Abstract
Species of the mussel genus Mytilus possess maternally and paternally transmitted mitochondrial genomes. In the interbreeding taxa Mytilus edulis and M. galloprovincialis, several genomes of both types have been fully sequenced. The genome consists of the coding part (which, in addition to protein and RNA genes, contains several small noncoding sequences) and the main control region (CR), which in turn consists of three distinct parts: the first variable (VD1), the conserved (CD), and the second variable (VD2) domain. The maternal and paternal genomes are very similar in gene content and organization, even though they differ by >20% in primary sequence. They differ even more at VD1 and VD2, yet they are remarkably similar at CD. The complete sequence of a genome from the closely related species M. trossulus was previously reported and found to consist of a maternal-like coding part and a paternal-like and a maternal-like CR. From this and from the fact that it was extracted from a male individual, it was inferred that this is a genome that switched from maternal to paternal transmission. Here we provide clear evidence that this genome is the maternal genome of M. trossulus. We have found that in this genome the tRNAGln in the coding region is apparently defective and that an intact copy of this tRNA occurs in the CR, that one of the two conserved domains is missing essential motifs, and that one of the two first variable domains has a high rate of divergence. These features may explain the large size and mosaic structure of the CR of the maternal genome of M. trossulus. We have also obtained CR sequences of the maternal and paternal genomes of M. californianus, a more distantly related species. We compare the control regions from all three species, focusing on the divergence among genomes of different species origin and among genomes of different transmission routes.
MUSSELS of the genus Mytilus have the doubly uniparental inheritance (Zouros et al. 1994a) mode of mitochondrial DNA (mtDNA): both genders have a maternally transmitted mtDNA, but a paternally transmitted mtDNA is found predominantly in males (Skibinski et al. 1994a,b; Zouros et al. 1994a,b; ; Hoeh et al. 1996; Liu et al. 1996; Passamonti and Scali 2001; Curole and Kocher 2002; Hoeh et al. 2002; Serb and Lydeard 2003). As a result, females are homoplasmic, even though small amounts of “leaked” paternal mtDNA can be detected in most of them (Stewart et al. 1995; Garrido-Ramos et al. 1998; Dalziel and Stewart 2002; Obata et al. 2006), and males are heteroplasmic, with their somatic tissues dominated by the maternal mtDNA and the gonad dominated by the paternal. There is good evidence that pure sperm contain only the paternal mtDNA (Venetis et al. 2006).
Mytilus edulis and M. galloprovincialis are closely related sibling species that hybridize freely in areas of sympatry and their separate species status is questionable (Gossling 1992; however, see Bierne et al. 2006). Their mitochondrial genomes, maternal or paternal, are indistinguishable. The maternal (F) and the paternal (M) genome of M. edulis/M. galloprovincialis have been fully sequenced and characterized (Boore et al. 2004; Mizi et al. 2005, 2006; Breton et al. 2006). The most notable differences are confined in the large unassigned region (LUR), first described by Hoffmann et al. (1992). It is convenient for this reason to divide the mitochondrial genome of Mytilus into two parts: the core that contains all protein, rRNA and tRNA coding genes and a few noncoding regions of <500 bp, and the LUR. The core is the same size in the F and M genomes and contains the same genes in the same order. Yet, it differs by ∼23% in primary sequence. The LUR varies between the two genomes in both primary sequence and size. Cao et al. (2004) have divided the LUR into three parts: the first variable domain (VD1), the conserved domain (CD), and the second variable domain (VD2). These domains closely parallel the three domains of human mtDNA and contain sequence motifs that are similar to those in sea urchins and in humans. In both species these motifs are known to be involved in the replication and transcription of the molecule. These observations reinforced previous suggestions (Hoffmann et al. 1992) that the LUR is the main control region of the genome. For this reason, the LUR is also referred to as the main control region (CR), a designation that we will use in this article.
A third type of mitochondrial genome, the C genome, was recently sequenced and described in M. galloprovincialis (Venetis et al. 2007). The sequence of its core is similar to that of the F genome, differing from it by ∼3%. But its CR is much larger than the CR of either the F or the M genome, ∼3.52 kb, compared to ∼1.15 kb for the F and ∼0.91 kb for the M genome. It consists of four tandem repeats of the CR of the M genome inserted within the CR of the F genome. The particular C genome that was sequenced was isolated from sperm that were forced to swim through a Percoll solution for the removal of any debris from somatic cells. In a previous study it was shown that sperm purified in this way contain only paternal mtDNA (Venetis et al. 2006). This produced firm evidence that the C genome is paternally inherited. From the fact that its core is of the F type, it was concluded that the C genome is a “masculinized” genome, i.e., a genome that descended from the F lineage and reversed its transmission route through the incorporation of sequences from the CR of the M genome. The phenomenon of masculinization was first proposed (Hoeh et al. 1997) to explain the presence in male gonads of genomes whose core was of the F type (Quesada et al. 1995, 1999; Hoeh et al. 1997; Saavedra et al. 1997). Masculinization also appears to be the most likely explanation for the clustering of maternal and paternal coding sequences from species from different genera and families of bivalves (Hoeh et al. 1996; Theologidis et al. 2008).
