Abstract
The ETS protein Spi-1/Pu.1 plays a pivotal and widespread role throughout hematopoiesis in many species. This study describes the identification, characterization, and functional analysis of a new zebrafish spi transcription factor spi-1–like (spi-1l) that is expressed in primitive myeloid cells, erythro-myelo progenitor cells, and in the adult kidney. Spi-1l functions genetically downstream of etsrp, scl, and spi-1/pu.1 in myeloid differentiation. Spi-1l is coexpressed in a subset of spi-1/pu.1 cells and its function is necessary and sufficient for macrophage and granulocyte differentiation. These results establish a critical role for spi-1l in zebrafish myeloid cell differentiation.
Introduction
Zebrafish (Danio rerio) and mammalian hematopoiesis share many similarities and zebrafish is therefore recognized as an excellent model system in which to study blood development.1 Similar primitive and definitive stages have been described for zebrafish and mammalian hematopoiesis, and they both share all major blood cell types related by common lineages and developmental pathways.2,3 Primitive hematopoiesis in zebrafish takes place in 2 locations: the anterior lateral plate mesoderm (A-LPM), which gives rise to myeloid cells, while the posterior lateral plate mesoderm (P-LPM) forms the intermediate cell mass (ICM) that gives rise to primitive erythrocytes.4,5 Hematopoietic stem cells (HSCs) capable of self-renewing and contributing to all the different blood lineages arise during definitive hematopoiesis. HSCs begin developing at approximately 32 hours after fertilization (hpf) from the ventral side of the dorsal aorta similar to that seen in the mammalian aorta-gonad-mesonephros (AGM) region.6 HSCs are marked by expression of runx1 and c-myb, and migrate to the caudal hematopoietic tissue (CHT) where they expand and further develop before moving to the third site of hematopoiesis, the kidney.7–9 The kidney is the adult hematopoietic site in zebrafish akin to the mammalian bone marrow.10
ETS domain–containing transcription factors are required for hematopoiesis across many species.11–13 Some of the most divergent members of the ETS domain subfamilies include SPI genes,14 the subject of this study. Mammals have 3 SPI genes: SPI-1/PU.1, SPI-B, and SPI-C, each of which plays an essential role in hematopoiesis.15,16 PU.1 is an important regulator that both activates and interacts with other transcription factors such as c-JUN, GATA-1, GATA-2, C/EBP, and RUNX-1,17,18 during cell proliferation, differentiation, lineage commitment, apoptosis, and homing, and is expressed in most hematopoietic lineages, from the HSC stage through differentiated cell types.13,15,19 The precise control of mammalian PU.1 is maintained by its upstream regulatory element (URE), which autoregulates PU.1 expression.20 The strict control of PU.1 expression is important for proper cell differentiation and in preventing cells from assuming a leukemic fate.
In zebrafish, the spi-1/pu.1 ortholog was the only member of the SPI class of genes thus far characterized.21 Embryonic expression of pu.1 in the A-LPM marks early primitive myeloid cells and is necessary for their development. Pu.1 is also expressed in the P-LPM where it has an antagonistic function with gata1 in the development of erythrocytes.22 Ectopic expression of pu.1 is able to partially rescue myelopoiesis in cloche mutants, which lack hematopoietic and vascular progenitors.22,23 Cells that express pu.1 are uncommon in the zebrafish adult kidney, the site of adult hematopoiesis, and its function in this tissue has not yet been fully characterized.
Given the diversity and importance of mammalian SPI function in hematopoiesis we decided to examine the zebrafish spi gene in some detail. We identified a new spi gene that we call spi-1–like (spi-1l), and show that spi-1l is expressed in cells derived from the A-LPM. We further demonstrate that spi-1l is a myeloid-specific gene that genetically functions downstream of Pu.1 and is necessary for proper myeloid cell differentiation.
Methods
Reverse-transcriptase PCR
Total RNA was purified from wild-type adult kidney and wild-type embryos using Trizol Reagent and protocol from Invitrogen (Frederick, MD). cDNA was generated using polyA primers and protocol from Invitrogen. Polymerase chain reaction (PCR) was performed using primers spi-1l forward ACCATGGAGAGCTGCGTTATTTC and spi-1l reverse TCACCCTGACCAGTTAAAGC and pfu Taq using a iCycler (Bio-Rad, Hercules, CA) with the following cycling conditions: 94°C for 4 minutes, then 30 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute, with a final extension of 72°C for 20 minutes.
Microinjection
Two spi-1l–specific MOs (MO1 [translation blocking], 5′-AACGCAGCTCTCCATTCTGTAATGC-3′ and MO2 [splice blocking] 5′-AGCGACTCACGCTGTGGAGGAACT-3′; Gene Tools, Philomath, OR) were used to inhibit the function of spi-1l mRNA. For analysis of spi-1l function, approximately 10 ng/embryo MO1 or MO2 was injected from 1- to 2-cell stage embryos. A standard control MO 5′-CCTCTTACCTCAGTTACAATTTATA-3′ was used from Gene Tools. A previously described pu.1 MO was used to assess loss of pu.1 function (kindly provided by J. P. Kanki, Department of Pediatric Oncology, Dana-Farber Cancer Institute, Boston, MA).22 Typically, 3 independent experiments were performed for each MO with a minimum of 15 to 20 embryos per experiment analyzed. Spi-1l mRNA was created by subcloning spi-1l open reading frame into the EcoR1 site of pCS2. Spi-1l mRNA was synthesized by digesting with Not1 and transcribing with the SP6 mMessage Machine Kit (Ambion, Austin, TX).
In situ hybridization
Whole-mount in situ hybridization was performed as described by Griffin et al.24 Double fluorescent in situ hybridization was performed as described by Schoenebeck25 and Sumanas et al.26 To synthesize dioxigenin (DIG)–labeled probe, spi-1l-pCR4-TOPO construct was digested with Spe1 and transcribed with T7 RNA polymerase. DNP probes were synthesized as described by Schoenebeck25 and Sumanas et al.26 The following probes were used spi-1l, mpx,27 l-plastin,5 c-myb,23 runx-1,28 spi-1,21 gata-1,29 gata-2,30 and scl.31
Zebrafish strains
Most of the spi-1l and pu.1 morpholino knockdown analysis was performed in wild-type zebrafish from Scientific Hatcheries (Huntington Beach, CA). We also used a clom39;Tg(kdr:EGFP)s843 zebrafish line for overexpression experiments. The UCLA Institutional Review Board approved the zebrafish research.
