Abstract
The collecting duct (CD) expresses considerable amounts of PPARδ. While its role is unknown in the CD, in other renal cells it has been shown to regulate both growth and apoptosis. We thus hypothesized that PPARδ reduces apoptotic responses and stimulates cell growth in the mouse CD, and examined the effect of GW501516, a synthetic PPARδ ligand, on these responses in mouse IMCD-K2 cells. High doses of GW501516 decreased both DNA and protein synthesis in these cells by 80%, but had no overall effect on cell viability. Although anisomycin treatment resulted in an increase of caspase-3 levels of about 2.59-fold of control, GW501516 did not affect anisomycin-induced changes in active caspase-3 levels. These results show that a PPARδ ligand inhibits growth but does not affect anisomycin-apoptosis in a mouse IMCD cell line. This could have therapeutic implications for renal diseases associated with increased CD growth responses.
1. Introduction
Traditional prostaglandin (PG) signaling in renal epithelial cells occurs through activation of prostanoid receptors [1–3]. In recent years, however, a novel signaling pathway for PGs has been found in which ligand-activated transcription factors called peroxisome proliferator-activated receptors (PPARs) [4] are activated at the nuclear membrane. One PPAR type, PPARδ, also known as PPARβ, is ubiquitously expressed in almost every tissue examined [5]. Both prostanoids PGI2 and PGE2 have been known to activate PPARδ [6, 7]. The main role of PPARδ is in cell survival and this has been well characterized in colon cancer [8–10].
In the kidney, PPARδ is found in every part of the nephron with high levels in the glomerulus, cortical collecting duct, and inner medullary collecting duct (IMCD) and low expression in the outer medullary collecting duct. This PPAR has been implicated in the renal complications of metabolic syndrome [11], but most studies describe a link between PPARδ and survival of renal cells. In a study by Hao et al. [6], PPARδ was shown to increase renal medullary interstitial cell survival during high levels of hypertonic stress, in response to PGI2. PPARδ activation also protected kidneys from renal failure after renal ischemia/reperfusion experiments via the antiapoptotic Akt pathway [12]. Although, most of the studies have suggested an antiapoptotic role of PPARδ, a few papers have shown that PPARδ can increase apoptosis [13]. However, PPARδ is recognized as an important protein in the survival of many renal cell types.
PPARδ has also been shown to promote growth responses in a variety of cells. PGE2, working through PPARδ, can stimulate proliferation of stem cells [14] and overexpression of PPARδ has been shown to reverse inhibitory growth signals in a prostate epithelial cell line [15]. Furthermore, upregulation of PPARδ in smooth muscle cells promotes cell cycle progression [16]. The effect of PPARδ activation on cell growth in the kidney, however, is unknown.
The collecting duct (CD), specifically the IMCD, survives in a harsh environment due to the hypertonic conditions in the interstitium needed to concentrate urine [17, 18]. In most cells, apoptosis would occur at the levels of stress at which they reside, but the CD manages to survive and resist any apoptotic activity [19]. It is known that cells react to high levels of tonicity by increasing the transport of several osmolytes into the cells aiding in their survival [20]. However, in cells where there is chronic hypertonicity, like the IMCD, the accumulation of these osmolytes may not be sufficient for their continued survival [21]. Cultured IMCD cells have been known to increase levels of cyclooxygenase-2 (COX-2) and stimulate PGE2 release up to 33-fold of control in response to hypertonic stress [22]. It is likely then that PGs may contribute to the inherent CD resistance to stress by targeting PPARδ to promote cell survival.
It would appear in most cell lines that PPARδ has opposing growth and apoptotic effects; however, its role in the CD has not been determined. Our study examines the role of PPARδ activation in the survival and growth of IMCD-K2 cells. The mouse IMCD-K2 cell line is derived from the initial portion of the IMCD from a mouse that is transgenic for Simian Virus 40 (SV40) and was shown to retain many characteristics of the intact IMCD, including amiloride-sensitive sodium absorption stimulated by aldosterone [23]. Therefore, we propose that PPARδ activation will regulate growth and apoptotic responses in the mouse IMCD-K2 cells.
2. Materials and Methods
2.1. IMCD-K2 Cell Culture
Cells were obtained from Dr. Bruce Stanton from Dartmouth Medical College (Hanover, NH, USA). The cells were maintained at 37°C and 5% CO2 in DMEM/F-12 supplemented with 10% fetal bovine serum, (FBS, Gibco, Carlsbad, Calif, USA), 1% penicillin/streptomycin (Gibco), ITS (5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL selenium; Sigma, St. Louis, Mo, USA), 2 mM L-glutamine (Sigma), and 5 μM dexamethasone (Sigma). Cell culture medium was replaced every 48 hours. Twenty-four hours prior to experiments, the culture medium was replaced with serum-free medium (DMEM/F-12).
2.2. Reverse Transcriptase-PCR
RNA was isolated from confluent 100 mm plates of IMCD-K2 cells using 1 mL of the TRIzol reagent (Gibco) according to the manufacturer's instructions and then treated with DNase I (Invitrogen, Carlsbad, Calif, USA). The RNA was reverse transcribed using MuLV reverse transcriptase and random hexamers from the GeneAmp RNA PCR Core Kit (Applied Biosystems, Foster City, Calif, USA) as per the manufacturer's instructions. Next, the RNA was amplified by polymerase chain reaction (PCR), using AmpliTaq DNA polymerase and specific primers for the prostanoid receptors (i.e., EP1–4 and IP) and PPARδ. The primers were obtained from the University of Ottawa Biotechnology Research Institute (Ottawa, ON, Canada). The PCR parameters used were 40 cycles of: 95°C for 30 seconds, 59°C for 45 seconds, and 72°C for 60 seconds, followed by 600 seconds at 72°C. The amplification was carried out using the GeneAMP PCR System 2400 (Applied Biosystems). The PCR products were run on an agarose gel and the bands were visualized using ethidium bromide under UV light.
2.3. 3H-cAMP Assay
Since prostanoids can alter cAMP, their effect on cAMP production in IMCD-K2 cells was determined. The cells were serum starved for 24 hours and stimulated with control (serum-free media), 10−5 M forskolin (Sigma) as a positive control, 10−6 M cicaprost and iloprost (prostacyclin analogs, Schering Ag Berlin, Berlin, Germany), or 10−9–10−6 M of prostaglandin E2 (PGE2, Cayman, Ann Arbor, Mich, USA) The cells were pretreated for fifteen minutes with 0.5 mM isobutylmethylxanthine (IBMX, Sigma), a cAMP phosphodiesterase inhibitor, and 10−5 M indomethacin (Sigma), a cyclooxygenase (COX) inhibitor. The stimulation was then stopped with cold 10% trichloroacetic acid for 30 minutes on ice. Four ether extractions were carried out, and a cAMP competitive binding assay was done using the 3H-cAMP DPC radioassay (Intermedico, Markham, ON, Canada) as per manufacturer's instructions. The amount of 3H-cAMP was measured using a scintillation counter (Beckman Coulter, Mississauga, ON, Canada) and the percent stimulation of cAMP levels over control levels of each sample was calculated.
