Abstract
The increasing use of molecular methods strongly motivates clinical laboratories to introduce automated nucleic acid extractors. We compared the easyMAG (bioMérieux) with a manual extraction method for hepatitis C virus (HCV) load quantification (RealTime HCV; Abbott). Both methods were comparable, and, therefore, the easyMAG is suitable to be implemented in our laboratory for the management of HCV-infected patients.
Hepatitis C virus (HCV) infects about 180 million people worldwide (11) and is the leading cause of chronic liver disease. Apart from diagnosis of infection (5), HCV load testing is routinely performed for baseline evaluation, treatment follow-up, and outcome assessment (2, 3, 6, 9, 12). Since molecular assays are also used for many other infectious diseases, many laboratories are moving to automated nucleic acid extraction. However, each assay manufacturer recommends using a certain extractor. Due to space limitations, ideally a single extractor should provide nucleic acids for any molecular application, regardless of the microorganism of interest and the specimen type.
Our laboratory uses a CE-marked real-time PCR assay (RealTime HCV; Abbott Molecular, Inc., Des Plaines, IL) that was originally evaluated with a magnetically based manual extraction procedure and, more recently, with different extractors, including Abbott m1000 and m2000sp, Tecan Freedom EVO Clinical, and Qiagen BioRobot EZ1 (7, 8, 10), but not easyMAG (bioMérieux, Boxtel, The Netherlands) (4). We have been using this instrument with success for other microorganisms and different clinical specimens. Since we wanted to change from manual to automated extraction without introducing other extractors, the aim of our study was to evaluate the easyMAG for the quantification of HCV load using the RealTime HCV assay in comparison with manual extraction.
The correlation between manual and automated extraction was compared in four runs, each including nine different sera from HCV-infected patients and three controls (negative, low positive, and high positive) for a total of 36 determinations. Sera were retrospectively selected spanning a wide HCV RNA virus load range, from <30 IU/ml—the lower quantification limit—up to 50,000,000 IU/ml, and negative specimens. The Clinical Research Ethics Committee at our institution approved this study. Several analytical parameters were assessed for the real-time PCR assay in combination with the extractor. (i) The first was lower detection limit. Twenty replicates of 25-, 12.5-, and 6.25-IU/ml dilutions obtained from the HCV RNA high-positive control (2 × 106 IU/ml; AcroMetrix Corporation, Benicia, CA) in negative plasma were tested per run. (ii) The second was linearity. Five replicates of five 10-fold dilutions (102 to 106 IU/ml) from the same standard were tested in one run. (iii) The third was intra-assay reproducibility. Five replicates of calibrators A and B (1,000 and 1.07 × 107 IU/ml, respectively; Abbott Molecular) and a dilution of the high-positive control (50,000 IU/ml) were tested. (iv) The fourth was interassay reproducibility. Five additional replicates of the same three dilutions were tested in another two runs performed on different days, adding up to a total of three runs. Negative, low-positive, and high-positive controls were included in each run.
Manual RNA extraction was performed as recommended by the manufacturer (Abbott RealTime HCV, Ref. 4J86; Abbott Molecular, Inc., Des Plaines, IL) from 200 μl of serum and elution in 88 μl. The internal control provided (unrelated armored RNA) was added to the lysis buffer at a proportion of 17 μl per sample. EasyMAG reagents are based on Boom's method for total nucleic acid extraction (1). Extraction was performed as recommended by the manufacturer (NucliSENS easyMAG user manual, v 1.1; BioMérieux, Boxtel, The Netherlands). A 250-μl sample volume and 110-μl elution volume were used to maintain the same sample/elution volume ratio as well as the proportion of internal control per sample. (A total of 235.7 μl of internal control was added to 314.3 μl of elution buffer and 550 μl of silica, and 100 μl of this premix was used per sample.)
Virus load quantification was performed using the RealTime HCV assay according to the manufacturer's instructions with the Abbott m2000rt instrument (Abbott RealTime HCV, Ref. 4J86). Separate calibration curves were obtained for manual and automated extraction methods with triplicate calibrators A and B.
Correlation between extraction methods was assessed through Deming regression and Bland-Altman plots with MedCalc software v9.6.2.0 (Mariakerke, Belgium). All other analyses were performed with SPSS v14.0 (SPSS, Inc., Chicago, IL), with a significance level of 0.05. Probit analysis was used to determine the lower limit of detection. Linearity was assessed through linear regression between expected and observed values and the Pearson correlation coefficient (r).