Image acquisition and processing
All images except the double fluorescent in situ hybridization were taken on a Zeiss Axioskop 2 plus microscope using a Zeiss 5×/0.15 NA dry objective (magnification, 50×) with a Zeiss Axiocam HRc digital camera, using AxioVision Rel. 4.3 software (all from Zeiss, Oberkochen, Germany). The double fluorescent in situ images were taken with a Zeiss LSM 510 Confocal Microscope Imager.Z1 using a 20×/0.50 NA dry objective (Zeiss; magnification, 200×) and LSM 510 software. Photoshop SC software (Adobe Systems, San Jose, CA) was used for further image processing. In general, images in different focal planes were combined using Photoshop to yield the highest clarity image.
Phylogenic analysis
Sequence identity values were determined by the EMBOSS pairwise alignment algorithm. The dendrogram was constructed using Clustal-x 232 for alignment and viewed with Treeview33 using human ELF5 as an outgroup. Bootstrap values were derived from 1000 bootstrap trials.
Tables and graphs
No less than 5 embryos per condition were counted from 3 separate experiments. The number of cells counted was averaged and a standard deviation was derived.
Results
Spi-1l, a novel ETS domain–containing transcription factor
Using an in silico approach, we identified a 2126-bp expressed sequence tag (EST; zgc:152991)34 encoding a novel ETS domain protein with high sequence similarity to PU.1. Using primers against the EST sequence, we performed a reverse-transcriptase–polymerase chain reaction (RT-PCR) on zebrafish adult kidney mRNA and identified a 2296-nucleotide transcript (GenBank accession no. EU68525335). The isolated cDNA contained an additional 170 bp ETS domain coding sequence over the annotated EST.
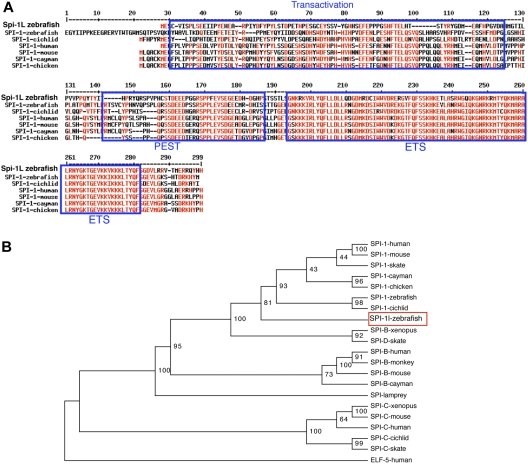
Sequence analysis of the RT-PCR product confirmed the coding potential to represent an ETS domain protein (Figure 1A). The protein sequence contains 252 amino acids with 3 predicted domains: an N-terminal presumptive transactivation (TA) domain, a PEST sequence, and a C-terminal ETS domain (Figure 1A). At the amino acid level, Spi-1l shares 45% overall identity with zebrafish Pu.1 and 49% identity with human PU.1. A lesser, 37% identity with human SPI-B and 25% with SPI-C was observed. The highest similarity between Spi-1l and human PU.1 is restricted to the ETS domain (77%), followed by the PEST domain (28%), and finally the TA domain (20%). A phylogenetic analysis of the SPI family members illustrates that spi-1l is most closely related to the to the SPI-1/PU.1 subfamily (Figure 1B). Zebrafish spi-1l has not been mapped to a linkage group yet.
Figure 1.
Spi-1l is a new SPI family member. (A) Alignment of Spi-1l and its closest homologous SPI-1/PU.1 proteins. Identical and similar amino acids are indicated in red. Blue boxes identify the different SPI protein domains. (B) Phylogenetic analysis of SPI family members based on the ETS DNA binding domain. GenBank GI (or accession) numbers: human SPI-1 4507175, mouse SPI-1 6755473, cayman SPI-1 8745406, chicken SPI-1 2369863, skate SPI-1 11245497, zebrafish SPI-1 AF321099, cichlid SPI-1 8745412, zebrafish Spi-1l EU685253, human SPI-B 36562, monkey SPI-B AF025395, mouse SPI-B 2586116, cayman SPI-B 8745407, Xenopus SPI-B 8745409, skate SPI-D 11245499, lamprey SPI 8745404, human SPI-C BC032317, Xenopus SPI-C BC077851, cichlid SPI-C 8745414, skate SPI-C 11245501, mouse SPI-C 6755618, and human EFL5 4557551. The phylogenetic tree is built using the neighbor joining method. Length of horizontal branches is proportional to the evolutionary distance between the protein molecules. The value at each node represents the probability that that branch length is not zero. This tree indicates that Spi-1l is most related to SPI-1 of all the SPI family members.
Spi-1l expression in early zebrafish development
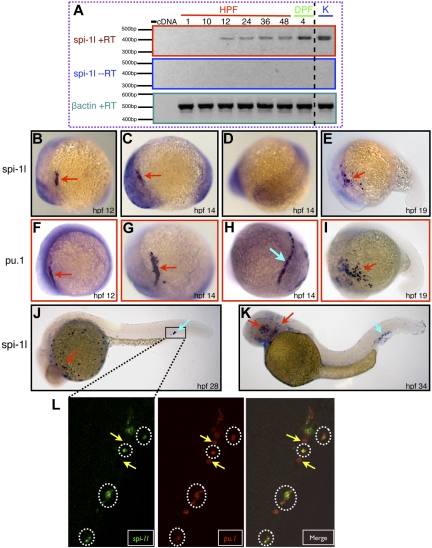
We first analyzed spi-1l expression by reverse-transcriptase PCR. We isolated total mRNA from different embryonic stages and determined that spi-1l expression begins at 12 hours after fertilization (hpf) and continues through 4 days after fertilization (dpf); it is also expressed in the adult kidney (Figure 2A). RT-PCR analysis also revealed that spi-1l is not expressed in the embryo at 1 hour after fertilization, indicating that it is not maternally inherited (Figure 2A). Whole-mount in situ hybridization revealed that spi-1l expression is similar to pu.1, but spi-1l expression is in a smaller number of cells than pu.1 at the same developmental stage (comparing Figure 2B-E with Figure 2F-I).