2.4. Western Blotting
IMCD-K2 lysates were obtained by sonicating samples in RIPA lysis buffer containing 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS, w/v), 4.5 mM NaCl, 2.5 mM Tris (pH 7.4), 8 μM EDTA, 0.2 mM sodium phosphate (pH 7.2), and fresh 0.5 mM PMSF, 1 : 100 protease inhibitor cocktail (Sigma), 1 mM sodium pyrophosphate, 10 mM sodium fluoride, and 100 μM sodium orthovanadate. The protein was quantified using the Bradford reagent method (Bio-Rad, Hercules, Calif, USA). The lysates were then resolved by SDS-PAGE on a polyacrylamide gel using a Mini-PROTEAN II apparatus (Bio-Rad) and transferred onto nitrocellulose membranes (Amersham, Amersham, UK). After blocking for 90 minutes in 10% milk/TBS-T (137 mM NaCl, 20 mM Tris base, 0.1% Tween20), the membranes were incubated overnight with the corresponding primary antibody. The membranes were then incubated with their respective secondary antibodies (i.e., anti-rabbit, anti-mouse, and anti-donkey) for 90 minutes and then washed for 60–90 minutes in TBS-T. The bands were then visualized by ECL. Detection of β-actin was used to normalize samples and densitometric analysis of the bands was performed using the Kodak 1D Image Analysis software (Eastman Kodak Company, Rochester, NY, USA).
2.5. Real-Time PCR
To determine the effect of PGE2 on PPARδ levels in IMCD-K2 cells, RNA was isolated and DNase-treated as described above. The cells were treated for 2, 8, or 24 hours with serum-free media as control, 10−5 M indomethacin, or 10−6 M PGE2. A fifteen-minute pretreatment with indomethacin was performed prior to stimulations with PGE2 and forskolin. The mRNA levels of PPARδ were ascertained by real-time PCR using TaqMan One-Step RT-PCR master mix reagents (Applied Biosystems) and an ABI Prism 7000 sequence detection system. Reactions were carried out by using 50 ng of total IMCD-K2 RNA under the following conditions: 48°C for 30 minutes, and 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The probe and primers used for mouse PPARδ were forward primer, 5′-GAGCCCAAGTTCGAGTTTGC-3′, reverse primer, 5′-TGAAGAGCGCCAGGTCACT-3′, and probe, FAM-AGTTCAATGCGCTGGAGCTCGATGA-TAM (Sigma Genosys, Oakville, ON, Canada). All values were normalized to GAPDH mRNA levels in the same sample, which was determined by the TaqMan Rodent GAPDH control reagent kit (AppliedBiosystems).
2.6. 3H-Thymidine Incorporation
To study the effect of the PPAR activation on cell proliferation, DNA synthesis was measured using the incorporation of 3H-thymidine. IMCD-K2 cells were cultured in 24-well plates, grown to ∼50% confluence, and then starved with serum-free media. Afterwards the cells were treated for 24 hours with vehicle-treated control (DMSO), or GW501516 (10−8, 10−7, 10−6, 10−5 M). GW501516 is among the more specific and most commonly used synthetic PPARδ ligands [24]. 3H-thymidine (Amersham, 0.5 μCi/mL) was added during the final four hours of stimulation. The plates were then washed four times in ice-cold PBS. Next, the cells were permeabilized in 500 μL of 1N NaOH at 37°C for ∼30 minutes, and the amount of 3H-thymidine in counts per minute (cpm) was measured using a scintillation counter. Samples were done in triplicate and thymidine incorporation is expressed as fold control.
2.7. 3H-Leucine Incorporation
To study the effect of the PPAR ligands on cell growth, protein synthesis was measured using the incorporation of 3H-leucine. IMCD-K2 cells were cultured in 24-well plates, grown to ∼50% confluence, and then starved with serum-free media for 24 hours. Next the cells were stimulated for 24 hours with vehicle-treated control (DMSO) and GW501516 (10−8, 10−7, 10−6, and 10−5 M). 3H-leucine (Amersham, Perkin Elmer, Waltham, Mass, USA, 0.5 μCi/mL) was added to each well during the 24-hour stimulation. The cells were then prepared as described above and the amount of 3H-leucine in counts per minute was measured using a scintillation counter. Samples were done in triplicate and leucine incorporation is expressed as fold control.
2.8. Cell Viability
To evaluate the effect of GW501516 on the viability of IMCD-K2 cells in both the presence and absence of a death response, a cell viability assay was performed. IMCD-K2 cells were grown to 70% confluence in 96-well plates and stimulated for 18 hours with control (1 μL of ethanol and 1 μL DMSO in DMEM/F-12), GW501516 (10−8, 10−7, 5 × 10−7, 10−6, 5 × 10−6, and 10−5 M), anisomycin (Sigma; 100, 1000 ng/mL), or cisplatin (Sigma; 50 ng/mL). Some cells were treated with both anisomycin and GW501516 (10−6 M) following a 24-hour pretreatment with GW501516 (10−6 M). Anisomycin and cisplatin were used to induce cell death. Following the stimulation, the cells were incubated in 100 μL of a 1x DNA dye-binding solution from the CyQAUNT NF kit (Invitrogen). The fluorescence intensity, representing the number of cells, was measured in a FLUOstar Galaxy plate reader (BMG Labtechnologies, Durham, NC, USA). Samples were done in triplicate and fluorescence intensity is expressed as fold control.
