Abstract
An outbreak of acute respiratory tract infection occurred in Shanxi Province, China, from March to April 2006. Of the 254 patients affected by this outbreak, 247 patients were students of a senior high school; 1 of these patients died during the outbreak. Serological tests and blood culture revealed no evidence of bacterial infection. The results of direct reverse transcription-PCR or PCR performed with clinical specimens collected from the patients, including the sole patient who died, were positive for human adenoviruses (HAdVs) but negative for influenza virus, measles virus, rubella virus, mumps virus, parainfluenza virus, respiratory syncytial virus, and human enteroviruses. These findings were confirmed by enzyme-linked immunosorbent assay for HAdV immunoglobulin A, the conventional neutralization test, and viral isolation and identification. Sequencing of the entire hexon gene revealed that HdAV type 11a (HAdV-11a) belonging to the B2 species of HAdV was the etiological agent responsible for the outbreak. However, both the analysis of the phylogenetic relationship and the similarity plot indicated that the sequence of the 3′ end of the hexon gene outside the hypervariable regions the HAdV-11a strain isolated in this outbreak may be a recombinant with the sequence of the HAdV-14 strain of species B2. Although isolates of HAdV species B2 seldom cause respiratory infections, they may pose a new global challenge with regard to acute respiratory diseases; this possibility cannot be overlooked and should be carefully considered. Hence, the need to establish and improve both epidemiological and virological surveillance of HAdV infections in China should be emphasized.
Human adenoviruses (HAdVs) are ubiquitous and are responsible for a broad spectrum of clinical diseases; furthermore, depending on the infections caused by the different serotypes, these viruses may cause uncomplicated acute respiratory and gastrointestinal infections in healthy individuals and chronic systemic infections in immunosuppressed hosts. The incidence of severe disease due to HAdV is the highest among young children and immunocompromised persons, especially transplant recipients (3, 15, 23).
Adenoviruses are nonenveloped particles with linear double-stranded DNA. The viral capsid is composed of three main proteins: hexons, penton bases, and fibers. Hexon proteins comprise 919 to 968 amino acids (22) containing serotype-specific epitopes encoded by seven hypervariable regions (HVRs) (6), which are recognized as the most important components of the viruses for serotype identification (24, 26).
Thus far, 52 serotypes of HAdVs have been characterized and classified into seven subgroups or species (subgroups A to G) of the genus Mastadenovirus on the basis of their biological properties and DNA sequence homology (1, 12). HAdV species B has been further classified into two subspecies, namely, B1 (HAdV type 3 [HAdV-3], HAdV-7, HAdV-16, HAdV-21, and HAdV-50) and B2 (HAdV-11, HAdV-14, HAdV-34, and HAdV-35). The B1 viruses are usually associated with respiratory tract infections, while the B2 viruses, except for HAdV-11a and HAdV-14, are associated with kidney and urinary tract infections (16, 21).
The occurrence of repeated outbreaks of HAdV-associated diseases has been described in young people, especially senior high school students, in China. These patients were reported to have an acute respiratory disease that was usually caused by HAdV-3 and HAdV-7 (B1 species) (8, 28, 31). Here, we describe an outbreak of acute respiratory tract infection in a senior high school (a combination of a boarding and commuter school) in Shanxi Province, China, from March to April 2006. Clinical specimens were collected from the patients and healthy people to identify the etiological agent responsible for this outbreak. A detailed analysis of the specimens was performed with respect to the etiology. The identification and molecular characterization of the HAdV strains of species B that were isolated from the specimens constituted the main objective of this study. We concluded that the outbreak of respiratory disease in the senior high school in the Qishan County, Shanxi Province, China, was caused by HAdV-11a, which belongs to species B2.
CASE REPORTS
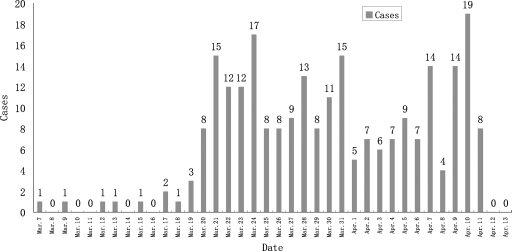
On 7 March 2006, a student from a senior high school (a combination of a boarding and commuter school) in Qishan County, Shanxi Province, China, experienced febrile symptoms accompanied by a body temperature of approximately 38.5°C. Antibiotic therapy with penicillin was initiated at a hospital, but the symptoms did not alleviate. The student recovered 6 days after the treatment was altered to virazole administered in combination with penicillin. Subsequently, students from the same senior high school presented with fever (body temperature, >38°C), cough, pharyngalgia, nasal obstruction, nasal discharge, headache, dizziness, and mild hemoptysis; fever was the initial symptom in all patients. The number of cases increased dramatically by 20 March. By 13 April 2006, a total of 254 patients (age range, 16 to 22 years), identified on the basis of a case definition and by conducting an active search, were affected by this outbreak; 247 patients were from the same senior high school as the first patient, and the other 7 patients were young adults from the same town. As seen in the bar graph in Fig. 1, the incidence of the relevant cases peaked three times within the duration of the outbreak. The findings of physical examinations and routine blood tests were unremarkable, while chest radiographs revealed increased lung markings and dense shadows indicative of plaque. The patients recovered quickly when they were provided with treatment for their symptoms, and the prognosis was good.