Figure 2.
Embryonic expression of spi-1l mRNA. (A) Spi-1l expression analysis by RT-PCR. −cDNA lane is a negative control in which cDNA was not added to the PCR. Lanes indicated with hours after fertilization (HPF) and days after fertilization (DPF) represent different developmental stages. K indicates adult kidney. −RT is a control reaction without the reverse-transcriptase enzyme added to the RT-PCR. β actin is used as a loading control. A dashed black line between the DPF and K lanes has been inserted to indicate a repositioned gel lane. (B-K) Whole-mount in situ hybridization analysis of spi-1l and pu.1 expression. The probe used is indicated to the left of the panels and the embryo's age marked by hours after fertilization (hpf). The embryos are oriented with anterior to the left and dorsal up. Red arrows point to the A-LPM region and blue arrows point to the P-LPM or PBI/CHT regions. All panels (B-K) except panels D and H show lateral views of embryos. (D,H) Posterior view of the same embryo as in panels C and G, respectively. Expression of spi-1l (B,C,E) and pu.1 (F,G,I) in the A-LPM. (D) spi-1l expression is absent in the P-LPM in contrast with pu.1, which is expressed in this region (H). (J) Expression of spi-1l throughout the yolk and in the posterior blood island (PBI). (K) Expression of spi-1l in the vessels of the eye and in the caudal hematopoietic tissue (CHT). (L) Single confocal section image at 28 hpf of fluorescent double in situ hybridization of spi-1l (green) and pu.1 (red). Right panel shows the merged image. Approximate region of the embryo scanned for the fluorescent double in situ hybridization is represented by the box in panel J. Hatched white circles indicate cells coexpressing spi-1l and pu.1 and yellow arrows indicate cells expressing only spi-1.
Expression of spi-1l is first observed in the anterior lateral plate mesoderm (A-LPM) at the 6-somite stage (Figure 2B), its expression expanding by the 10-somite stage (Figure 2C). Unlike pu.1, spi-1l is not expressed in the posterior lateral plate mesoderm (P-LPM; Figure 2D,H). By the 19-somite stage, spi-1l–expressing cells begin to disperse and spread over the yolk (Figure 2E), their number steadily increases, enveloping the yolk as the embryo develops. By 28 hpf, there are spi-1l cells all around the yolk and also in the posterior blood island (PBI), where erythro-myelo progenitors (EMPs) are located (Figure 2J). At 34 hpf, spi-1l–expressing cells appear in the posterior cardinal vein in the vessels of the head and retina, as well as in the venous plexus CHT (Figure 2K), which serves as a temporary residence for HSCs and/or EMP cells.7–9,36 We further determined that this expression of spi-1l in EMP cells was not due solely to migration by examining silent heart37,38 mutant embryos. These embryos do not have blood circulation due to a heart defect, but they still express spi-1l in the PBI/CHT (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), supporting recent findings that the EMP cells arise de novo in this tissue.7,36
Double fluorescent in situ hybridization using probes for zebrafish pu.1, and spi-1l, revealed that the expression pattern of the 2 genes was partially overlapping. We determined that spi-1l and pu.1 are coexpressed in a subset of pu.1-positive cells in the CHT (Figure 2L). In addition to double-positive spi-1l/pu.1 cells, we also identified single-positive pu.1 cells (Figure 2L), however single-positive spi-1l cells were rarely observed. Our expression data are consistent with either only a specific subset of pu.1-positive cells expressing spi-1l, or pu.1-expressing cells activating spi-1l expression during differentiation.
Morpholino knockdown of spi-1l function
To address the role of spi-1l in hematopoiesis, we decided to determine whether there is a phenotype due to loss of spi-1l function. The expression pattern of spi-1l in the A-LPM and the CHT prompted us to elucidate its function through the use of antisense morpholino oligonucleotides (MOs) to knock down its expression.39,40 Two different MOs, one a splice blocker and another a translation blocker, were used to assess the function of spi-1l. For the embryos injected with the splice-blocking morpholino at 8 to 10 ng/embryo, the spi-1l transcript is 114 bp shorter compared with wild-type embryos (Figure S2A), consistent with an exon being skipped during splicing due to spi-1l MO function. Further, we used a translation-blocking MO to reduce Spi-1l function by also injecting with 8 to 10 ng/embryo of the MO. The expression of spi-1l in embryos injected with the translation-blocking MO was severely reduced (not shown). The morphants appear morphologically normal and the MOs used seem to be specific, effective, and nontoxic at the dosage at which they were used. (For the subsequent text, where spi-1l MO is used, the splice-blocking morpholino was used except where noted otherwise.)