2.9. Colorimetric Caspase-3 Activity Assay
To determine if GW501516 is involved in apoptosis, a caspase-3 activity assay was carried out using the colorimetric CaspACE Assay System kit (Promega, Madison, Wis, USA), as per manufacturer's instructions. IMCD-K2 cells were grown to ∼50% confluence in 100 mm plates and starved for 24 hours. Next the cells were treated with control (1 μL of ethanol and 1 μL of DMSO in 1 mL of DMEM/F-12), 10% FBS, GW501516 (10−6 M), anisomycin (250 ng/mL), or Z-VAD-FMK (2 × 10−5 M), a pan-caspase inhibitor supplied in the kit. Some cells were treated with both anisomycin and GW501516 following a 24-hour pretreatment with GW501516. Also, some plates were treated with anisomycin and Z-VAD-FMK following a 24-hour pretreatment with the inhibitor. Following stimulations, the cells were resuspended in 100–200 μL of the caspase lysis buffer subjected to two freeze-thaw cycles (at −20°C), and a 15-minute incubation on ice. The lysates were then centrifuged for 20 minutes at 13 100 revolutions per minute and protein was quantified using the Bradford method. 50 μg of each sample was incubated with the colorimetric caspase-3 substrate Ac-DEVD-pNA (2 × 10−4 M) in a 96-well plate at room temperature overnight. The endogenous active caspase-3 bound to the substrate was determined by the absorbance at 405 nm (FLUOstar Galaxy). Samples were done in duplicate and the caspase activity is expressed as relative absorbance.
2.10. Tunel Assay
To further evaluate the effect of GW501516 on apoptosis, a terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling (TUNEL) assay was performed. IMCD-K2 cells were grown to ∼60% confluency on glass coverslips in a 24-well plate and treated in duplicate for 24 hours with control (1 μL of ethanol and 1 μL of DMSO in 1 mL of DMEM/F-12), GW501516 (10−8, 10−6 M), or anisomycin (250 ng/mL). Some cells were treated with both GW501516 (10−8, 10−6 M) and anisomycin, following a 3-hour pretreatment with the respective concentrations of GW501516. After the stimulation, the cells was fixed in 4% paraformaldehyde and 0.2% picric acid in 0.16 M sodium phosphate buffer (pH 6.9). The coverslips were then washed with PBS three times and incubated with 2 μL CoCl2, 5 μL 5X TDT buffer, 0.1 μL terminal transferase (all taken from the Roche Terminal Transferase recombinant kit), 0.17 μL biotin-16-dUTP (Roche, Mississauga, ON, Canada), and 17.73 μL of H2O for 1 hour at 37°C in a humid chamber. Next, the cells were washed with 4X SSC (0.6 M NaCl, 76 mM Na Citrate, pH 7), blocked for 30 minutes with 1% milk/4X SSC at room temperature, and incubated with streptavidin-Cy2 (Molecular Probes,Carlsbad, Calif, USA, 1.0 mg/mL) at 1 : 1000 for 45 minutes at room temperature in the dark. The coverslips were then washed with PBS and incubated with DAPI for 45 minutes at 1 : 1000. Following three more washes, the coverslips were inverted onto Fluoromount mounting media (Electron Microscopy Sciences, Hatfield, Pa, USA) on glass slides. The cells were visualized with the fluorescent microscope Axioskop Mot 2 (Zeiss, Oberkochen, Germany) and captured with the camera AxioCam (Zeiss). For each coverslip, 8 fields of view at 200x magnification were examined using pictures from both the DAPI filter (excitation: ∼360 nm and emission: ∼450 nm) and the green filter (excitation: ∼480 nm and emission: ∼540 nm). The pictures were processed using the Axiovision software (Zeiss). The terminal transferase and biotin-16-dUTP target-free DNA fragments of apoptotic cells produce a fluorescent green signal. The number of apoptotic cells was counted, and from the DAPI filter a percentage of TUNEL-positive cells were determined. A total of 16 fields of view, or two coverslips, were used for each sample.
2.11. Statistics
The GraphPad Prism software (La Jolla, Calif, USA) for Windows (version 4.02) was used to analyze data. Results are expressed as means ± standard error of the mean (S.E.M). Either an unpaired t-test or a one-sample t-test (with a hypothetical value of 1) was used to evaluate the statistical significance between data points. A P-value < .05 was considered statistically significant.
3. Results
3.1. EP Receptor Subtypes and PPARδ Expression
PGE2 is known to activate prostanoid receptors, EP1, EP2, EP3, and EP4 as well as PPARδ. Thus, the presence of these receptors in IMCD-K2 cells was determined. PCR was performed and bands corresponding to the prostanoid and PPARδ receptors were visualized as shown in Figures 1(a) and 1(b), respectively. Previous studies in our laboratory demonstrated that at higher stringency the band in the EP2 lane disappears and that the product detected with the IP primers, present at approximately 200 base pair (bp) greater than the predicted size of 407 bp, does not correspond to the IP receptor, as determined by cloning and sequencing. The band seen between 700 and 800 bp in the EP1 lane represents protein kinase N (PKN), as the genes for PKN and EP1 overlap [25]. The multiple bands seen in the EP3 are most likely due to alternative splicing of the EP3 gene [26]. The products were not present when the reverse transcriptase was omitted (negative control; data not shown). These results indicate that IMCD-K2 cells express EP1, EP3, EP4, and PPARδ.
Figure 1.
EP receptor subtypes and PPARδ are expressed in IMCD-K2 cells. RNA was isolated from IMCD-K2 cells using TRIzol, reverse transcribed, amplified by polymerase chain reaction using primers for (a) the prostanoid receptors and (b) PPARδ receptor, and visualized on an agarose gel using ethidium bromide under ultraviolet light. As shown, specific bands for each PCR product were obtained at the predicted size for EP1 (336 base pair), EP3 (437 base pair; plus bands at ∼350 base pair and ∼600 base pair), and EP4 (423 base pair), as well as PPARδ (161 base pair).
3.2. Effect of PGE2 on Camp Stimulation
Both the EP3 and EP4 receptors elicit changes in intracellular cAMP; therefore, the effect of different prostanoids on cAMP stimulation was examined. Forskolin was used as a positive control. As shown in Figure 2(a), prostacyclin (PGI2) analogs cicaprost (CCP) and iloprost (ILP) did not alter cAMP levels. PGE2, however, did cause a significant increase in percent stimulation, at about 70% above control, comparable to forskolin. As shown in Figure 2(b), treatment with increasing amounts of PGE2 resulted in a concentration-dependent increase in cAMP, from 14.0 to 85.5%.
Figure 2.
PGE2 stimulates cAMP in IMCD-K2 cells.Serum-starved cells were stimulated for ten minutes with (a) 10−6 M of different prostanoids (cicaprost (CCP), iloprost (ILP), and PGE2) or 10−5 M forskolin (FOR), and with (b) differing concentrations of PGE2 (10−9 to 10−6 M). A competitive binding assay with 3H-cAMP was performed and the percent cAMP stimulation was determined using a scintillation counter. Values are means ± S.E.M.; n = 3–4. *P < .05 compared to control.