M. trossulus is the most closely related species to the M. edulis/M. galloprovincialis sibling pair with which it forms the M. edulis species group. M. trossulus is known to occur in the American Atlantic coast north of Maine and in the northeast Pacific region from central California to Alaska. It has also been reported to occur in the Baltic Sea, where it has apparently introgressed with M. edulis. The Baltic populations of M. trossulus can be considered as a taxon whose nuclear genome is dominated by M. trossulus sequences, but whose mitochondrial genomes, both maternal and paternal, are of M. edulis origin. American M. trossulus populations, on the other hand, have their own species-specific maternal and paternal mtDNA. This was evident from early studies of coding sequences that demonstrated the existence of four distinct mtDNAs in M. edulis and M. trossulus, two in each, and showed that these sequences affiliate according to mode of transmission rather than according to origin of species (Rawson and Hilbish 1995; Stewart et al. 1995). More recently, Breton et al. (2006) have reported the complete sequence of a mitochondrial genome that they have extracted from a male M. trossulus gonad from Atlantic Canada. The primary sequence of the core of this genome resembled closely the F genome of M. edulis/M. galloprovincialis, but its CR was much larger than the CR of the F or M genomes of M. edulis/M. galloprovincialis. It consisted of two CRs of which one resembled the CR of the M genome and the other that of the F genome of M. edulis/M. galloprovincialis. Breton et al. (2006) concluded that this was a “recently masculinized” genome, i.e., a maternal genome with a reversed transmission route, similar to the C genome of M. galloprovincialis that was subsequently reported. This interpretation by Breton et al. (2006) did not, however, agree with a previous study by Rawson (2005) in which a genome with an identical CR was identified as the sole mtDNA in M. trossulus females.
The alleged masculinized genome in M. trossulus invites its comparison with the maternal genome of this species, from which it is supposed to have derived, and with the paternal genome, whose role it is supposed to perform. This comparison was indeed done with the C genome and its conspecific F and M genomes of M. galloprovincialis (Venetis et al. 2007). The comparison showed that the differences among the three genomes are confined in the CR. But the CR sequence of the maternal and paternal genomes of M. trossulus remain unknown. We have, therefore, obtained the sequences of the CR of genomes isolated from eggs and pure sperm from several M. trossulus individuals. This gave us firm evidence that the genome sequenced by Breton et al. (2006) is in fact the maternal genome of M. trossulus. We then obtained the sequence of the CR of the maternal and paternal genomes of another mytilid, M. californianus, a distant relative of the M. edulis species group. The complete set of CRs of both genomes from three species of varying phylogenetic distance provided us with some information about the evolution of CR in mytilids. It also provided hints about the functionality of some parts of the duplicated CR of the maternal mtDNA of M. trossulus, which may explain the unusual structure and length of this genome.
MATERIALS AND METHODS
Sample collection and identification:
Mussels were collected in Canada (Lunenburg Bay, Nova Scotia: individuals w2, w22, w65, w143, w193, and w202; Eskosoni, Nova Scotia: individuals 11, 12, 17, and 23; Lemmens Inlet, British Columbia: individuals BC8 and BC9) and in the United States (Neah Bay, WA: individuals 15 and 26; Terrace Point, CA: individuals P1 and P2) (Table 1) and transported alive to the laboratory. Sex was determined by examining the gonad under a light microscope for the presence of sperm or eggs. DNA of P1 and P2 was extracted as described in Ort and Pogson (2007). Total DNA of other mussel samples was extracted using a modified saturated-salt extraction procedure (Miller et al. 1988). Species identification of mussels from Canada was performed by genotyping samples at two nuclear markers, ITS and Glu5, using the methods described by Heath et al. (1995) and Rawson et al. (1996), respectively. Only individuals that were homozygous for M. edulis or M. trossulus alleles at both markers were used.
TABLE 1.