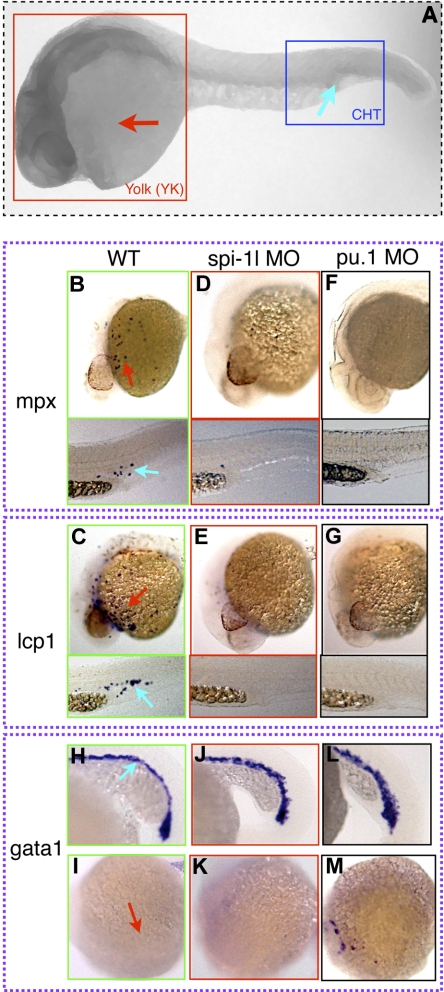
We subsequently analyzed whether the loss of spi-1l would affect hematopoiesis by examining a variety of molecular markers in spi-1l morphants. Among the markers tested, mpx and lcp1 are affected, whereas pu.1, scl, gata1, gata2, runx1, rag1, and c-myb appear normal. The expression of 2 different markers of myeloid differentiation, myeloid-specific peroxidase (mpx), which is expressed in granulocytes,5,27 and L-plastin (lcp1), which is expressed in macrophages41 and neutrophils,42 are largely eliminated in spi-1l morphants (higher magnification view in Figure 3B-E; full image view Figure S2B-E). Embryos injected with spi-1l translation-blocking MO show a consistent phenotype (Figure S2H-K) to that of the splice-blocking MO, and embryos injected with a control MO did not show a decrease in mpx or lcp1 expression (Figure S2L-O). To further demonstrate the specificity of the spi-1l MO, a rescue experiment was performed by coinjection of the spi-1l splice-blocking MO with spi-1l mRNA. This resulted in almost complete rescue of mpx in 68% (47/69) and lcp1 in 64% (41/64) of the injected embryos. The loss of granulocyte and macrophage markers in spi-1l MO embryos indicates that spi-1l functions in myeloid cell differentiation. In addition, the loss of mpx and lcp1 expression in the PBI/CHT (Figure 3B-E lower panel) indicates the need for spi-1l function in the development of the EMPs, which reside in this tissue.7 These myeloid markers are also eliminated in pu.1 morphants (Figure 3F,G),22 suggesting an overlapping function of Spi-1l and Pu.1 in myeloid differentiation. We also determined whether lack of spi-1l expression would affect erythropoiesis by examining gata1 expression. No change in gata1 expression in the ICM, or ectopic anterior expression of gata1 at 20 hpf, was seen in spi-1l morphants (Figure 3H-K), as was previously seen with pu.1 MO (Figure 3L,M).22 Thus Spi-1l is not required for the repression of the erythroid fate in the A-LPM, where it is coexpressed with pu.1, or in the P-LPM, consistent with the observed lack of spi-1l expression in the ICM.
Figure 3.
Effect of spi-1l loss on hematopoiesis. (A) Image of a zebrafish embryo for orientation of the rest of the figure panels. Red box surrounds the yolk (YK) area and blue box surrounds the caudal hematopoietic tissue (CHT). (B-M) Red and blue arrows indicate mRNA expression in the YK and the CHT regions, respectively, of wild-type embryos. Probes for genes and morpholino used are as indicated. (B-G) Lateral view of 28-hpf embryos. Expression of (B) mpx and (C) lcp1 in wild-type sibling embryo. Spi-1l and pu.1 morphant embryos exhibit an almost complete absence of granulocyte (mpx) (D,F) and macrophage (lcp1) (E,G) markers. (H-M) Twenty-two–hpf embryos. (H) Expression of gata1 in control uninjected embryo in the ICM, (I) the head region, demonstrating wild-type gata1 expression in the ICM and absence of expression in the A-LPM. (J) gata1 expression is unchanged in spi-1l morphants in the ICM; (K) there is no expression in the head region. (L) Expression of gata1 in the ICM and (M) ectopic expression of gata1 in the head region of pu.1 morphant.
We next examined whether early hematopoietic genes, definitive hematopoiesis, or lymphoid genes are affected in spi-1l morphants. No effect was seen in the expression of 2 of the earliest markers of HSC formation, runx1 or c-myb (Figure S3B-E). Similarly, the expression of the early hematopoietic genes scl and gata2 is not affected in spi-1l morphant embryos (Figure S3F-I). The expression of rag1, a lymphoid-specific gene was also not affected due to the loss of spi-1l function (Figure S3J,K). These results further demonstrate that spi-1l functions in later myeloid differentiation, but not in early primitive hematopoiesis, in the formation of HSCs or during embryonic lymphopoiesis.
We analyzed whether the loss of both spi-1l and pu.1 genes in zebrafish would cause a greater defect in hematopoiesis than just the loss of any one alone. The spi-1l/pu.1 double morphants lost mpx and lcp1 expression, as was seen in the individual morphant, and gata2 expression is not affected either in the double or the single morphant (not shown). We saw no increase in the loss of runx1 expression in the spi-1l/pu.1 double morphant than what is seen in the pu.1 morphant alone; also expression of c-myb remains unchanged in the double morphants. Thus, the loss of both pu.1 and spi-1l does not cause a more robust phenotype than the loss of the individual genes.
Regulation of spi-1l expression
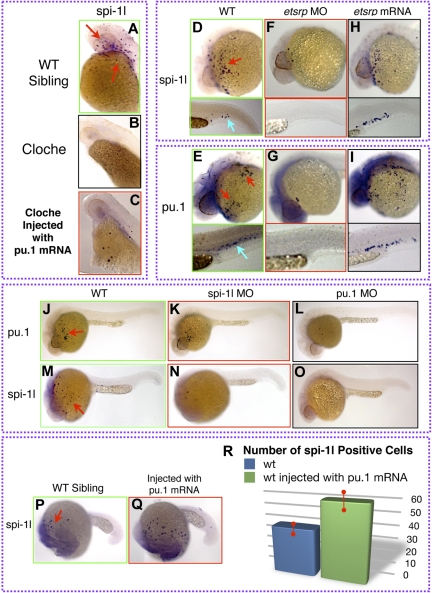
Zebrafish cloche mutant embryos have been used extensively in assessing both hematopoietic and vascular development, since these mutants fail to develop differentiated myeloid, erythroid, and most of the vascular cells. We used a cloche flk-GFP transgenic zebrafish line to further understand spi-1l function during hematopoiesis. Through expression analysis, we established that spi-1l is not detected in homozygous cloche mutants (Figure 4A,B). Next, we injected embryos from a heterozygous cross of cloche zebrafish at the 1- to 2-cell stage with pu.1 mRNA. In the homozygous cloche embryos (as identified by morphologic criteria and by the lack of flk1-GFP expression), spi-1l expression was partially restored (Figure 4A-C; Figure S4A-C). This result shows that spi-1l expression can be activated by exogenous pu.1 mRNA expression in cloche mutants.
Figure 4.
