Demirkan, I., S. D. Carter, C. Winstanley, K. D. Bruce, N. M. McNair, M. Woodside, and C. A. Hart. 2001. Isolation and characterisation of a novel spirochaete from severe virulent ovine foot rot. J. Med. Microbiol. 501061-1068. [DOI] [PubMed] [Google Scholar]
- 8.Demirkan, I., H. F. Williams, A. Dhawi, S. D. Carter, C. Winstanley, K. D. Bruce, and C. A. Hart. 2006. Characterization of a spirochaete isolated from a case of bovine digital dermatitis. J. Appl. Microbiol. 101948-955. [DOI] [PubMed] [Google Scholar]
- 9.Dhawi, A., C. A. Hart, I. Demirkan, I. H. Davies, and S. D. Carter. 2005. Bovine digital dermatitis and severe virulent ovine foot rot: a common spirochaetal pathogenesis. Vet. J. 169232-241. [DOI] [PubMed] [Google Scholar]
- 10.Dopfer, D., A. Koopmans, F. A. Meijer, I. Szakall, Y. H. Schukken, W. Klee, R. B. Bosma, J. L. Cornelisse, A. J. van Asten, and A. A. ter Huurne. 1997. Histological and bacteriological evaluation of digital dermatitis in cattle, with special reference to spirochaetes and Campylobacter faecalis. Vet. Rec. 140620-623. [DOI] [PubMed] [Google Scholar]
- 11.Ehresmann, C., P. Stiegler, G. A. Mackie, R. A. Zimmermann, J. P. Ebel, and P. Fellner. 1975. Primary sequence of the 16S ribosomal RNA of Escherichia coli. Nucleic Acids Res. 2265-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, N. J., J. M. Brown, I. Demirkan, R. D. Murray, R. J. Birtles, C. A. Hart, and S. D. Carter. Treponema pedis sp. nov., a novel spirochete isolated from bovine digital dermatitis lesions. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 13.Evans, N. J., J. M. Brown, I. Demirkan, R. D. Murray, W. D. Vink, R. W. Blowey, C. A. Hart, and S. D. Carter. 2008. Three unique groups of spirochetes isolated from digital dermatitis lesions in UK cattle. Vet. Microbiol. 130141-150. [DOI] [PubMed] [Google Scholar]
- 14.Glauert, A. M. 1975. Fixation, dehydration and embedding of biological specimens. Elsevier, North-Holland Biomedical Press, Amsterdam, The Netherlands.
- 15.Hernandez, J., J. K. Shearer, and D. W. Webb. 2002. Effect of lameness on milk yield in dairy cows. J. Am. Vet. Med. Assoc. 220640-644. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez, J., J. K. Shearer, and D. W. Webb. 2001. Effect of lameness on the calving-to-conception interval in dairy cows. J. Am. Vet. Med. Assoc. 2181611-1614. [DOI] [PubMed] [Google Scholar]
- 17.Klitgaard, K., M. Boye, N. Capion, and T. K. Jensen. 2008. Evidence of multiple Treponema phylotypes involved in bovine digital dermatitis as shown by 16S rRNA gene analysis and fluorescence in situ hybridization. J. Clin. Microbiol. 463012-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koniarova, I., A. Orsag, and V. Ledecky. 1993. The role anaerobes in dermatitis digitalis and interdigitalis in cattle. Vet. Med. (Praha) 38589-596. (In Slovak.) [PubMed] [Google Scholar]
- 19.Laven, R. A., and D. N. Logue. 2006. Treatment strategies for digital dermatitis for the UK. Vet. J. 17179-88. [DOI] [PubMed] [Google Scholar]
- 20.Losinger, W. C. 2006. Economic impacts of reduced milk production associated with papillomatous digital dermatitis in dairy cows in the USA. J. Dairy Res. 73244-256. [DOI] [PubMed] [Google Scholar]