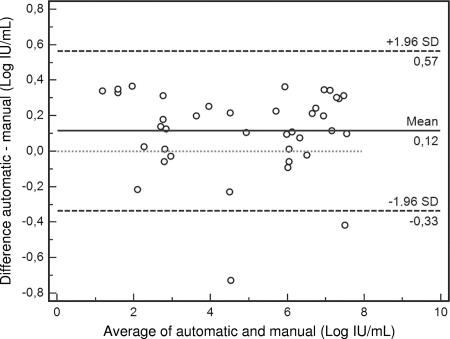
For the correlation study, a total of 48 sera from HCV-infected patients and controls were extracted in parallel by both manual and automated methods. Three clinical specimens were negative by both methods, as were the four negative controls, which confirmed the absence of carryover contamination. Another three specimens were positive at <30 IU/ml by manual extraction; one of them was also positive at <30 IU/ml, and the other two had virus loads of 55 and 58 IU/ml by automatic extraction. The real-time PCR was inhibited in two specimens: one of them after both manual and automated extractions and the other only after automatic extraction. A total of 36 specimens and controls with viral loads quantified after both manual and automatic extractions were included in the regression analysis. Deming regression showed that the correlation between the two methods was remarkable (y = 0.01379 + 1.0163 x, r = 0.993, P < 0.01): they did not differ by a constant amount (y intercept not significantly different from 0; 95% confidence interval, −0.2135 to 0.2411), and there was not a proportional difference between them either (slope not significantly different from 1; 95% confidence interval, 0.9749 to 1.0577). Bland-Altman analysis (Fig. 1) only identified two outliers, possibly due to a sporadic technical error; the mean difference between the two methods differed from the line of equality by 0.12 log10 IU/ml, which does not have a significant effect on therapeutic decisions.
FIG. 1.
Bland-Altman plot of differences versus average of values obtained after automatic and manual extraction with 95% limits of agreement and the line of equality (dotted line).
Regarding the lower limit of detection of the PCR assay with the automated extraction method, a virus load of 14.5 IU/ml (1.16 log10 IU/ml) could be detected with a 95% probability according to Probit analysis. This result is similar to the lower detection limit reported by the manufacturer for this assay with manual extraction (30 IU/ml or 1.48 log10 IU/ml). We were unable to obtain clinical specimens with virus loads of >7.6 log10 IU/ml to assess the upper linear limit of the assay (8 log10 IU/ml). However, linearity through the range of viral loads studied was remarkable (y = 0.415 + 0.949 x, r = 0.992, P < 0.01).
Results for the inter- and intra-assay reproducibility experiments are shown in Table 1. The variation observed after automated extraction was similar to that reported by the manufacturer for the real-time PCR technique with manual extraction over the range of viral loads tested (0.05 to 0.09 and 0.01 to 0.08 standard deviations in log10 IU/ml for the intra-assay and interassay reproducibility studies, respectively) (Abbott RealTime HCV, Ref. 4J86). Our results were comparable to those obtained with m1000 and EZ1 extractors in recent studies (7, 8), with intra- and interassay coefficients of variation of ≤2.4% for both instruments.
TABLE 1.
Intra- and interassay reproducibility of the real-time PCR method using specimens extracted with easyMAG
| Reproducibility | Sample testeda | No. of replicates | Expected HCV virus load (log10 IU/ml) | Observed mean HCV virus load ± SD (log10 IU/ml) | Coefficient of variation (%) |
|---|---|---|---|---|---|
| Intra-assay | Calibrator A | 5 | 3.00 | 3.03 ± 0.06 | 1.83 |
| Standard | 5 | 4.70 | 4.77 ± 0.02 | 0.23 | |
| Calibrator B | 5 | 7.03 | 7.01 ± 0.02 | 0.35 | |
| Interassay | Calibrator A | 15 | 3.00 | 3.01 ± 0.05 | 1.66 |
| Standard | 15 | 4.70 | 4.64 ± 0.10 | 2.06 | |
| Calibrator B | 15 | 7.03 | 6.96 ± 0.04 | 0.59 |
The calibrators were supplied by Abbott Molecular: calibrator A, 1,000 IU/ml; calibrator B, 1.07 × 107 IU/ml. The standard represents the dilution of the HCV RNA high-positive control with a virus load of 5.0 ×104 IU/ml.
The combination of real-time PCR and nucleic acid extraction platforms results in almost full automation, better reproducibility, and manual labor savings. Our study demonstrates that the combination of nucleic acid extraction with the easyMAG system followed by HCV RNA quantification with the Abbott RealTime HCV assay is suitable to be implemented in our laboratory for the management of HCV-infected patients without modifying therapeutic decisions with respect to the manual extraction method. Furthermore, easyMAG is an easy to use platform that extracts total nucleic acids from 24 specimens in an hour, providing the high-throughput processing needed in our laboratory.