Regulation of spi-1l expression. (A-C) Lateral view of the yolk (YK) region at 36 hpf of cloche wild-type sibling, cloche, and injected cloche embryo, respectively. (A) Expression of spi-1l in a wild-type sibling from a cloche intercross. (B) Expression of spi-1l is absent in cloche mutant embryo. (C) Partial rescue of spi-1l expression in cloche mutant injected with pu.1 mRNA. (D-Q) Lateral view of embryos. Probes for genes, morpholino, and mRNA used are as indicated. Red and blue arrows indicate mRNA expression on the YK and in the CHT regions, respectively, of wild-type embryo. (D-I) Etsrp as an upstream component of spi-1l and pu.1. (D) There are 87 plus or minus 14 spi-1l cells and (E) 106 plus or minus 14 pu.1 cells expressed in control uninjected embryos at 28 hpf. Etsrp morphants lack spi-1l (F) and pu.1 (E) expression. Ectopic etsrp mRNA expression causes increase in the number of spi-1l cells to 114 plus or minus 27 (H), and pu.1 cells to 152 plus or minus 30 (I). (J-O) Lateral view of spi-1l and pu.1 morphants at 28 hpf. Wild-type expression of pu.1 (J) and spi-1l (M) in control uninjected embryos. (K) Expression of pu.1 is unaffected in spi-1l morphants. (L) As a control, expression of pu.1 is lost in pu.1 morphants. (N) As a control, expression of spi-1l is severely reduced in spi-1l morphants. (O) Expression of spi-1l is nearly absent in pu.1 morphants. (P) Wild-type expression of spi-1l at 22 hpf. (Q) Wild-type embryo injected with pu.1 mRNA has increased number of spi-1l–expressing cells. (R) Graph demonstrating the increase in the number of spi-1l–positive cells in pu.1-injected embryos compared with wild-type siblings. We determined that the number of spi-1l–positive cells was 59 plus or minus 9 in the injected embryos compared with 37 plus or minus 4 in uninjected siblings. A total of 30 embryos from 3 independent experiments were counted for each condition.
We further examined the regulation of spi-1l by determining whether its expression can be affected by ets1-related protein (etsrp), an ETS domain transcription factor with an essential role in zebrafish primitive myelopoiesis and vasculogenesis.26,43 Etsrp is thought to function at the hemangioblast stage and the loss of its function results in the loss of most primitive myeloid cells and angiogenesis, but has no effect on erythropoiesis.26 We examined whether Etsrp is also involved in regulating spi-1l expression. To knock down etsrp expression, we analyzed wild-type embryos that were injected at the 1- to 2-cell stage with an etsrp MO.43 This resulted in almost a complete loss of spi-1l and pu.1 expression (Figure 4F,G, Figure S4D-G), compared with wild-type embryos (Figure 4D,E). Wild-type embryos injected with etsrp mRNA show an increased number of spi-1l–expressing (114 ± 27) and pu.1-expressing (152 ± 30) cells, both on the yolk cell as well as in the CHT (Figure 4H,I; Figure S4H,I), compared with uninjected siblings (spi-1l: 87 ± 14; pu.1: 106 ± 14) (Figure 4D,E). The overlap in expression between spi-1l and pu.1, as well as the involvement of etsrp in the regulation of spi-1l cell numbers, strongly suggests that Spi-1l is involved in differentiation of primitive myeloid cells.
The expression of pu.1 is not affected in spi-1l morphants (Figure 4J,K), suggesting that Pu.1 functions genetically upstream of spi-1l expression. In contrast, spi-1l expression was found to be missing in pu.1 morphants (Figure 4M,N). This further confirmed that spi-1l is genetically downstream of Pu.1. Consistent with this result, wild-type embryos injected with pu.1 mRNA at the 1-cell stage cause an increase in spi-1l expression (Figure 4P-R). We also observed by in situ hybridization that in spi-1l morphant embryos there is a large decrease in the number of spi-1l–expressing cells (Figure 4N,P). Taken together, the genetic and expression analysis are consistent with Spi-1l functioning in myeloid differentiation downstream of Cloche, Etsrp, and Pu.1.
Ectopic spi-1l expression partially rescues myeloid development in pu.1 morphants and cloche mutants
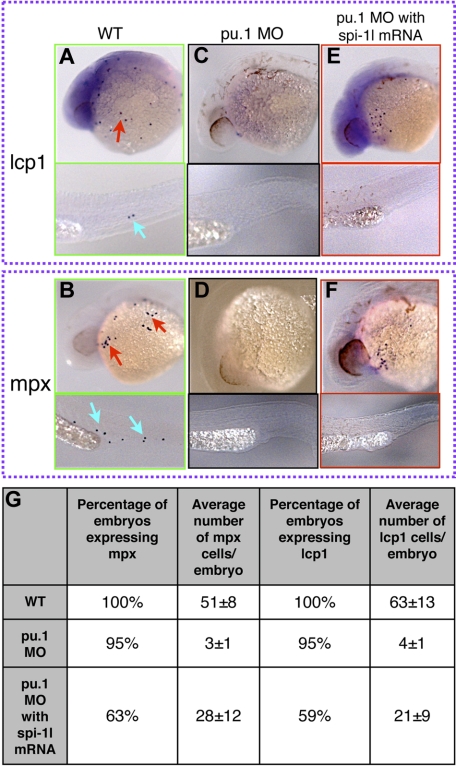
To gain further understanding of the relation between spi-1l and other myeloid genes, we determined whether spi-1l is capable of rescuing myelopoiesis in pu.1 morphants and cloche mutants, which both have a block in myeloid development. We coinjected spi-1l mRNA with pu.1 MO and assessed mpx and lcp1 expression. As a control, pu.1 MO was injected with and without spi-1l mRNA. In the case of pu.1 MO injection alone, there was almost complete loss of mpx and lcp1 expression (Figure 5A-D, Figure S5A-F). When spi-1l mRNA was coinjected with the pu.1 MO, mpx and lcp1 expression was rescued (Figure 5E,F). This rescue was incomplete: 63% and 59% of the injected embryos, respectively, re-expressed mpx and lcp1 (Figure 5G); thus Spi-1l is able to cause myeloid differentiation in the absence of Pu.1 function, but not with wild-type efficiency.
Figure 5.