3.3. The Effect of PGE2 on PPARδ Expression
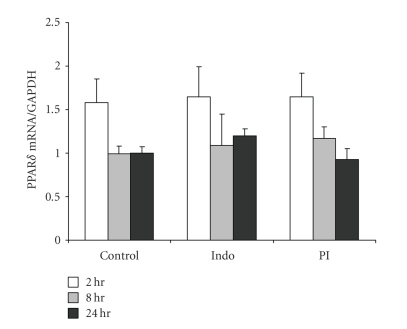
Since PGE2 has been known to activate PPARδ to regulate several growth processes [7, 14], it was ascertained that if PGE2 stimulation could affect PPARδ expression in IMCD-K2 cells. Indomethacin, a COX inhibitor, was used to inhibit endogenous PG synthesis. As shown in Figures 3(a) and 3(b), treatment with PGE2, in the presence of indomethacin, for 8 hours resulted in a significant increase, of ∼2-fold, in PPARδ protein expression compared to control or indomethacin alone, as measured by Western Blotting. Stimulation for 24 hours also resulted in increased PPARδ protein expression, but this increase was not significant when compared with indomethacin alone. A representative blot for 24 hours is shown in Figure 3(a). However, two-hour stimulation did not alter the protein levels of PPARδ. We also examined the effect of PGE2 on PPARδ mRNA expression by real-time PCR. As shown in Figure 4, PPARδ mRNA remained unchanged with any treatment group or exposure time. Forskolin had no effect on expression either (data not shown) indicating that cAMP does not alter PPARδ protein or RNA expression.
Figure 3.
PGE2 increases PPARδ protein expression. Protein was isolated from IMCD-K2 cells that had been treated with serum-free media (control, C), 10−5 M indomethacin (Indo, I), or 10−6 M PGE2 in the presence of I (PI) 2, 8, and 24 hours. The proteins were (a) run on an SDS-PAGE gel and Western blotted using an anti-PPARδ antibody (1 : 500). A representative blot of 24-hour treatment is shown. The membranes were stripped and β-actin was detected to normalize samples for (b) densitometric analysis. Expression is presented as fold of control. Values are means ± S.E.M.; n = 3–6. *P < .05 compared to control; §P < .05 compared to indomethacin.
Figure 4.
PGE2 does not affect PPARδ mRNA expression. RNA was isolated from IMCD-K2 cells using the TRIzol method. The cells had been treated for 2, 8, and 24 hours with serum-free media (control), 10−5 M indomethacin (Indo), or 10−6 M PGE2 in the presence of 10−5 M indomethacin (PI). The RNA samples were quantified using real-time PCR using primers and probes for PPARδ. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. Expression is presented as the amount of PPARδ mRNA expression divided by GAPDH expression. Values are means ± S.E.M.; n = 5.
3.4. The Effect of GW501516 on Cell Proliferation/Growth
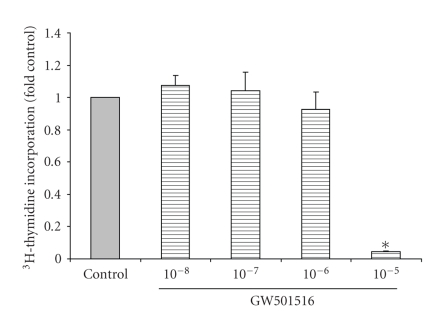
PPARδ has been known to affect cell proliferation; therefore, we studied the effect of the PPARδ agonist, GW501515, on the proliferation/growth of IMCD-K2 cells. As shown in Figure 5, 10−5 M GW501516 reduced 3H-thymidine incorporation to about 0.05-fold control. PPARδ also regulates cell growth and as shown in Figure 6, 10−5 M GW501516 produced a significant reduction in 3H-leucine incorporation to approximately 0.19 of control. At lower concentrations of GW501516, neither DNA nor protein synthesis was altered.
Figure 5.
GW501516 causes a decrease in DNA synthesis at high doses. Serum-starved cells were stimulated for 24 hours with serum-free media (control) or increasing concentrations of GW501516 (10−8 to 10−5 M) 3H-thymidine was added to IMCD-K2 cells while they were being stimulated and thymidine incorporation was measured in counts per minute using a scintillation counter, and expressed as fold control. Values are mean ± S.E.M.; n = 3. *P < .05 compared to control.
Figure 6.
GW501516 causes a decrease in protein synthesis at high doses. Serum-starved cells were stimulated for 24 hours with serum-free media (control) or increasing concentrations of GW501516 (10−8 to 10−5 M) 3H-leucine was added to IMCD-K2 cells while they were being stimulated and leucine incorporation was measured in counts per minute using a scintillation counter, and expressed as fold control. Values are mean ± S.E.M.; n = 3–5. *P < .05 compared to control.
3.5. The Effect of GW501516 on Cell Viability
To further characterize the effect of GW501516 on IMCD-K2 cells and determine if it has a role in cell survival, a cell viability assay was carried out. As shown in Figure 7, GW501516 (10−8, 10−7, 5 × 10−7, 10−6, 5 × 10−6, and 10−5 M) caused a decrease in cell viability between 0.8 and 0.6-fold of control but none of the values was significant. On its own, GW501516 did not induce death in the IMCD-K2 cells. To examine if the PPARδ agonist could protect cells from an induced death, IMCD-K2 cells were treated with both anisomycin and GW501516. Cisplatin (cis-diamminedichloridoplatinum[II]) was used as an additional positive control for cell death. As shown in Figure 8, anisomycin (100 and 1000 ng/mL) caused cell viability to decrease to approximately 0.46- and 0.37-fold of control, respectively. However, 10−6 M GW501516 did not significantly alter the decrease in cell viability seen with the addition of either 100 or 1000 ng/mL of anisomycin.
Figure 7.
GW501516 does not significantly alter IMCD-K2 cell viability. Serum-starved cells were stimulated for 24 hours with serum-free media (control) or increasing concentrations of GW501516 (10−8 to 10−5 M). The cells were then incubated for one hour with the DNA dye-binding solution from the CyQUANT NF assay kit (Invitrogen). Fluorescence intensity, representing number of cells, was then measured as per the manufacturer's instructions. Values (fold control) are means ± S.E.M.; n = 3.
Figure 8.
GW501516 has no effect on anisomycin-induced cell death. Serum-starved cells were stimulated with serum-free media (control), 10−6 M GW501516 (GW), 100 and 1000 ng/mL anisomycin (A100 and A1000, resp.), or 50 ng/mL cisplatin (Cis) for 18 hours. Some of the cells treated with anisomycin were pretreated with 10−6 GW501516 for 24 hours and cotreated with the agonist and anisomycin for 18 hours (GW/A100 and GW/A1000). The cells were then incubated for one hour with the DNA dye-binding solution from the CyQUANT NF assay kit (Invitrogen). Fluorescence intensity, representing number of cells, was then measured as per the manufacturer's instructions. Values are means ± S.E.M.; n = 3. *P < .05 compared to control; §P < .05 compared to GW.