FIG. 1.
Case distribution in the senior high school during the outbreak. On 7 March 2006, a student from a senior high school in Qishan County, Shanxi Province, presented with febrile symptoms. The number of similar cases increased dramatically by 20 March. By 13 April 2006, the outbreak affected a total of 247 students of the same senior high school where the first patient attended school; these cases were identified on the basis of a case definition and by conducting an active search. The duration of the outbreak consisted of three phases of peak incidence, with approximately 1-week intervals between the phases.
During the outbreak, an 18-year-old student from the senior high school concerned died. On 26 March 2006, this patient presented with fever and a body temperature of approximately 37.8°C. He was hospitalized 5 days later, and antibiotics and antiviral therapy were administered. A chest radiograph revealed a patchy shadow over the lung. On 3 April, the patient's condition deteriorated and he developed shock, respiratory failure, and heart failure. The patient was continuously provided with systemic treatment consisting of antibiotics and antiviral therapy; however, this treatment did not prevent the worsening of his condition and he died 3 days later. A postmortem examination found a condition of bone marrow megaloblastic anemia complicated by the HAdV-11-induced acute respiratory disease, which was the probable cause of death.
MATERIALS AND METHODS
Specimen collection.
During the outbreak of disease in the senior high school patients, 18 throat swab specimens and 18 serum samples were collected from 18 patients with acute-phase infection after onset; 3 patients were also found to have convalescent-phase sera. One sample each of hydrothorax fluid, sputum, and serum was collected from the sole patient who died. The throat swab specimens were collected and transferred to 2 ml viral transport medium.
Fifty apparently healthy students from a different senior high school in a different town of the same county were selected as the control population, and serum samples were collected from each student (total number of samples, 50). That senior high school is also a combination of a boarding and commuter school; the distance between the two high schools is more than 40 kilometers, and no case of infection was reported at the school from which the control population was selected.
An enzyme-linked immunosorbent assay (ELISA) was performed to detect bacterial antibodies, and the specimens were cultured to assess them for bacterial growth. They were then prepared for viral detection. All the specimens were dispatched in sterile containers through a cold chain with a controlled low temperature of between 2 and 8°C to the Institute for Viral Disease Control and Prevention in the Chinese Center for Disease Control and Prevention, where the sera and other specimens were stored at −20°C and −80°C, respectively, for further analysis.
Extraction of viral nucleic acid and direct RT-PCR or PCR.
The viral nucleic acid was directly extracted from the clinical specimens by using a QIAamp mini-viral RNA extraction kit or a QIAamp DNA minikit (Qiagen, Valencia, CA). Direct reverse transcription-PCR (RT-PCR) or PCR was performed by using different special primer pairs for adenoviruses (forward primer, 5′-ACCCACGATGTGACCACCGA-3′, nucleotides [nt] 157 to 176; reverse primer, 5′-TGTCAAAGAATGTGCTGGCC-3′, nt 286 to 305; the nucleotide positions are according to the hexon gene of HAdV-3 [GenBank accession number AF542126], and the primers were designed at the China Center for Disease Control and Prevention and are specific for species B of HAdV), influenza virus (27), measles virus (29), rubella virus (33), mumps virus (11), parainfluenza virus (7), respiratory syncytial virus (32), and human enteroviruses (30).
Cell culture and virus isolation.
The 21 clinical specimens, including 18 throat swab specimens collected from the patients and each of the hydrothorax fluid, sputum, and serum samples from the patient who died, were separately inoculated into HEp-2 cells and were cultured in a maintenance medium (minimal essential medium containing 2% fetal calf serum, 100 U/ml penicillin G, 100 μg/ml streptomycin) at 37°C in a closed system in the absence of a CO2 incubator. At the end of the observation period, if no cytopathic effect (CPE) was observed in the cultures, they were further cultured for another 7 days. Cultures exhibiting an adenovirus-like CPE were passaged again to confirm the presence of the virus. Primary identification of positive isolates was performed by PCR with adenovirus-specific primers.
Neutralization test.
The entire virion of the HAdV strain isolated and identified was used as the neutralization virus, and the 50% cell culture infective dose (CCID50)/50 μl was calculated by using the formula of Kärber (14). The stored serum samples were inactivated at 56°C for 30 min, diluted 20 times with the maintenance medium, and filtered through a 0.22-μm-pore-size filter membrane. Several dilutions of the serum samples (from 1:20 to 1:2,560) were prepared, and 50 μl of each dilution was added to four wells of a 96-well microplate. Furthermore, 50 μl of the viral antigen, diluted to a CCID50 of 100, was added to each well, the contents were mixed well, and the plate was incubated for 2 h in an open system in the presence of 5% CO2. The HEp-2 cell suspension was then added to each well. Positive and negative controls were prepared, the plate was incubated once again in the open system in the presence of 5% CO2, and the CPE was observed daily.