Ectopic expression of spi-1l mRNA restores myeloid markers in pu.1 morphants. (A-F) Lateral view of 26-hpf embryos. Probes for genes, morpholino, and injection mixture used are as indicated. Red and blue arrows indicate mRNA expression in the YK region and the CHT, respectively. Wild-type expression of lcp1 (A) and mpx (B) in uninjected embryos. Spi-1 MO eliminates lcp1 (C) and mpx (D) expression. Embryos coinjected with pu.1 MO and spi-1l mRNA recover some lcp1 (E) and mpx (F) expression. (G) Table demonstrating percentage of embryos and the number of cells per embryo expressing mpx and lcp1 in wild-type, pu.1 MO, and embryos coinjected with spi-1l mRNA and pu.1 MO.
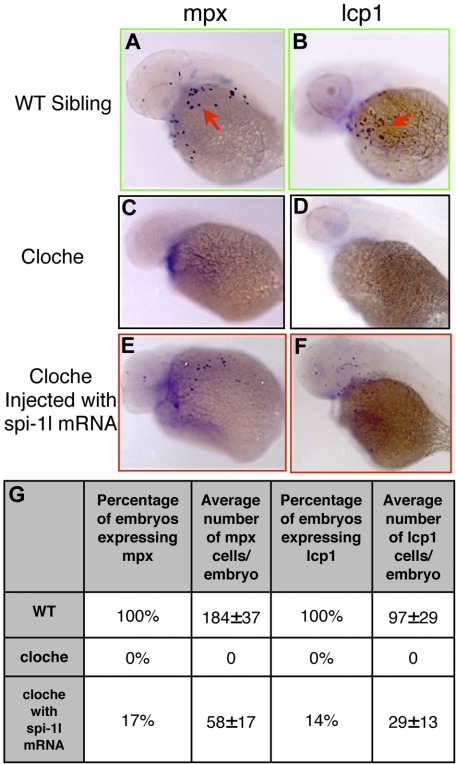
Subsequently, we tested whether ectopic spi-1l mRNA expression would be adequate to rescue myelopoiesis in cloche mutants. One- to 2-cell embryos from a heterozygous cross of cloche mutant zebrafish were injected with spi-1l mRNA. Both the macrophage/neutrophil marker, lcp1, and the granulocyte marker, mpx, were restored in spi-1l–injected homozygous cloche embryos (Figure 6A-F; Figure S6A-F). expressing The rescue was partial with mpx and lcp1 markers restored in a subset of cells in 17% and 14%, respectively, of the injected embryos (Figure 6G). Expression of lcp1 and mpx was not affected in wild-type embryos injected with spi-1l mRNA. We also determined that coinjection of spi-1l and pu.1 mRNA did not lead to a more significant rescue of mpx or lcp1 in cloche mutants. The coinjected cloche embryos had on average only 7 more mpx-positive cells and 3 more lcp1-positive cells. The cloche rescue results once again demonstrate that high level of spi-1l expression can drive myeloid differentiation, and taken together with the epistasis data, indicate an important role for Spi-1l in myeloid differentiation.
Figure 6.
Ectopic expression of spi-1l mRNA is sufficient to rescue myeloid markers in cloche mutants. (A-F) Lateral view of the yolk (YK) region of embryos from a heterozygous cloche intercross at 36 hpf. Probes for genes, embryo genotype, and mRNA injected are as indicated. Red arrows indicate mRNA expression in the YK region of a wild-type sibling from a cloche intercross. Wild-type sibling of cloche mutants express mpx (A) and lcp1 (B). Cloche mutant embryos lack expression of mpx (C) and lcp1 (D). Cloche embryos injected with spi-1l mRNA partially recover mpx (E) and lcp1 (F) expression. (G) Table demonstrating percentage and number of cells per embryo of cloche wild-type sibling, cloche mutant, and cloche mutant embryos injected with spi-1l mRNA, expressing mpx and lcp1.
Discussion
In this study, we identified, characterized, and functionally analyzed a new zebrafish SPI transcription factor that we call Spi-1l. Spi-1l–expressing cells develop from the A-LPM, then expand and move across the yolk, and later can be seen in the CHT/PBI where EMPs7 and HSCs8,9 are temporarily located. We also determined that spi-1l is expressed in a subset of the pu.1-positive cells in the CHT, which have recently been shown to have EMP potential,7 suggesting a role for Spi-1l in EMP function. Although, we demonstrate spi-1l and pu.1 colocalization at 28 hpf for earlier developmental stages due to technical reasons, we can only infer rather than directly demonstrate colocalization. In addition, the temporal expression of spi-1l and its similarity in expression pattern to pu.1 point to spi-1l function in zebrafish hematopoiesis. Furthermore, our results demonstrate that spi-1l expression is limited exclusively to myeloid cells during hematopoiesis.
Our data suggest that Spi-1l functions genetically downstream of Cloche, Etsrp, and Pu.1 during myeloid differentiation. Loss of spi-1l expression results in a lack of primitive macrophages and granulocytes but does not affect early hematopoietic genes such as gata2 or scl. The expression of pu.1, an early marker of myeloid cells, is also unaffected in spi-1l morphants, even though spi-1l is expressed in a subset of pu.1-positive cells. We also determined that spi-1l expression is lost in pu.1 morphants. The loss of both spi-1l and pu.1 expression did not cause an exacerbation of the individual phenotypes or a novel phenotype. These results are consistent with a model in which Spi-1l and Pu.1 function in the same pathway in determining myeloid differentiation.
Injection of cloche mutants with pu.1 mRNA can rescue spi-1l expression, and overexpression of pu.1 mRNA in wild-type embryos can cause an increase in the number of spi-1l–positive cells. These results establish that pu.1 can activate spi-1l expression, which is subsequently necessary for myeloid cell differentiation. Ectopic spi-1l mRNA expression in pu.1 morphants as well as in cloche mutant embryos causes partial recovery of macrophages and granulocytes, suggesting that Spi-1l is sufficient to activate myeloid cell differentiation. However, it is possible that in the case of spi-1l overexpression in cloche mutants, the myeloid differentiation recovery is partially mediated by Spi-1l binding to some genes that are ordinarily regulated by Pu.1 alone. In fact, in mammalian cell culture experiments, it has been demonstrated that overexpression of either SPI-B and SPI-C can bind and activate PU.1 target genes.44,45 Significantly, partial rescue of myeloid markers can be seen when pu.1 mRNA is overexpressed in spi-1l morphants. This result could be due to high ETS domain sequence similarity between Pu.1 and Spi-1l such that at very high expression levels, Pu.1 could bypass the need for Spi-1l in activating myeloid cell differentiation. In this situation, it is also possible that the combination of a high level of pu.1 expression with a very low level of spi-1l expression in spi-1l morphants allows for a partial rescue of myelopoiesis.