Sequences used in this study
| Length (bp)
|
Sequence characterization | GenBank accession no. | |||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Sequence codea | Species | Gender | Tissue | Genome regionb | CR | Totalc | ||
| 1 | ef_w22_eF | M. edulis | Female | Gill + gonad | 12S-cytb | 1159 | 4334 | M. edulis, maternal (F-ed) | AY350784 |
| 2 | em_w143_eM | Male | Gill + gonad | 12S-cytb | 928 | 4082 | M. edulis, paternal (M-ed) | AY350791 | |
| GB | gm_S35_gC | M. galloprovincialis | Male | Sperm | 12S-cytb | 3522 | 6697 | M. galloprovincialis, paternal (C-ga) | DQ399833 |
| 3 | tf_w2_VL | M. trossulus | Female | Gill + gonad | 12S-cytb | 3061 | 6237 | M. trossulus, maternal (F-tr) | AY515230 |
| 4 | tf_w193_VL | Gill + gonad | 16S-Y | 3075 | 3314 | M. trossulus, maternal (F-tr) | EU826073 | ||
| 5 | tf_BC8_VL | Gonad | 16S-Y | 3043 | 3282 | M. trossulus, maternal (F-tr) | EU826074 | ||
| 6 | tf_w65_VL | Gill + gonad | 16S-Y | 3060 | 3299 | M. trossulus, maternal (F-tr) | EU826072 | ||
| 7 | tf_17_VL | Gonad | 16S-Y | 3063 | 3302 | M. trossulus, maternal (F-tr) | EU826075 | ||
| 8 | tf_23_tM | Gonad | 16S-Y | 1070 | 1309 | M. trossulus, paternal (M-tr) | EU826079 | ||
| GB | tm_RM_VL | Male | Gonad | 12S-cytb | 3062 | 6239 | M. trossulus, maternal (F-tr) | AY823625 | |
| 9 | tm_w202_tM | Gill + gonad | 12S-cytb | 1073 | 4186 | M. trossulus, paternal (M-tr) | AY515231 | ||
| 10 | tm_12_VL | Foot | 16S-Y | 3061 | 3300 | M. trossulus, maternal (F-tr) | EU826076 | ||
| 11 | tm_11_tM | Gonad | 16S-Y | 1072 | 1311 | M. trossulus, paternal (M-tr) | EU826078 | ||
| 12 | tm_BC9_tM | Gonad | 16S-Y | 1070 | 1309 | M. trossulus, paternal (M-tr) | EU826077 | ||
| 13 | cf_26_cF | M. californianus | Female | Gonad | 12S-cytb | 1107 | 4295 | M. californianus, maternal (F-ca) | AY515227 |
| 14 | cf_P1_cF | Gill | 12S-Y | 1107 | 3447 | M. californianus, maternal (F-ca) | EU826123 | ||
| 15 | cm_15_cM | Male | Gonad | 12S-cytb | 695 | 3717 | M. californianus, paternal (M-ca) | AY515226 | |
| 16 | cm_P2_cM | Gonad | 12S-Y | 759 | 2934 | M. californianus, paternal (M-ca) | EU826124 | ||
Sequences were extracted from 16 animals, listed according to species and gender. Two sequences (DQ399833 and AY823625) were extracted from GenBank (GB). On six occasions gonads could not be separated from the gill.
The first letter refers to the species (e, edulis; g, galloprovincialis; t, trossulus; c, californianus), the second letter to gender (f, female; m, male), followed by animal code, followed by the sequence type (F, F type; M, M type; C, C type; VL, CR of M. trossulus very long type).
12S-cytb, the amplified fragment extends from part of 12S-rRNA to part of Cyt b; 16S-Y, from part of 16S-rRNA to part of tRNATyr; and 12S-Y, from part of 12S-rRNA to part of tRNATyr.
Total length contains the length of the CR and of the flanking sequences.
PCR amplification, cloning, and sequencing:
Extracted DNA was used as template to amplify parts of the mitochondrial genome using primers listed in Table 2. The primer pairs used in each species and the primer binding sites in the mitochondrial genome are illustrated in supplemental Figure 1. mtDNA amplifications of all samples, except P1 and P2, were carried out in a 25-μl PCR reaction containing ∼50–100 ng of template DNA, 0.8 μm of each primer, 200 μm of each dNTP, 2.5 mm MgCl2, and 1 unit Taq polymerase (MBI Fermentas) in the buffer supplied by MBI Fermentas. The reactions were heated initially at 94° for 3 min and then incubated at 94° for 20 sec–1 min, 46°–53° for 20 sec–1.5 min, and 72° for 45 sec–3 min depending on fragment size for 40 cycles and 72° for 6 min for a final extension. The amplified products were visualized on a 1% agarose gel. DNA was recovered using a gel band purification kit (Biomio) and cloned on pGEM-T vector (Promega) following the procedure provided by the supplier. Positive clones from each individual were confirmed by PCR amplification with the same primers and conditions as above. One or two positive clones from each individual were sequenced commercially from both directions using either a LICOR 4200 or ABI 373 automated sequencer. Primer walking was applied when cloned fragments were >1.2 kb.