ELISA.
The 18 samples from the acute phase collected from the 18 patients and the 50 serum samples collected from the control population were tested to detect HAdV immunoglobulin A (IgA) by using an ELISA Classic adenovirus IgA kit (Institute Virion/Serion GmbH, Würzburg, Germany). This kit enables the detection of serum antibodies against all serotypes of HAdV that are pathogenic for humans.
Extraction of viral DNA and conventional PCR.
DNA was extracted from the clinical specimens by using the QIAamp DNA minikit (Qiagen, Valencia, CA). PCR was performed with a 50-μl reaction mixture containing 1× PCR buffer, 2 mM MgSO4, 0.2 mM each deoxynucleoside triphosphate (Promega), 1 U high-fidelity Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA), 0.2 μM each primer (listed in Table 1), and 2.5 μl of the template DNA. An initial denaturation of 94°C for 1 min was followed by 35 cycles of dentauration at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 68°C for 5 min in a GeneAmp 9700 thermal cycler (Applied Biosystems). The amplification products were analyzed by electrophoresis of the samples in 1% agarose gels and were visualized with ethidium bromide under UV light.
TABLE 1.
Primers used for amplification and sequencing of the entire hexon gene
| Primera | Sequence (5′-3′ orientation) | Positionsb | Ampliconc |
|---|---|---|---|
| Hexon-s | CCCGTCACCTTGGATTTGC | 17942-17960 | 3,449 bp |
| Hexon-as | CGATCATCCGAGAATCCAAA | 21372-21391 | |
| H-1s | GCTTAACTTGCCTATCTGTG | 18156-18175 | Seq |
| H-2s | AATGCTCCTGTAAAAGCT | 18741-18758 | Seq |
| H-3s | CAAGTTCCGAAGCTAAT | 19171-19187 | Seq |
| H-4s | AGATGAACTTCCCAACTACTGT | 19466-19487 | Seq |
| H-5s | CGGACGTTATGTGCCTTTC | 19898-19916 | Seq |
| H-6s | CCCATTTCCATTCCTTCTC | 20232-20250 | Seq |
| H-7s | CAGGCAGGTGGTTGATG | 20648-20664 | Seq |
| H-1as | CCTATTGGGAGTCCTTCTTT | 18774-18793 | Seq |
| H-2as | TGTAGTTGGGTCTGTTGG | 19207-19224 | Seq |
| H-3as | ACCCTGTCCGATCTCAC | 19591-19607 | Seq |
| H-4as | ACTTGTATGTGGAAAGGCAC | 19908-19927 | Seq |
| H-5as | GAGGGAGTTTCTTTGGTTT | 20293-20311 | Seq |
| H-6as | AATTGACCTCATCAACCACC | 20654-20673 | Seq |
| H-7as | ATGGCACAGGCGAGCTTAT | 21262-21280 | Seq |
s, sense orientation; as, antisense orientation.
The nucleotide positions indicated are those according to the prototype strain of HAdV-11 (Slobitski strain; GenBank accession number AF532578).
Length of the PCR product or use of the primer for sequencing (Seq).
Sequence determination.
The PCR products were sequenced directly after purification (QIA gel extraction kit; Qiagen, KK, Japan) by the dye terminator method (BigDye Terminator, version 3.1, cycle sequencing kit; Applied Biosystems) in an ABI Prism 3100 genetic analyzer (Applied Biosystems). The primers used for sequencing are listed in Table 1.
Sequence analysis.
The sequence data were stored as standard chromatogram format (.ab1) files and were analyzed with Sequencher software (version 4.0.5; GeneCodes, Ann Arbor, MI). The nucleotide sequence homology was inferred from the identity scores obtained by using the BLASTn program (National Center for Biotechnology Information, Bethesda, MD). Recombination analysis was performed by using SimPlot software (Johns Hopkins University School of Medicine, Baltimore, MD) (17). Sequence alignments were created with BioEdit sequence alignment editor software (version 5.0.9; Tom Hall, North Carolina State University) (9), and a phylogenetic dendrogram was constructed by the neighbor-joining method with the MEGA program (Sudhir Kumar, Arizona State University); the reliability of the tree was estimated with 1,000 bootstrap pseudoreplicates (25).
Nucleotide sequence accession numbers.
The nucleotide sequence of the entire hexon gene for strain HAdV11-QS/ShX/CHN/2006, which was determined in this study, has been deposited in the GenBank nucleotide sequence database under accession number DQ874353.
RESULTS
Adenovirus is the pathogenic agent.