- 21.Moore, L. J., M. J. Woodward, and R. Grogono-Thomas. 2005. The occurrence of treponemes in contagious ovine digital dermatitis and the characterisation of associated Dichelobacter nodosus. Vet. Microbiol. 111199-209. [DOI] [PubMed] [Google Scholar]
- 22.Moter, A., G. Leist, R. Rudolph, K. Schrank, B. K. Choi, M. Wagner, and U. B. Gobel. 1998. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology 1442459-2467. [DOI] [PubMed] [Google Scholar]
- 23.Nordhoff, M., A. Moter, K. Schrank, and L. H. Wieler. 2008. High prevalence of treponemes in bovine digital dermatitis—a molecular epidemiology. Vet. Microbiol. 131293-300. [DOI] [PubMed] [Google Scholar]
- 24.Read, D. H., and R. L. Walker. 1996. Experimental transmission of papillomatous digital dermatitis (footwarts) in cattle. Vet. Pathol. 33607. [Google Scholar]
- 25.Read, D. H., R. L. Walker, A. E. Castro, J. P. Sundberg, and M. C. Thurmond. 1992. An invasive spirochaete associated with interdigital papillomatosis of dairy cattle. Vet. Rec. 13059-60. [DOI] [PubMed] [Google Scholar]
- 26.Rocas, I. N., J. F. Siqueira, Jr., A. F. Andrade, and M. Uzeda. 2003. Oral treponemes in primary root canal infections as detected by nested PCR. Int. Endod. J. 3620-26. [DOI] [PubMed] [Google Scholar]
- 27.Rurangirwa, F. R., P. M. Dilbeck, T. B. Crawford, T. C. McGuire, and T. F. McElwain. 1999. Analysis of the 16S rRNA gene of micro-organism WSU 86-1044 from an aborted bovine foetus reveals that it is a member of the order Chlamydiales: proposal of Waddliaceae fam. nov., Waddlia chondrophila gen. nov., sp. nov. Int. J. Syst. Bacteriol. 49577-581. [DOI] [PubMed] [Google Scholar]
- 28.Sabo, J., A. Hudac, and E. Fendtova. 1988. Ecology of anaerobic non-sporulating bacteria in relation to dermatitis digitalis in beef cattle. Vet. Med. (Praha) 33265-272. (In Slovak.) [PubMed] [Google Scholar]
- 29.Schrank, K., B. K. Choi, S. Grund, A. Moter, K. Heuner, H. Nattermann, and U. B. Gobel. 1999. Treponema brennaborense sp. nov., a novel spirochaete isolated from a dairy cow suffering from digital dermatitis. Int. J. Syst. Bacteriol. 4943-50. [DOI] [PubMed] [Google Scholar]
- 30.Siqueira, J. F., Jr., and I. N. Rocas. 2004. Treponema species associated with abscesses of endodontic origin. Oral Microbiol. Immunol. 19336-339. [DOI] [PubMed] [Google Scholar]
- 31.Stamm, L. V., H. L. Bergen, and R. L. Walker. 2002. Molecular typing of papillomatous digital dermatitis-associated Treponema isolates based on analysis of 16S-23S ribosomal DNA intergenic spacer regions. J. Clin. Microbiol. 403463-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trott, D. J., M. R. Moeller, R. L. Zuerner, J. P. Goff, W. R. Waters, D. P. Alt, R. L. Walker, and M. J. Wannemuehler. 2003. Characterization of Treponema phagedenis-like spirochetes isolated from papillomatous digital dermatitis lesions in dairy cattle. J. Clin. Microbiol. 412522-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker, R. L., D. H. Read, K. J. Loretz, and R. W. Nordhausen. 1995. Spirochetes isolated from dairy cattle with papillomatous digital dermatitis and interdigital dermatitis. Vet. Microbiol. 47343-355. [DOI] [PubMed] [Google Scholar]
- 34.Whay, H. R., A. E. Waterman, and A. J. Webster. 1997. Associations between locomotion, claw lesions and nociceptive threshold in dairy heifers during the peri-partum period. Vet. J. 154155-161. [DOI] [PubMed] [Google Scholar]