3.6. The Effect of GW501516 on Caspase Activity
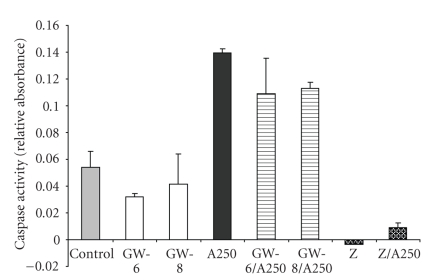
Besides having effects in cell growth, PPARδ has also been linked to changes in apoptosis. Therefore, the antiapoptotic effect of GW501516 was evaluated in IMCD-K2 cells. Caspase-3 activity was examined because it is a downstream effector protease that is common to all apoptotic pathways. As shown in Figure 9, anisomycin treatment resulted in an increase of caspase-3 levels of about 2.59-fold of control. Pre/cotreatment with GW501516 showed a decrease that was 0.79-fold (10−6 M) and 0.81-fold (10−8 M) of anisomycin alone. GW501516 alone showed no considerable change in caspase activity. The caspase inhibitor, Z-VAD completely abolished caspase activity in unstimulated cells, and in anisomycin-treated cells it decreased caspase activity to 0.06-fold of anisomycin alone.
Figure 9.
Anisomycin increases caspase activity in IMCD-K2 cells, independent of GW501516. Serum-starved cells were treated with serum-free media (control) for 24 hours, 250 ng/mL anisomycin (A250) for 24 hours, 10−6 M GW501516, and 10−8 M GW501516 for 24 hours pretreatment followed by 10−6 M and 10−8 M GW501516 for 24 hours with (GW-6/A250 and GW-8/A250) or without anisomycin (GW-6 and GW-8), Z-VAD-FMK, a pan-caspase inhibitor with (Z/A250) or without (Z) anisomycin. Values are mean ± S.E.M.
Activation of caspase was also measured by Western Blotting using a cleaved caspase-3 antibody. As shown in Figure 10(a), a 17 kDa band corresponding to cleaved caspase-3 was observed for the lysates that had been treated with anisomycin. The intensity of the bands in the lanes representing control and GW501516 alone was too low to be analyzed in the majority of the experiments; therefore, densitometric analysis was not performed for those samples. The levels of active caspase-3 (see Figure 10(b)) increased to 1.48 and 1.18 of anisomycin alone with both cotreatments (10−6 and 10−8 M GW501516, resp.) but the difference was not significant.
Figure 10.
Pretreatment with GW501516 does not significantly alter anisomycin-induced cleaved caspase-3 protein in IMCD-K2 cells. Protein was isolated from IMCD-K2 cells that had been treated with control serum-free media, 10−6 or 10−8 M GW501516, 250 ng/mL anisomycin (A250 or A) for 24 hours, 10−6 M GW501516 for 3 hours pretreatment followed by 10−6 M GW501516 for 24 hours with A250 (GW-6/A250), or 10−8 M GW501516 for 3 hours pretreatment followed by 10−8 M GW501516 for 24 hours with A250 (GW-8/A250). (a) The lysates were run on an SDS-PAGE gel and incubated with an anticleaved caspase-3 antibody (1 : 1000). (b) The membranes were stripped and incubated with anti-β-actin antibody (1 : 10000) to normalize for densitometry. Expression is presented as fold A250. Values are mean ± S.E.M.; n = 8.
3.7. The Effect of GW501516 on Apoptosis
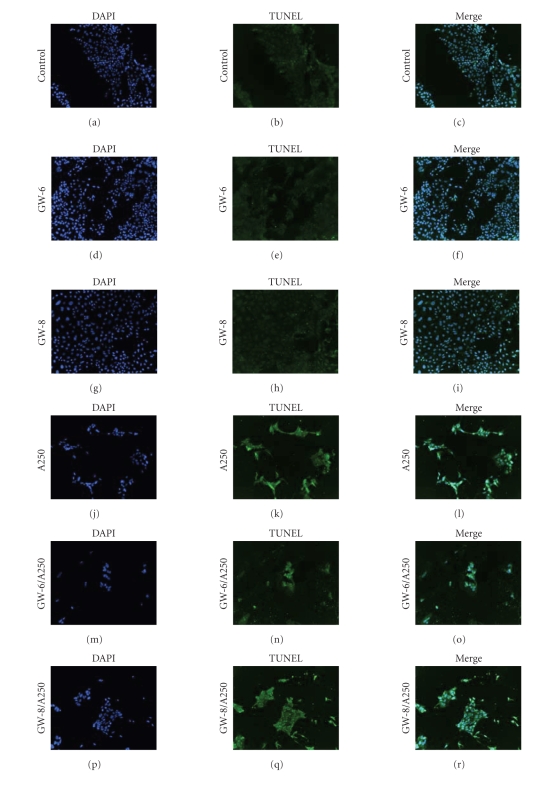
In addition to looking at the effect of GW501516 on caspase activity, TUNEL assays were performed to further evaluate the apoptotic response. As shown in Figure 11, the proportion of TUNEL-positive cells increased while the total number of cells decreases with the addition of 250 ng/mL anisomycin when compared to control or the PPARδ agonist (at both concentrations). Pre/cotreatment of 10−6 M or 10−8 M GW501516 with anisomycin was comparable to anisomycin alone.
Figure 11.
GW501516 pretreatment has no effect on anisomycin-induced apoptosis. Serum-starved cells were stimulated with (a)–(c) serum-free media (control), (d)–(f) 10−6 M GW501516 (GW-6), (g)–(i) 10−8 M GW501516 (GW-8), (j)–(l) 250 ng/mL anisomycin (A250) for 24 hours, (m)–(o) 10−6 M, or (p)–(r) 10−8 M GW501516 for 3 hours pretreatment followed by 10−6 M or 10−8 M GW501516 for 24 hours with A250 (GW-6/A250 and GW-8/A250). The cells were incubated with terminal transferase and biotin-16-dUTP for one hour, followed by two 45-minute incubations with streptavidin-Cy2 and anti-DAPI. The cells were then mounted on slides, visualized with a fluorescent microscope, and captured with a camera. Apoptotic cells are shown in green, while the blue represents DAPI-positive cells.