At the beginning of the outbreak described here, the disease was suspected to be a mild form of acute respiratory infection. A series of serological tests and routine blood cultures were performed for the detection of Mycoplasma spp., Streptococcus spp., Coxiella spp., typhoid fever, etc.; but the results of all tests and cultures were negative. When the results of the virus detection test proved to be positive, the focus was shifted to confirmation of the viral infection.
PCR or RT-PCR was performed with a total of 21 clinical specimens (18 throat swab specimens collected from the 18 patients and 1 sample each of hydrothorax fluid, sputum, and serum from the sole patient who died) and primer sets specific for viruses, including species B of HAdV, influenza virus (27), measles virus (29), rubella virus (33), mumps virus (11), parainfluenza virus (7), respiratory syncytial virus (32), and human enteroviruses (30). Positive results were obtained only for species B of HAdV. Of all the clinical specimens examined, DNA from 13 specimens, including 3 specimens obtained from the sole patient who died, was successfully amplified by PCR with primers specific for 149 bp of the partial hexon gene of HAdV species B. Further identification by sequence determination and BLAST sequence analysis revealed that all the sequences had the highest grade of homology (100%) with species B of HAdV (strain RKI-2797/04, HAdV-11a; GenBank accession number AY972815). Thus, no evidence of bacterial or viral infections except adenovirus infection was found.
Molecular analysis of the HAdV strain.
All 21 clinical specimens were separately inoculated into HEp-2 cells, and a characteristic adenovirus-like CPE was observed in the HEp-2 cells for 5 throat swab samples and 1 hydrothorax fluid sample. In all these cases, a CPE was observed within two passages after inoculation, and the entire hexon gene was successfully amplified by PCR from all samples with adenovirus-specific primers to obtain the predicted products of 3,449 bp. Furthermore, sequence determination revealed that six viral isolates exhibited 99.9% to 100% homology in their nucleic acid sequences. Therefore, it was concluded that all the patients were infected with the same virus. A viral strain designated HAdV11-QS was isolated from the hydrothorax fluid sample of the sole patient who died, and the strain was used for phylogenetic analysis.
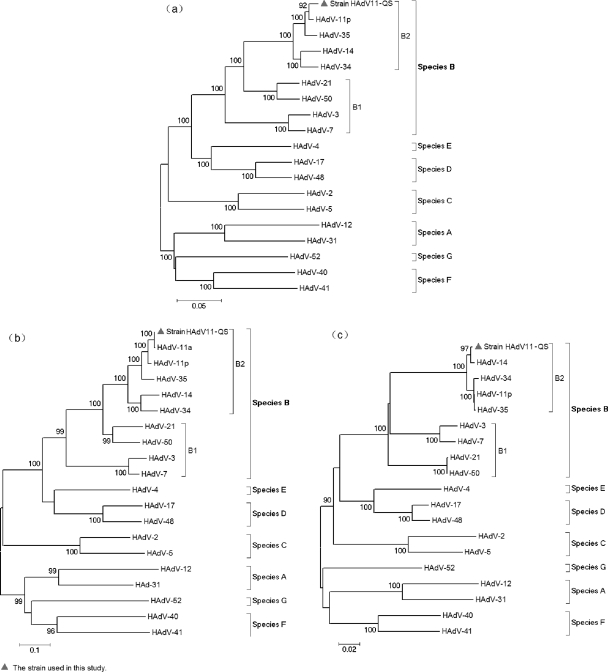
In order to investigate the genetic relationships between strain HAdV11-QS and other HAdV strains, a phylogenetic analysis was conducted on the basis of the entire hexon gene (nt 18255 to 21101, Slobitski strain) with 18 virus strains representing the seven HAdV species (Fig. 2a). All HAdVs can be classified into seven species, i.e., species A to G, on the basis of bootstrap support values. Strain HAdV11-QS could definitely be clustered within HAdV-11 since it exhibited the highest degree of nucleotide sequence identity with the HAdV-11 strain (Slobitski strain, HAdV-11p; identity, 2,794/2,848 nucleotides [98%]; gaps, 6/2,848 nucleotides [0%]).
FIG. 2.
Phylogenetic analysis of HAdV: (a) the entire hexon gene (nt 18255 to 21101, Slobitski strain), (b) the 5′ end of the partial hexon gene covering HVRs 1 to 7 (nt 18361 to 19801, Slobitski strain), and (c) the 3′ end of the partial hexon gene outside the HVRs (nt 19802 to 21101, Slobitski strain). The phylogenetic tree was generated by using the neighbor-joining method; bootstrap values are provided for the basal nodes of each species (species A to G). The GenBank accession numbers for each HAdV are as follows: HAdV-12, X73487; HAdV-31, DQ149611; HAdV-3, X76549; HAdV-7, AF515814; HAdV-11a, AY972815; HAdV-11p, AF532578; HAdV-14, AY803294; HAdV-21, AY008279; HAdV-34, AB052911; HAdV-35, AB330116; HAdV-50, DQ149643; HAdV-2, AJ293903; HAdV-5, AF542130; HAdV-17, AB330098; HAdV-48, U20821; HAdV-4, AF065063; HAdV-40, X51782; HAdV-41, AB330122; and HAdV-52, DQ923122. Strain HAdV11-QS could definitely be identified as an HAdV-11a strain belonging to the B2 species, but recombination with the sequence of HAdV-14 was found at the 3′ end of the hexon gene.