TABLE 2.
Oligonucleotides used as primers in PCR reactions
| Code | Sequence | Location in sequence AY497292 (F-ga) | Reference |
|---|---|---|---|
| UNFOR1 | 5′-TTGCGACCTCGATGTTGGC-3′ | 16,497–16,515 | Caoet al. (2004a) |
| UNREV1 | 5′-AGCTCACCACCTATTCCTC-3′ | 1170–1188 | Caoet al. (2004a) |
| 12SF1134 | 5′-ACCAGGATTAGATACCCTGT-3′ | 14,412–14,431 | This study |
| 16SBR | 5′-CCGGTCTGAACTCAGATCACGT-3′ | 16,577–16,598 | Rawson and Hilbish (1995) |
| UCYTB270R | 5′-AANAGGAARTAYCAYTCNGGYTG-3′ | 2075–2097 | Merritt et al. (1998) |
| UARW2 | 5′-GAGAAAGTGGTGAATTTGACC-3′ | 759–780a | This study |
| cytbtfR | 5′-CATACATAGCGGAGCATACTC-3′ | 1744–1764 | This study |
| UCYTB151F | 5′-TGTGGRGCNACYGTWATYACTAA-3′ | 1691–1713 | Merritt et al. (1998) |
| 16SF3160 | 5′-AGCTACTCTAGGGATAACAGC-3′ | 16,440–16,460 | This study |
| CytbCfR | 5′-ACTTTGCATTACAGACTGTC-3′ | 1774–1793 | This study |
| UCYTB144F | 5′-TGAGSNCARATGTCNTWYTG-3′ | 1673–1692 | Merritt et al. (1998) |
| UCYTB272R | 5′-GARTGGTAYTTYCTNTTYGC-3′ | 2081–2100 | Merritt et al. (1998) |
| 12SF1186 | 5′-CTAAACATTTGGTTGTCAAAGAG-3′ | 14,464–14,486 | This study |
| 16sCmR2 | 5′-AGGTCTTGCTTGTGAGCAA-3′ | 16,401–16,421b | This study |
| Ucytb1R | 5′-CCYARRGGRTTATTDCTHCC-3′ | 1880–1899 | This study |
| 16SAR | 5′-CGCCTGTTTATCAAAAACAT-3′ | 16,071–16,090 | Rawson and Hilbish (1995) |
| M-nad5F | 5′-CTCTATGTATGGCTGTTTGTGAGGC-3′ | 11,697–11,721 | Ort and Pogson (2007) |
| M-nad2R | 5′-CAGAAAAGTTTACCGCCAARACC-3′ | 7978–8000 | Ort and Pogson (2007) |
| F-nad5F2 | 5′-ATTAGCTGGTTCAAGCGCAGTG-3′ | 12,566–12,587 | This study |
| F-nad2R | 5′-ATGCCGCTGGCCTAAAAGC-3′ | 8038–8056 | This study |
The target sequence in M. trossulus is shorter by one nucleotide.
The target sequence in M. californianus is longer by two nucleotides.
For mussels P1 and P2 of M. californianus, primer pairs F-nad5F2/F-nad2R and M-nad5F/M-nad2R, specific to F and M genomes, were used to amplify an ∼12-kb product extending from the ND5 to the 3′-end of the ND2, encompassing the entire CR. PCR reactions for these “F12” and “M12” fragments contained 20 mm Tris–HCl (pH 8.8), 10 mm KCl, 10 mm (NH4)2SO4, 3.0 (F12) or 2.0 (M12) mm MgSO4, 0.1% Triton X-100, 100 ng/μl bovine serum albumin, 200 μm of each dNTP, 0.5 μm forward and reverse primers, 0.6 units of Taq 2000 DNA polymerase (Stratagene), 0.6 units Taq Extender PCR additive (Stratagene), and 150 ng template DNA. Reactions were performed in 10-μl sealed glass capillary tubes in an Idaho Technology A1605 Air Thermo-Cycler. Cycling conditions consisted of an initial denaturation step of 45 sec at 94°, 35 cycles of denaturation at 94° for 1 sec, primer annealing at 52° for 1 sec, primer extension at 72° for 10–14 min, and a final hold at 72° for 10–14 min. Multiple replicate PCRs were pooled and gel purified using Zymoclean columns (Zymo Research). Purified PCR products were cloned and shotgun sequenced to 7.8–11.5× coverage (Macrogen, South Korea).