4. Discussion
4.1. PGE2 and PPARδ
It has been shown previously that prostaglandins, including PGE2, and PPARs influence one another's activity [27–30]. Although our studies showed that the PGE2 receptors EP1, EP3, and EP4 are present in the IMCD-K2 cells and PGE2 produces an increase in cAMP production, consistent with the expression of EP4, PGE2 had no effect on PPARδ mRNA expression. However, treatment with PGE2 for eight hours did elicit an increase in PPARδ protein levels. There are two likely explanations for this. The first reason is that the PPARδ protein could be stabilized. A previous study in a prostate cancer cell line has shown that PGE2 can stabilize hypoxia-inducible factor 1 alpha (HIF-1α) protein levels, without affecting mRNA levels [31]. This would most likely be due to PGE2 preventing proteolysis of PPARδ. Interestingly, it has been revealed that unlike most nuclear receptors, degradation of PPARδ does not occur upon ligand binding but is in fact inhibited by it [32]. This would indicate that PGE2 could be causing an increase in PPARδ activation. The second possible role of PGE2 is that it could be enhancing the translation of basal PPARδ mRNA levels or stabilizing it. The ability of PGE2 to stabilize mRNA (interleukin 8) has been previously described in a paper by Yu and Chadee [33]. Therefore, from the results reported here and the literature, the effect of PGE2 on PPARδ is most likely a posttranscriptional event. We also observed an increase in PPARδ protein in response to 24 hours indomethacin. It is not clear at this time whether this effect is due to a direct increase in expression, but it has previously been shown that PPAR expression is altered by NSAIDs [34].
4.2. PPARδ Agonist and Cell Proliferation/Growth in IMCDK2 Cells
It has been previously reported that activation of PPARδ and the use of PPARδ ligands promote cell survival and proliferation. One recent study, using both PPARδ short interfering RNA (siRNA) and a PPAR antagonist, showed that downregulation or blocking the activation of PPARδ, respectively, inhibited the PGE2-induced proliferation of mouse embryonic stem cells [14]. Another study revealed that a PPARδ agonist can stimulate propagation in both human and mouse aortic endothelial cells [35]. However, the effects of PPARδ on renal cell growth responses have not been characterized. Here we show that a highly specific PPARδ ligand, GW501516, decreased DNA and protein synthesis in IMCD-K2 cells, but only at high concentrations. Although most studies indicate that PPARδ activation results in stimulation of cell growth, mostly in human and mouse colon cancer cells [7, 36], there have been previous findings showing that PPARδ agonists, including GW501516, can hinder or have no effect on cell growth in certain human breast cancer cells and melanomas [37, 38]. These differences may be due to species differences or diversity among cell lines.
4.3. PPARδ Agonist and Anisomycin-Induced Apoptosis in IMCD-K2 Cells
Another finding in our study is that treatment with GW501516 does not reverse the reduction in cell survival due to exposure to anisomycin. These results are inconsistent with the literature that demonstrates that activation of PPARδ, or the use of a PPARδ agonist, protects many different cell types, including renal cells [12]. It is possible that the agonist may not actually be activating PPARδ in IMCD-K2 cells, as its efficacy in this cell line has never been tested. In addition, PPARδ activation may not be affecting cell survival if the death response is mostly through necrosis because studies only show a survival role for PPARδ with regard to apoptosis. To evaluate if the agonist is having an effect on PPARδ activation, experiments can be performed with cells transfected with a PPRE-driven reporter plasmid [39]. To completely assess death pathways activated by anisomycin, flow cytometry using the annexin V-FITC and propidium iodide stains should be used [40].
Even though the use of the agonist, in our experiments, elicits no response in overall cell survival in IMCD-K2 cells, the effect on apoptosis had yet to be investigated. When the CD is in a disease state due to urinary tract obstruction, ischemia-reperfusion injury or other insults apoptosis levels tend to increase [41, 42]. Thus, we wished to examine if PPARδ could protect the CD from apoptosis. Using a variety of experiments assessing apoptosis (caspase activity assay, Western blotting, TUNEL), we found no significant change in the levels of apoptosis in cells treated with GW501516 compared to those stimulated with just anisomycin. The variation between our data and the majority of the literature could be due to differences in cell types and the condition of the cells, as most studies have focussed on colorectal cancer tissues [7, 8, 43, 44]. It seems unlikely that the discrepancy in PPARδ effects is due to species differences as most of the cancer studies were done using mouse tissues. However, a few studies clearly indicate that PPARδ may not provide any prevention in certain types of apoptosis, such as in the López et al. [45] study where overexpression of PPARδ did not protect against aspirin-induced apoptosis in Jurkat cells. Thus, it is possible that PPARδ may only protect cells from apoptosis in certain conditions and cell types. From the data we collected, GW501516 has no effect on cell survival or anisomycin-induced apoptosis. The IMCD and possibly the IMCD-K2 cells are resistant to cell death because their environment in vivo is one of hypertonicity [46]. This inherent resiliency may make them resistant to different treatments, similar to that previously observed in a mouse IMCD cell line [47].
4.4. Summary
The main objective of this study was to determine whether PPARδ regulates growth and apoptosis in IMCD-K2 cells. PPARδ is highly expressed in the CD, and thus may be responsible for the protection of the CD. We showed that growth responses, including DNA and protein synthesis, in these cells are reduced by PPARδ ligands, but only at high doses. GW501516 had no effect on anisomycin-induced apoptosis in the IMCD-K2 cell line.