Only the partial sequence of the HAdV-11a hexon gene (the HVR-spanning region) was available in the GenBank database. Thus, for further analysis of the genotypes and recombination, two other phylogenetic trees were constructed on the basis of the sequence of the 5′ end of the partial hexon gene covering HVRs 1 to 7 (nt 18361 to 19801, Slobitski strain) (Fig. 2b) and the 3′ end of the partial hexon gene outside the HVRs (nt 19802 to 21101, Slobitski strain) (Fig. 2c). In the HVR-spanning region, the sequence of strain HAdV11-QS was nearly identical to the corresponding sequence of HAdV-11a strain RKI-2797/04 (GenBank accession number AY972815; identity, 1,434/1,435 nucleotides [99%]; gaps, 0/1,435 nucleotides [0%]), but the sequence exhibited less homology with HAdV-11p strain Slobitski (identity, 1,405/1,442 nucleotides [97%]; gaps, 6/1,442 nucleotides [0%]). However, in the regions outside the HVR-spanning regions, the sequence of strain HAdV11-QS clustered with that of HAdV-14; this finding indicated that recombination may occur between strain HAdV11-QS and HAdV-14 within HAdV species B2.
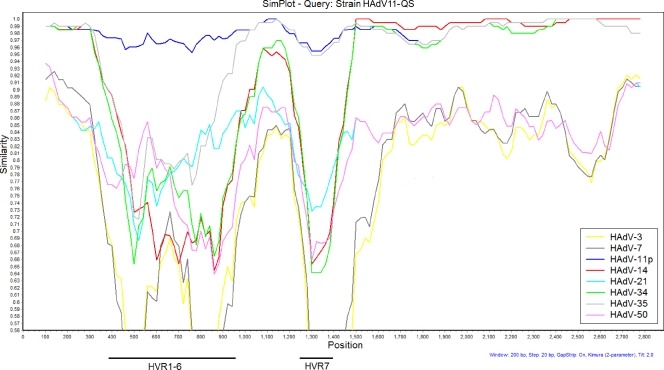
In order to clarify the possible recombination events, a similarity plot with a sliding window of 200 residues was used to analyze the relationship of strain HAdV11-QS to species B strains of HAdV (Fig. 3). When the sequence of the entire hexon gene of strain HAdV11-QS was used as the query sequence and compared to the sequences of eight strains representing the eight serotypes of HAdV species B, the 5′ end of the strain HAdV11-QS hexon gene covering HVRs 1 to 7 exhibited the highest degree of similarity to the sequence of HAdV-11p (prototype strain Slobitski), while the 3′ end of the strain HAdV11-QS hexon gene outside the HVRs exhibited the highest degree of similarity to the sequence from HAdV-14 (prototype strain de Wit), although all strains of species B2 (HAdV-11p, HAdV-14, HAdV-34, and HAdV-35) exhibited higher degrees of homology to each other; this finding shows that the sequence of the 3′ end of the strain hexon gene outside the HVRs of strain HAdV11-QS may be a recombinant with the sequence of HAdV-14.
FIG. 3.
Similarity of the entire hexon sequence (nt 18255 to 21101, Slobitski strain) of strain HAdV11-QS compared with the sequences of species B strains of HAdV. Windows, 200 bp; step, 20 bp. The Kimura model with the Jukes-Cantor correction was used. The vertical axis indicates the percent nucleotide identities between strain HAdV11-QS and eight strains representing eight serotypes of HAdV species B. The horizontal axis indicates the nucleotide positions of the entire hexon gene. GenBank accession numbers for each HAdV are as follows: HAdV-3, X76549; HAdV-7, AF515814; HAdV-11p, AF532578; HAdV-14, AY803294; HAdV-21, AY008279; HAdV-34, AB052911; HAdV-35, AB330116; and HAdV-50, DQ149643. The horizontal axis at the bottom indicates the positions of HVRs 1 to 7.
Results of serological assays indicating that infection was caused by HAdV-11.
The ELISA for HAdV IgA was performed with the 18 serum samples from the acute phase collected from 18 patients; 15 of these samples yielded positive results for IgA, with the proportion of positive samples being 83.3%. Among the 50 individual serum samples collected from the control population, 6 yielded positive results, with the proportion of positive samples being 12.0%. Thus, the proportion of samples positive for HAdV IgA was significantly higher for the patients than for the controls (P < 0.01); these results support the conclusion that the outbreak was caused by an adenovirus infection.