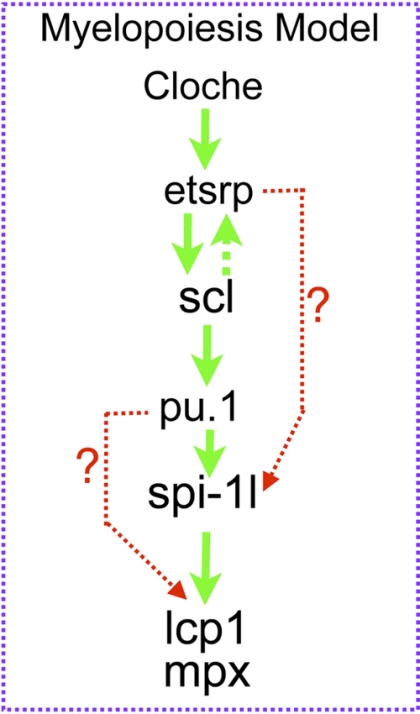
Our data suggest a model in which spi-1l is activated by Pu.1 and is required for myeloid differentiation (Figure 7). We believe that spi-1l may be functioning to increase the overall level of SPI transcription factors, thereby resulting in further differentiation of myeloid lineages. Our data point to spi-1l and pu.1 possibly functioning in a cooperative manner during myeloid differentiation. In mammalian hematopoiesis, the exact control of PU.1 level is crucial for the differentiation of multiple lineages. In erythrocytes and pro T cells, PU.1 levels must be low, allowing precursor cells to differentiate.15 In granulocyte/monocyte progenitors (GMPs), PU.1 levels must rise for the differentiation of macrophages and granulocytes to occur. An important way in which the level of mammalian PU.1 expression is controlled is through its upstream regulatory element (URE).20 However, such a URE has not been detected in the zebrafish genome, and appears to be a mammalian feature. The discovery of a second myeloid-specific SPI gene may therefore present an alternative way to fine-tune SPI levels in zebrafish, without the need for the autoregulation found in mammalian hematopoiesis. We speculate that in zebrafish Pu.1 activates spi-1l expression to increase the total level of Spi transcription factors at the GMP stage, which in turn causes further differentiation to take place during myelopoiesis in a manner similar to what occurs in mammals, but using an alternative strategy (Figure 7). During zebrafish myelopoiesis, Spi-1l and Pu.1 function together, either sequentially or to activate target genes, possibly depending on the cell type and stage of differentiation. Spi-1l and Pu.1 can perhaps activate some of the same genes such as mpx and lcp1, but they also activate a subset of unique genes such as runx1 and gata1, likely depending on their expression pattern and the cofactors with which they interact.
Figure 7.
Function of spi-1l in myelopoiesis. Proposed model of spi-1l function during myelopoiesis. Spi-1l functions downstream of cloche, etsrp, and pu.1 and is regulated by these transcription factors. Spi-1l is necessary and sufficient for mpx and lcp1 expression. At this time, we cannot differentiate whether spi-1l activates mpx and lcp1 expression together or sequentially with spi-1.
Clearly, certain aspects of Spi functional diversity result from the restricted expression pattern of spi-1l to the A-LPM and the PBI/CHT regions compared with pu.1, which is also expressed in the ICM. This explains the inability of spi-1l in affecting gata1 expression, whereas pu.1 MO causes ectopic gata1 expression. An as yet unexplored difference in the function between spi-1l and pu.1 may also lie in their roles in adult hematopoiesis, which will be followed up in later studies. Here we have identified a new player in zebrafish myeloid differentiation and determined the genetic hierarchy to explain its role in this important developmental process.
Supplementary Material
Acknowledgments
We greatly appreciate and acknowledge the help of Dr Saulius Sumanas for providing etsrp morphant– and etsrp mRNA–injected embryo. We thank Dr Julian Martinez for critical reading of the paper.
This work was funded by an American Cancer Research Society Scholar Grant (Oakland, CA; K.J.P.G.), USPHS National Research Service Award GM07104 grant (Bethesda, MD; A.B.), and Hamburger Estate fund and Bergman fund of the Jonsson Comprehensive Cancer Center (Los Angeles, CA; U.B.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.B. designed and performed experiments, analyzed data, and wrote the draft of the paper; K.J.P.G. conceived of the project; Y.Z. performed experiments; and U.B. and S.L. supervised research and helped write the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Utpal Banerjee, Department of Molecular, Cell and Developmental Biology, UCLA, 621 Charles E. Young Drive South, Los Angeles, CA 90095-1606; e-mail: banerjee@mbi.ucla.edu.