References
- 1.Hébert RL, Breyer RM, Jacobson HR, Breyer MD. Functional and molecular aspects of prostaglandin E receptors in the cortical collecting duct. Canadian Journal of Physiology and Pharmacology. 1995;73(2):172–179. doi: 10.1139/y95-026. [DOI] [PubMed] [Google Scholar]
- 2.Breyer MD, Davis L, Jacobson HR, Breyer RM. Differential localization of prostaglandin E receptor subtypes in human kidney. American Journal of Physiology. 1996;270(5, part 2):F912–F918. doi: 10.1152/ajprenal.1996.270.5.F912. [DOI] [PubMed] [Google Scholar]
- 3.Breyer MD, Breyer RM. G protein-coupled prostanoid receptors and the kidney. Annual Review of Physiology. 2001;63:579–605. doi: 10.1146/annurev.physiol.63.1.579. [DOI] [PubMed] [Google Scholar]
- 4.Lim H, Dey SK. PPARδ functions as a prostacyclin receptor in blastocyst implantation. Trends in Endocrinology and Metabolism. 2000;11(4):137–142. doi: 10.1016/s1043-2760(00)00243-5. [DOI] [PubMed] [Google Scholar]
- 5.Guan Y. Targeting peroxisome proliferator-activated receptors (PPARs) in kidney and urologic disease. Minerva Urologica e Nefrologica. 2002;54(2):65–79. [PubMed] [Google Scholar]
- 6.Hao C-M, Redha R, Morrow J, Breyer MD. Peroxisome proliferator-activated receptor δ activation promotes cell survival following hypertonic stress. The Journal of Biological Chemistry. 2002;277(24):21341–21345. doi: 10.1074/jbc.M200695200. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Wang H, Shi Q, et al. Prostaglandin E2 promotes colorectal adenoma growth via transactivation of the nuclear peroxisome proliferator-activated receptor δ . Cancer Cell. 2004;6(3):285–295. doi: 10.1016/j.ccr.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Gupta RA, Tan J, Krause WF, et al. Prostacyclin-mediated activation of peroxisome proliferator-activated receptor δ in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(24):13275–13280. doi: 10.1073/pnas.97.24.13275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harman FS, Nicol CJ, Marin HE, Ward JM, Gonzalez FJ, Peters JM. Peroxisome proliferator-activated receptor-δ attenuates colon carcinogenesis. Nature Medicine. 2004;10(5):481–483. doi: 10.1038/nm1026. [DOI] [PubMed] [Google Scholar]
- 10.Wang D, Wang H, Guo Y, et al. Crosstalk between peroxisome proliferator-activated receptor δ and VEGF stimulates cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(50):19069–19074. doi: 10.1073/pnas.0607948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan Y. Peroxisome proliferator-activated receptor family and its relationship to renal complications of the metabolic syndrome. Journal of the American Society of Nephrology. 2004;15(11):2801–2815. doi: 10.1097/01.ASN.0000139067.83419.46. [DOI] [PubMed] [Google Scholar]
- 12.Letavernier E, Perez J, Joye E, et al. Peroxisome proliferator-activated receptor β/δ exerts a strong protection from ischemic acute renal failure. Journal of the American Society of Nephrology. 2005;16(8):2395–2402. doi: 10.1681/ASN.2004090802. [DOI] [PubMed] [Google Scholar]
- 13.Hatae T, Wada M, Yokoyama C, Shimonishi M, Tanabe T. Prostacyclin-dependent apoptosis Mediated by PPARδ . The Journal of Biological Chemistry. 2001;276(49):46260–46267. doi: 10.1074/jbc.M107180200. [DOI] [PubMed] [Google Scholar]
- 14.Kim YH, Han HJ. High-glucose-induced prostaglandin E2 and peroxisome proliferator-activated receptor δ promote mouse embryonic stem cell proliferation. Stem Cells. 2008;26(3):745–755. doi: 10.1634/stemcells.2007-0786. [DOI] [PubMed] [Google Scholar]
- 15.Jarvis MC, Gray TJB, Palmer CNA. Both PPARγ and PPARδ influence sulindac sulfide-mediated p21WAF1/CIP1 upregulation in a human prostate epithelial cell line. Oncogene. 2005;24(55):8211–8215. doi: 10.1038/sj.onc.1208983. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Fu M, Zhu X, et al. Peroxisome proliferator-activated receptor δ is up-regulated during vascular lesion formation and promotes post-confluent cell proliferation in vascular smooth muscle cells. The Journal of Biological Chemistry. 2002;277(13):11505–11512. doi: 10.1074/jbc.M110580200. [DOI] [PubMed] [Google Scholar]
- 17.Khandrika L, Kim FJ, Campagna A, Koul S, Meacham RB, Koul HK. Primary culture and characterization of human renal inner medullary collecting duct epithelial cells. The Journal of Urology. 2008;179(5):2057–2063. doi: 10.1016/j.juro.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi J, Imai M. Computer analysis of the significance of the effective osmolality for urea across the inner medullary collecting duct in the operation of a single effect for the counter-current multiplication system. Clinical and Experimental Nephrology. 2006;10(4):236–243. doi: 10.1007/s10157-006-0436-5. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Cai Q, Michea L, Dmitrieva NI, Andrews P, Burg MB. Proliferation and osmotic tolerance of renal inner medullary epithelial cells in vivo and in cell culture. American Journal of Physiology. 2002;283(2):F302–F308. doi: 10.1152/ajprenal.00038.2002. [DOI] [PubMed] [Google Scholar]
- 20.Burg MB. Renal osmoregulatory transport of compatible organic osmolytes. Current Opinion in Nephrology and Hypertension. 1997;6(5):430–433. doi: 10.1097/00041552-199709000-00004. [DOI] [PubMed] [Google Scholar]
- 21.Burg MB, Ferraris JD, Dmitrieva NI. Cellular response to hyperosmotic stresses. Physiological Reviews. 2007;87(4):1441–1474. doi: 10.1152/physrev.00056.2006. [DOI] [PubMed] [Google Scholar]
- 22.Yang T, Schnermann JB, Briggs JP. Regulation of cyclooxygenase-2 expression in renal medulla by tonicity in vivo and in vitro. American Journal of Physiology. 1999;46(1):F1–F9. doi: 10.1152/ajprenal.1999.277.1.F1. [DOI] [PubMed] [Google Scholar]
- 23.Kizer NL, Lewis B, Stanton BA. Electrogenic sodium absorption and chloride secretion by an inner medullary collecting duct cell line (mIMCD-K2) American Journal of Physiology. 1995;268(2, part 2):F347–F355. doi: 10.1152/ajprenal.1995.268.2.F347. [DOI] [PubMed] [Google Scholar]
- 24.Wei Z-L, Kozikowski AP. A short and efficient synthesis of the pharmacological research tool GW501516 for the peroxisome proliferator-activated receptor δ . Journal of Organic Chemistry. 2003;68(23):9116–9118. doi: 10.1021/jo035140g. [DOI] [PubMed] [Google Scholar]