We used the entire virion of the HAdV strain isolated and identified (strain HAdV11-QS) as the neutralization virus; the CCID50 was determined by using the formula of Kärber (14) to be approximately 105.5 CCID50s/50 μl. Conventional neutralization tests were performed with three pairs of paired serum samples (with each pair consisting of one sample from the acute phase and one sample from the convalescent phase from the same patient). The results (Table 2) revealed that the neutralization titers of the convalescent-phase samples were four times higher than those of the acute-phase samples for all three pairs. This finding also suggests that HAdV-11 was associated with this outbreak.
TABLE 2.
Results of a conventional neutralization test performed with three pairs of serum samples and strain HAdV11-QS
| Serum sample no. | Date of onset | Serum in acute phase
|
Serum in convalescent phase
|
||
|---|---|---|---|---|---|
| Interval from onset (days) | Neutralization titer | Interval from onset (days) | Neutralization titer | ||
| 1 | 10 April 2006 | 1 | <1:20 | 30 | 1:450 |
| 2 | 10 April 2006 | 1 | <1:20 | 30 | 1:112 |
| 3 | 20 March 2006 | 22 | <1:20 | 51 | 1:112 |
DISCUSSION
Numerous outbreaks of acute respiratory infection caused by HAdV have been reported during the last decade in many countries, including China (10, 28, 31). In spring 2006, an acute respiratory infection broke out in Shanxi Province, China. The results of the present study indicate that HAdV-11a, which belongs to the B2 species, was responsible for this outbreak. This postulation is supported by the clinical manifestations of the affected patients; epidemiological data from the outbreak; and the results of direct PCR experiments and sequence determination, conventional neutralization assays, IgA serological assays, viral isolation assays, and molecular typing and analysis.
In recent years, molecular techniques have played a key role in disease control because they enable rapid and early diagnosis. In this study, the serotype and the genotype of the HAdV strain involved were identified by performing PCR and sequencing of the entire hexon gene. HVRs 1 to 7 of the HAdV hexon gene encode serotype-specific epitopes and have been described for use for the serotype identification of HAdV (24, 26). Furthermore, the results of the sequencing of HVRs 1 to 7 of the HAdV hexon gene appeared to correlate well with those of serological typing and other molecular typing methods (20). For these reasons, use of the 1,441-bp sequence (nt 18361 to 19801, Slobitski strain) covering HVRs 1 to 7, which was used in this study, should be considered for precise molecular typing. On the basis of such molecular typing results, the viruses isolated from this outbreak belonged to HAdV-11a. However, recombination is a well-known feature in HAdV genetics and an important force driving the evolution of HAdV; in general, intraspecies recombination is observed much more frequently than interspecies recombination (19). In this study, both the analysis of the phylogenetic relationship and the similarity plot indicated that the sequence of the 3′ end of the hexon gene outside the HVRs of strain HAdV11-QS may be a recombinant with the sequence of the HAdV-14 strain of species B2; this is an instance of intraspecies recombination.
In this study, we obtained detailed clinical descriptions of the outbreak. The clinical course and the available epidemiological data were well consistent with the clinical course and epidemiological data described in previous reports on HAdV-associated acute respiratory diseases. In the outbreak studied here, most of the affected individuals (97.2%) were students of a senior high school. The commonly recognized route of transmission is through respiratory droplets and close contact, which leads to the rapid and widespread dissemination of HAdV in crowded places. Similar events have been reported to occur in military camps and day care centers in China and other countries (4). The early detection of HAdV infections should receive more attention from clinicians and public health officials when they are evaluating and responding to outbreaks.
HAdV-11 was first isolated from a fecal sample from a child diagnosed with poliomyelitis in 1954; poliovirus type I was also recovered from that patient. HAdV-11 has been reported to be associated with upper and lower respiratory illnesses; with hemorrhagic cystitis in children and young adults, especially in renal transplant recipients; and occasionally with endemic hemorrhagic conjunctivitis (16). In 1991, HAdV-11 strains were classified into at least two genome types, designated HAdV-11p and HAdV-11a, on the basis of similarities in fragment comigration patterns during restriction genome typing (16). These two types exhibit different tropisms: HAdV-11a infects respiratory epithelial cells, while HAdV-11p infects renal cells. HAdV-11a strains were more frequently isolated in China from 1965 to 1985 (16). Although HAdV-11 infections have not been reported for more than 20 years, the etiological analysis of this outbreak reveals that HAdV-11a has continued to exist and circulate in China. Furthermore, phylogenetic analysis revealed that this strain is also circulating in other countries, such as Turkey (4). This assumption is based on the evidence that the nucleotide sequence of strain HAdV11-QS was exactly identical to the partial hexon gene sequence of strain RKI-2797/04 (GenBank accession number AY972815) isolated in Turkey. HAdV-11a was also isolated from patients with acute respiratory tract infections in Spain and Latin America (13).
Most children and young adults infected with HAdV have only mild illnesses with respiratory symptoms, and this virus seldom causes severe illness. However, the host immune response may be the key determinant of the clinical course of HAdV infection. The sole patient who died during the outbreak had bone marrow megaloblastic anemia as an underlying disease, and HAdV coinfection may have contributed to the onset of a life-threatening disease.