Acknowledgments
This work was partially funded by bioMérieux; grant CD05/00258 (E.M.) (Contratos Postdoctorales de Perfeccionamiento) from the Ministerio de Sanidad y Consumo, within the Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica (I+D+I); and grant 2006FI-00534 (V.S.) from the Comissionat per a Universitats i Recerca del Departament d'Innovació, Universitats i Empresa de la Generalitat de Catalunya i del Fons Social Europeu (Spain).
Footnotes
Published ahead of print on 7 January 2009.
REFERENCES
- 1.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halfon, P., M. Bourlière, G. Pénaranda, H. Khiri, and D. Ouzan. 2006. Real-time PCR assays for hepatitis C virus (HCV) RNA quantitation are adequate for clinical management of patients with chronic HCV infection. J. Clin. Microbiol. 442507-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee, S. S., E. J. Heathcote, K. R. Reddy, S. Zeuzem, M. W. Fried, T. L. Wright, P. J. Pockros, D. Haussinger, C. I. Smith, A. Lin, and S. C. Pappas. 2002. Prognostic factors and early predictability of sustained viral response with peginterferon alfa-2a (40KD). J. Hepatol. 37500-506. [DOI] [PubMed] [Google Scholar]
- 4.Loens, K., K. Bergs, D. Ursi, H. Goossens, and M. Ieven. 2007. Evaluation of NucliSens easyMAG for automated nucleic acid extraction from various clinical specimens. J. Clin. Microbiol. 45421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Institutes of Health. 2006. National Institutes of Health Consensus Development Conference statement. Management of hepatitis C: 2002-2004. National Institutes of Health, Bethesda, MD. http://consensus.nih.gov.
- 6.Sanchez-Tapias, J. M., M. Diago, P. Escartin, J. Enriquez, M. Romero-Gomez, R. Barcena, J. Crespo, R. Andrade, E. Martinez-Bauer, R. Perez, M. Testillano, R. Planas, R. Sola, M. Garcia-Bengoechea, J. Garcia-Samaniego, M. Munoz-Sanchez, and R. Moreno-Otero. 2006. Peginterferon-alfa2a plus ribavirin for 48 versus 72 weeks in patients with detectable hepatitis C virus RNA at week 4 of treatment. Gastroenterology 131451-460. [DOI] [PubMed] [Google Scholar]
- 7.Schneider, G., K. Kuper, K. Abravaya, C. Mullen, M. Schmidt, and M. Sprenger-Haussels. 2007. Performance evaluation of the QIAGEN EZ1 DSP virus kit with Abbott RealTime HIV-1, HCV and HBV assays. Abstr. 17th Eur. Congr. Clin. Microbiol. Infect. Dis. European Society of Clinical Microbiology and Infectious Diseases, Paris, France. [DOI] [PubMed]
- 8.Schutten, M., E. Fries, C. Burghoorn-Maas, and H. G. Niesters. 2007. Evaluation of the analytical performance of the new Abbott RealTime RT-PCRs for the quantitative detection of HCV and HIV-1 RNA. J. Clin. Virol. 4099-104. [DOI] [PubMed] [Google Scholar]
- 9.Shiffman, M. L., F. Suter, B. R. Bacon, D. Nelson, H. Harley, R. Sola, S. D. Shafran, K. Barange, A. Lin, A. Soman, and S. Zeuzem. 2007. Peginterferon alfa-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3. N. Engl. J. Med. 357124-134. [DOI] [PubMed] [Google Scholar]
- 10.Tiemann, C., D. Knoop, C. Dunn, C. Huber, and S. Schaffer. 2007. Performance evaluation of a new instrument for genomic viral DNA or RNA extraction with HIV-1 and HCV viral load assays, abstr. PCR007. Abstr. 10th Annu. Eur. Soc. Clin. Virol. Meet. European Society for Clinical Virology, Leuven, Belgium.
- 11.World Health Organization. 2005. State of the art vaccine research development, p.81-88. World Health Organization, Geneva, Switzerland. http://www.who.int/vaccine_research/documents/stateoftheart/en/index.html.
- 12.Zeuzem, S., M. Buti, P. Ferenci, J. Sperl, Y. Horsmans, J. Cianciara, E. Ibranyi, O. Weiland, S. Noviello, C. Brass, and J. Albrecht. 2006. Efficacy of 24 weeks treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C infected with genotype 1 and low pretreatment viremia. J. Hepatol. 4497-103. [DOI] [PubMed] [Google Scholar]