- 25.Båtshake B, Sundelin J. The mouse genes for the EP1 prostanoid receptor and the PKN protein kinase overlap. Biochemical and Biophysical Research Communications. 1996;227(1):70–76. doi: 10.1006/bbrc.1996.1469. [DOI] [PubMed] [Google Scholar]
- 26.Breyer RM, Emeson RB, Tarng J-L, et al. Alternative splicing generates multiple isoforms of a rabbit prostaglandin E2 receptor. The Journal of Biological Chemistry. 1994;269(8):6163–6169. [PubMed] [Google Scholar]
- 27.Yu K, Bayona W, Kallen CB, et al. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. The Journal of Biological Chemistry. 1995;270(41):23975–23983. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- 28.Nosjean O, Boutin JA. Natural ligands of PPARγ: are prostaglandin J2 derivatives really playing the part? Cellular Signalling. 2002;14(7):573–583. doi: 10.1016/s0898-6568(01)00281-9. [DOI] [PubMed] [Google Scholar]
- 29.Xu L, Han C, Wu T. A novel positive feedback loop between peroxisome proliferator-activated receptor-δ and prostaglandin E2 signaling pathways for human cholangiocarcinoma cell growth. The Journal of Biological Chemistry. 2006;281(45):33982–33996. doi: 10.1074/jbc.M600135200. [DOI] [PubMed] [Google Scholar]
- 30.Han S, Ritzenthaler JD, Wingerd B, Roman J. Activation of peroxisome proliferator-activated receptor β/δ (PPARβ/δ) increases the expression of prostaglandin E2 receptor subtype EP4. The roles of phosphatidylinositol 3-kinase and CCAAT/enhancer-binding protein β . The Journal of Biological Chemistry. 2005;280(39):33240–33249. doi: 10.1074/jbc.M507617200. [DOI] [PubMed] [Google Scholar]
- 31.Liu XH, Kirschenbaum A, Lu M, et al. Prostaglandin E2 induces hypoxia-inducible factor-1α stabilization and nuclear localization in a human prostate cancer cell line. The Journal of Biological Chemistry. 2002;277(51):50081–50086. doi: 10.1074/jbc.M201095200. [DOI] [PubMed] [Google Scholar]
- 32.Genini D, Catapano CV. Block of nuclear receptor ubiquitination: a mechanism of ligand-dependent control of peroxisome proliferator-activated receptor δ activity. The Journal of Biological Chemistry. 2007;282(16):11776–11785. doi: 10.1074/jbc.M609149200. [DOI] [PubMed] [Google Scholar]
- 33.Yu Y, Chadee K. Prostaglandin E2 stimulates IL-8 gene expression in human colonic epithelial cells by a posttranscriptional mechanism. The Journal of Immunology. 1998;161(7):3746–3752. [PubMed] [Google Scholar]
- 34.Schwab M, Reynders V, Loitsch S, et al. PPARγ is involved in mesalazine-mediated induction of apoptosis and inhibition of cell growth in colon cancer cells. Carcinogenesis. 2008;29(7):1407–1414. doi: 10.1093/carcin/bgn118. [DOI] [PubMed] [Google Scholar]
- 35.Piqueras L, Reynolds AR, Hodivala-Dilke KM, et al. Activation of PPARβ/δ induces endothelial cell proliferation and angiogenesis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2007;27(1):63–69. doi: 10.1161/01.ATV.0000250972.83623.61. [DOI] [PubMed] [Google Scholar]
- 36.Yang L, Zhou Z-G, Zheng X-L, et al. RNA interference against peroxisome proliferator-activated receptor δ gene promotes proliferation of human colorectal cancer cells. Diseases of the Colon & Rectum. 2008;51(3):318–326. doi: 10.1007/s10350-007-9145-8. [DOI] [PubMed] [Google Scholar]
- 37.Hollingshead HE, Killins RL, Borland MG, et al. Peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) ligands do not potentiate growth of human cancer cell lines. Carcinogenesis. 2007;28(12):2641–2649. doi: 10.1093/carcin/bgm183. [DOI] [PubMed] [Google Scholar]
- 38.Girroir EE, Hollingshead HE, Billin AN, et al. Peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) ligands inhibit growth of UACC903 and MCF7 human cancer cell lines. Toxicology. 2008;243(1-2):236–243. doi: 10.1016/j.tox.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sumanasekera WK, Tien ES, Davis JW, II, Turpey R, Perdew GH, Vanden Heuvel JP. Heat shock protein-90 (Hsp90) acts as a repressor of peroxisome proliferator-activated receptor-α (PPARα) and PPARβ activity. Biochemistry. 2003;42(36):10726–10735. doi: 10.1021/bi0347353. [DOI] [PubMed] [Google Scholar]
- 40.Chen S, Cheng A-C, Wang M-S, Peng X. Detection of apoptosis induced by new type gosling viral enteritis virus in vitro through fluorescein annexin V-FITC/PI double labeling. World Journal of Gastroenterology. 2008;14(14):2174–2178. doi: 10.3748/wjg.14.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen T, Loutochin O, Amin M, Capolicchio J-P, Goodyer P, Jednak R. PAX2 is reactivated in urinary tract obstruction and partially protects collecting duct cells from programmed cell death. American Journal of Physiology. 2007;292(4):F1267–F1273. doi: 10.1152/ajprenal.00281.2006. [DOI] [PubMed] [Google Scholar]
- 42.Han K-H, Kim H-Y, Croker BP, et al. Effects of ischemia-reperfusion injury on renal ammonia metabolism and the collecting duct. American Journal of Physiology. 2007;293(4):F1342–F1354. doi: 10.1152/ajprenal.00437.2006. [DOI] [PubMed] [Google Scholar]
- 43.Shureiqi I, Jiang W, Zuo X, et al. The 15-lipoxygenase-1 product 13-S-hydroxyoctadecadienoic acid down-regulates PPAR-δ to induce apoptosis in colorectal cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(17):9968–9973. doi: 10.1073/pnas.1631086100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liou J-Y, Ghelani D, Yeh S, Wu KK. Nonsteroidal anti-inflammatory drugs induce colorectal cancer cell apoptosis by suppressing 14-3-3ε . Cancer Research. 2007;67(7):3185–3191. doi: 10.1158/0008-5472.CAN-06-3431. [DOI] [PubMed] [Google Scholar]
- 45.López JM, Fernández MA, Piqué M, Gil J. Aspirin-induced apoptosis in Jurkat cells is not mediated by peroxisome proliferator-activated receptor delta. Molecular and Cellular Biochemistry. 2004;266(1-2):57–63. doi: 10.1023/b:mcbi.0000049138.67699.7b. [DOI] [PubMed] [Google Scholar]
- 46.Rauchman MI, Nigam SK, Delpire E, Gullans SR. An osmotically tolerant inner medullary collecting duct cell line from an SV40 transgenic mouse. American Journal of Physiology. 1993;265(3, part 2):F416–F424. doi: 10.1152/ajprenal.1993.265.3.F416. [DOI] [PubMed] [Google Scholar]
- 47.Maroni PD, Koul S, Meacham RB, Chandhoke PS, Koul HK. Effects of oxalate on IMCD cells: a line of mouse inner medullary collecting duct cells. Annals of the New York Academy of Sciences. 2004;1030:144–149. doi: 10.1196/annals.1329.018. [DOI] [PMC free article] [PubMed] [Google Scholar]