Since little is known about the dynamics of HAdV-11, which was involved in the emergence of pathogens causing acute febrile respiratory disease in the senior high school students, studies investigating the persistence and transmission of this agent within populations should be conducted by the use of continuous surveillance. In this study, few people (12%) in the control population also tested positive for HAdV IgA; this may have been due to the presence of an inapparent infection, which may also contribute to HAdV transmission. Thus, it is not possible to predict whether HAdV-11 will persist as a significant cause of acute respiratory disease outbreaks in high schools.
HAdV-14, which also belongs to the B2 species, has recently emerged and has caused outbreaks of severe respiratory disease in the United States. From March to June 2007, cases of HAdV-14-induced respiratory illness were confirmed in 140 patients in Oregon, Washington, and Texas. Of these 140 patients, 53 (38%) patients were hospitalized, 24 (17%) patients were admitted to intensive care units; and 9 (5%) patients died (2, 18). In this study, recombination between HAdV-11a and HAdV-14 was found. However, it was very difficult to identify the time of recombination and the manner in which it occurred, but it seemed that HAdV-14 also circulated in China at some time.
Acute respiratory diseases due to HAdV are primarily attributed to HAdV-4, HAdV-7, HAdV-3, and HAdV-21. Thus, HAdV species B2, including HAdV-11 and HAdV-14, may pose a new global challenge with regard to acute respiratory diseases. Although HAdV isolates of this species seldom cause respiratory infections, this possibility cannot be overlooked and should be carefully considered. Additionally, such HAdV infections may be underreported, especially if the infections are mild or asymptomatic; hence, the need for the establishment and improvement of both epidemiological and virological surveillance for HAdV infections in China should be emphasized.
Acknowledgments
We thank all the staff members of the Shanxi Microbiology Laboratory and Epidemiology Department for collecting the specimens and performing some of the bacterial detection studies. Without their valuable assistance, it would have been impossible to successfully complete this study.
This work was supported by grant 2007AA02Z463 from the Ministry of Science and Technology of the People's Republic of China.
None of the authors reports a conflict of interest.
Footnotes
Published ahead of print on 24 December 2008.
REFERENCES
- 1.Benko, M., B. Harrach, and W. C. Russell. 2000. Adenoviridae, p. 227-237. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. Mcgeoch, C. R. Pringle, and R. B. Wickner (ed.), Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, San Diego, CA.
- 2.CDC. 2007. Acute respiratory disease associated with adenovirus serotype 14—four states, 2006-2007. MMWR Morb. Mortal. Wkly. Rep. 561181-1184. [PubMed] [Google Scholar]
- 3.Chakrabarti, S., V. Mautner, H. Osman, K. E. Collingham, C. D. Fegan, P. E. Klapper, P. A. Moss, and D. W. Milligan. 2002. Adenovirus infections following allogeneic stem cell transplantation: incidence and outcome in relation to graft manipulation, immunosuppression, and immune recovery. Blood 1001619-1627. [DOI] [PubMed] [Google Scholar]
- 4.Chmielewicz, B., J. Benzler, G. Pauli, G. Krause, F. Bergmann, and B. Schweiger. 2005. Respiratory disease caused by a species B2 adenovirus in a military camp in Turkey. J. Med. Virol. 77232-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Crawford-Miksza, L. K., and D. P. Schnurr. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 701836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echevarria, J. E., D. D. Erdman, E. M. Swierkosz, B. P. Holloway, and L. J. Anderson. 1998. Simultaneous detection and identification of human parainfluenza viruses 1, 2, and 3 from clinical samples by multiplex PCR. J. Clin. Microbiol. 361388-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu, W. Y., D. Liang, Y. C. Zheng, W. M. Liu, Z. Xu, H. J. Guo, and Z. L. Wang. 1989. A study of molecular epidemiology of adenovirus of types 3 and 7 on infant pneumonia in northern China. Chin. Med. J. (Engl.) 102857-861. [PubMed] [Google Scholar]
- 9.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 10.Harley, D., B. Harrower, M. Lyon, and A. Dick. 2001. A primary school outbreak of pharyngoconjunctival fever caused by adenovirus type 3. Commun. Dis. Intell. 259-12. [PubMed] [Google Scholar]
- 11.Jin, L., S. Beard, and D. W. Brown. 1999. Genetic heterogeneity of mumps virus in the United Kingdom: identification of two new genotypes. J. Infect. Dis. 180829-833. [DOI] [PubMed] [Google Scholar]