References
- 1.Davidson AJ, Zon LI. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene. 2004;23:7233–7246. doi: 10.1038/sj.onc.1207943. [DOI] [PubMed] [Google Scholar]
- 2.Amatruda JF, Zon LI. Dissecting hematopoiesis and disease using the zebrafish. Dev Biol. 1999;216:1–15. doi: 10.1006/dbio.1999.9462. [DOI] [PubMed] [Google Scholar]
- 3.de Jong JL, Zon LI. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet. 2005;39:481–501. doi: 10.1146/annurev.genet.39.073003.095931. [DOI] [PubMed] [Google Scholar]
- 4.Carradice D, Lieschke GJ. Zebrafish in hematology: sushi or science? Blood. 2008;111:3331–3342. doi: 10.1182/blood-2007-10-052761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett CM, Kanki JP, Rhodes J, et al. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98:643–651. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- 6.Hsia N, Zon LI. Transcriptional regulation of hematopoietic stem cell development in zebrafish. Exp Hematol. 2005;33:1007–1014. doi: 10.1016/j.exphem.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murayama E, Kissa K, Zapata A, et al. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Kissa K, Murayama E, Zapata A, et al. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 2008;111:1147–1156. doi: 10.1182/blood-2007-07-099499. [DOI] [PubMed] [Google Scholar]
- 10.Traver D, Zon LI. Walking the walk: migration and other common themes in blood and vascular development. Cell. 2002;108:731–734. doi: 10.1016/s0092-8674(02)00686-4. [DOI] [PubMed] [Google Scholar]
- 11.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 12.Bartel FO, Higuchi T, Spyropoulos DD. Mouse models in the study of the Ets family of transcription factors. Oncogene. 2000;19:6443–6454. doi: 10.1038/sj.onc.1204038. [DOI] [PubMed] [Google Scholar]
- 13.Gallant S, Gilkeson G. ETS transcription factors and regulation of immunity. Arch Immunol Ther Exp (Warsz) 2006;54:149–163. doi: 10.1007/s00005-006-0017-z. [DOI] [PubMed] [Google Scholar]
- 14.Sharrocks AD, Brown AL, Ling Y, Yates PR. The ETS-domain transcription factor family. Int J Biochem Cell Biol. 1997;29:1371–1387. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- 15.Gangenahalli GU, Gupta P, Saluja D, et al. Stem cell fate specification: role of master regulatory switch transcription factor PU. 1 in differential hematopoiesis. Stem Cells Dev. 2005;14:140–152. doi: 10.1089/scd.2005.14.140. [DOI] [PubMed] [Google Scholar]
- 16.Anderson MK, Sun X, Miracle AL, Litman GW, Rothenberg EV. Evolution of hematopoiesis: three members of the PU. 1 transcription factor family in a cartilaginous fish, Raja eglanteria. Proc Natl Acad Sci U S A. 2001;98:553–558. doi: 10.1073/pnas.021478998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oikawa T, Yamada T, Kihara-Negishi F, et al. The role of Ets family transcription factor PU. 1 in hematopoietic cell differentiation, proliferation and apoptosis. Cell Death Differ. 1999;6:599–608. doi: 10.1038/sj.cdd.4400534. [DOI] [PubMed] [Google Scholar]
- 18.Singh H, DeKoter RP, Walsh JC. PU. 1, a shared transcriptional regulator of lymphoid and myeloid cell fates. Cold Spring Harb Symp Quant Biol. 1999;64:13–20. doi: 10.1101/sqb.1999.64.13. [DOI] [PubMed] [Google Scholar]
- 19.Iwasaki H, Somoza C, Shigematsu H, et al. Distinctive and indispensable roles of PU. 1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okuno Y, Huang G, Rosenbauer F, et al. Potential autoregulation of transcription factor PU. 1 by an upstream regulatory element. Mol Cell Biol. 2005;25:2832–2845. doi: 10.1128/MCB.25.7.2832-2845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieschke GJ, Oates AC, Paw BH, et al. Zebrafish SPI-1 (PU. 1) marks a site of myeloid development independent of primitive erythropoiesis: implications for axial patterning. Dev Biol. 2002;246:274–295. doi: 10.1006/dbio.2002.0657. [DOI] [PubMed] [Google Scholar]
- 22.Rhodes J, Hagen A, Hsu K, et al. Interplay of pu. 1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 23.Thompson MA, Ransom DG, Pratt SJ, et al. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197:248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- 24.Griffin K, Patient R, Holder N. Analysis of FGF function in normal and no tail zebrafish embryos reveals separate mechanisms for formation of the trunk and the tail. Development. 1995;121:2983–2994. doi: 10.1242/dev.121.9.2983. [DOI] [PubMed] [Google Scholar]
- 25.Schoenebeck JJ, Keegan BR, Velon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sumanas S, Gomez G, Zhao Y, Park C, Choi K, Lin S. Interplay between Etsrp/ER71, scl and alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008;111:4500–4510. doi: 10.1182/blood-2007-09-110569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001;98:3087–3096. doi: 10.1182/blood.v98.10.3087. [DOI] [PubMed] [Google Scholar]
- 28.Kalev-Zylinska ML, Horsfield JA, Flores MV, et al. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129:2015–2030. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- 29.Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- 30.Meng A, Tang H, Ong BA, Farrell MJ, Lin S. Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc Natl Acad Sci U S A. 1997;94:6267–6272. doi: 10.1073/pnas.94.12.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 1998;12:621–626. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.European Bioinformatics Institute. [Accessed December 5, 2007];ClustalW. http://www.ebi.ac.uk/Tools/clustalw2/index.html.
- 33.European Bioinformatics Institute. TreeView X. [Accessed December 5, 2007]; http://www.ebi.ac.uk/ebisearch/search.
- 34.Strausberg RL, Feingold EA, Grouse LH, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Center for Biotechnology Information. [Accessed June 4, 2008];GenBank. http://www.ncbi.nlm.nih.gov/Genbank.
- 36.Bertrand JY, Kim AD, Teng S, Traver D. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development. 2008;135:1853–1862. doi: 10.1242/dev.015297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stainier DY, Fouquet B, Chen JN, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- 38.Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 39.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 40.Sumanas S, Larson JD. Morpholino phosphorodiamidate oligonucleotides in zebrafish: a recipe for functional genomics? Brief Funct Genomic Proteomic. 2002;1:239–256. doi: 10.1093/bfgp/1.3.239. [DOI] [PubMed] [Google Scholar]
- 41.Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- 42.Le Guyader D, Redd MJ, Colucci-Guyon E, et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2008;111:132–141. doi: 10.1182/blood-2007-06-095398. [DOI] [PubMed] [Google Scholar]
- 43.Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray D, Bosselut R, Ghysdael J, Mattei MG, Tavitian A, Moreau-Gachelin F. Characterization of Spi-B, a transcription factor related to the putative oncoprotein Spi-1/PU. 1. Mol Cell Biol. 1992;12:4297–4304. doi: 10.1128/mcb.12.10.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweitzer BL, Huang KJ, Kamath MB, Emelyanov AV, Birshtein BK, DeKoter RP. Spi-C has opposing effects to PU. 1 on gene expression in progenitor B cells. J Immunol. 2006;177:2195–2207. doi: 10.4049/jimmunol.177.4.2195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.