- 12.Jones, M. S., B. Harrach, R. D. Ganac, M. M. Gozum, W. P. dela Cruz, B. Riedel, C. Pan, E. L. Delwart, and D. P. Schnurr. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 815978-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kajon, A. E., A. S. Mistchenko, C. Videla, M. Hortal, G. Wadell, and L. F. Avendaño. 1996. Molecular epidemiology of adenovirus acute lower respiratory infections of children in the south cone of South America (1991-1994). J. Med. Virol. 48151-156. [DOI] [PubMed] [Google Scholar]
- 14.Kärber, G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch. Exp. Pathol. Pharmakol. 162480-483. [Google Scholar]
- 15.Larrañaga C., H. J. Martínez, M. A. Palomino, C. M. Peña, A. F. Carrión, and C. L. Avendaño. 2007. Molecular characterization of hospital-acquired adenovirus infantile respiratory infection in Chile using species-specific PCR assays. J. Clin. Virol. 39175-181. [DOI] [PubMed] [Google Scholar]
- 16.Li, Q. G., J. Hambraeus, and G. Wadell. 1991. Genetic relationship between thirteen genome types of adenovirus 11, 34, and 35 with different tropisms. Intervirology 32338-350. [DOI] [PubMed] [Google Scholar]
- 17.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louie, J. K., A. E. Kajon, M. Holodniy, L. Guardia-LaBar, B. Lee, A. M. Petru, J. K. Hacker, and D. P. Schnurr. 2008. Severe pneumonia due to adenovirus serotype 14: a new respiratory threat? Clin. Infect. Dis. 46421-425. [DOI] [PubMed] [Google Scholar]
- 19.Lukashev, A. N., O. E. Ivanova, T. P. Eremeeva, and R. D. Iggo. 2008. Evidence of frequent recombination among human adenoviruses. J. Gen. Virol. 89380-388. [DOI] [PubMed] [Google Scholar]
- 20.Madisch, I., G. Harste, H. A. Pommer, and A. Heim. 2005. Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy. J. Virol. 7915265-15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mei, Y. F., J. Skog, K. Lindman, and G. Wadell. 2003. Comparative analysis of the genome organization of human adenovirus 11, a member of the human adenovirus species B, and the commonly used human adenovirus 5 vector, a member of species C. J. Gen. Virol. 842061-2071. [DOI] [PubMed] [Google Scholar]
- 22.Shenk, T., and M. S. Horwitz. 2001. Adenoviridae: the viruses and their replication, adenoviruses, p. 2265-2326. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 23.Slatter, M. A., S. Read, C. E. Taylor, B. N. Crooks, M. Abinun, T. J. Flood, A. J. Cant, C. Wright, and A. R. Gennery. 2005. Adenovirus type F subtype 41 causing disseminated disease following bone marrow transplantation for immunodeficiency. J. Clin. Microbiol. 431462-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi, S., N. Itoh, E. Uchio, K. Aoki, and S. Ohno. 1999. Serotyping of adenoviruses on conjunctival scrapings by PCR and sequence analysis. J. Clin. Microbiol. 371839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 26.Wohlfart, C. 1988. Neutralization of adenoviruses: kinetics, stoichiometry, and mechanisms. J. Virol. 622321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright, K. E., G. A. Wilson, D. Novosad, C. Dimock, D. Tan, and J. M. Weber. 1995. Typing and subtyping of influenza viruses in clinical samples by PCR. J. Clin. Microbiol. 331180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu, W., A. Cui, Z. Shi, H. Wang, Y. Zhang, and Z. Tang. 2005. Etiological research of the unknown mild respiratory tract infectious disease in an outbreak in Jiangsu Province. Chin. J. Virol. 21325-331. [Google Scholar]
- 29.Xu, W., A. Tamin, J. S. Rota, L. Zhang, W. J. Bellini, and P. A. Rota. 1998. New genetic group of measles virus isolated in the People's Republic of China. Virus Res. 54147-156. [DOI] [PubMed] [Google Scholar]
- 30.Yang, C. F., L. De, S. J. Yang, J. Ruiz Gomez, J. R. Cruz, B. P. Holloway, M. A. Pallansch, and O. M. Kew. 1992. Genotype-specific in vitro amplification of sequences of the wild type 3 polioviruses from Mexico and Guatemala. Virus Res. 24277-296. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, Q., X. Su, S. Gong, Q. Zeng, B. Zhu, Z. Wu, T. Peng, C. Zhang, and R. Zhou. 2006. Comparative genomic analysis of two strains of human adenovirus type 3 isolated from children with acute respiratory infection in southern China. J. Gen. Virol. 871531-1541. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Y., W. Xu, K. Shen, Z. Xie, L. Sun, Q. Lu, C. Liu, G. Liang, J. A. Beeler, and L. J. Anderson. 2007. Genetic variability of group A and B human respiratory syncytial viruses isolated from 3 provinces in China. Arch. Virol. 1521425-1434. [DOI] [PubMed] [Google Scholar]
- 33.Zhu, Z., W. Xu, E. S. Abernathy, M. H. Chen, Q. Zheng, T. Wang, Z. Zhang, C. Li, C. Wang, W. He, S. Zhou, and J. Icenogle. 2007. Comparison of four methods using throat swabs to confirm rubella virus infection. J. Clin. Microbiol. 452847-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]