Search of mtDNA sequences in eggs, purified sperm, and somatic tissues of M. trossulus:
Mussels from Bras D'Or (Cape Breton, Canada) were placed in individual containers with sea water and induced to spawn by increasing the incubation temperature from 8° to 20°. Pure spermatozoa were obtained from 10 males by forcing sperm to swim through a solution of Percoll as described in Venetis et al. (2006). Eggs from five females were washed thoroughly with sterile sea water. These procedures were followed to avoid contamination with mtDNA of somatic cell origin. Total DNA was extracted from gametes and the foot according to Douris et al. (1998) and used as PCR template with the primers UNFOR1/UNREV1. PCR amplification was carried out in 25-μl reaction volumes containing 50–100 ng of template DNA, 0.5 μm of each primer, 200 μm of each dNTP, 2.5 mm MgCl2, and 0.125 μl of enzyme mix (Invitrogen elongase enzyme mix) in buffer B supplied by the company. Reaction conditions were in accordance with the supplier's recommendations. PCR products were visualized after agarose gel electrophoresis.
Sequence data analysis:
Cao et al. (2004) defined the CR of M. edulis/M. galloprovincialis as the part of the genome contained between the 16S rRNA and the tRNATyr and divided it into three domains, VD1, CD, and VD2. The definition of the three domains in the M. trossulus genomes was straightforward because the CR extracted from sperm and the two joint CRs extracted from eggs show a high similarity with the CD of the F and M genome of M. edulis. To delineate the three domains of the F and M genomes of M. californianus, we aligned their CR and determined the part that contained the highest sequence similarity. The approximate limits of the so-defined CDs were determined after alignment with the CD of the F and M genomes of M. edulis using ClustalX v.1.83 (Thompson et al. 1997; parameters: gap opening = 12; gap extension = 1).
Sequences from M. trossulus and M. californianus upstream of the 3′-end of 16S-rRNA and downstream of the 5′-end of tRNATyr were aligned on the basis of gene similarity using the default parameters of ClustalX v.1.83 (Thompson et al. 1997) followed by eye correction. All F-type CR sequences were multiply aligned (gap opening = 12; gap extension = 1), and the same was done with all M-type sequences (gap opening = 8; gap extension = 1). Selection of the optimal parameters (opening and extension gap penalties) was performed according to Lecanidou et al. (1994). The first alignment resulted in “profile 1” and the second in “profile 2.” Profile 2 was subsequently aligned to profile 1 using the default parameters of ClustalX v.1.83 (Thompson et al. 1997). Further correction of the alignment was performed manually to maximize sequence similarity, particularly for the two-size variable regions that flanked the relatively conserved CD. This alignment was finally concatenated with the alignment of sequences upstream and downstream of CR. The whole alignment is in the supplemental material. The program MEGA v3.1 (Kumar et al. 2004) was used for the estimation of p-distances, and the standard error computation was performed by the bootstrap method for 10,000 replications.
RESULTS
The two CR types of M. trossulus:
We have extracted 10 sequences from various tissues of M. trossulus (Table 1). Two of these (nos. 3 and 9) extended from around the beginning of 12S-rRNA to near the end of Cyt b (corresponding to 14,478–2074 bp of the complete F-type sequence given by Mizi et al. 2005), and the other eight extended from the end of 16S-rRNA to tRNATyr (corresponding to 16,518–1169 bp of the complete F-type sequence; Mizi et al. 2005). Using the definition of the CR of mytilids as the part of the mitochondrial genome contained between 16S-rRNA and tRNATyr (Cao et al. 2004), six sequences (nos. 3, 4, 5, 6, 7, 10) were found to contain a long CR (∼3.1 kb), and four sequences (nos. 8, 9, 11, 12) to contain a short CR (∼1.1 kb). The CR in the short sequence resembled closely the CR of the F and M genomes of M. edulis/M. galloprovincialis. The long type, on the other hand, was almost identical to that reported by Breton et al. (2006).
Assignment of the two CR types of M. trossulus to maternal and paternal genomes:
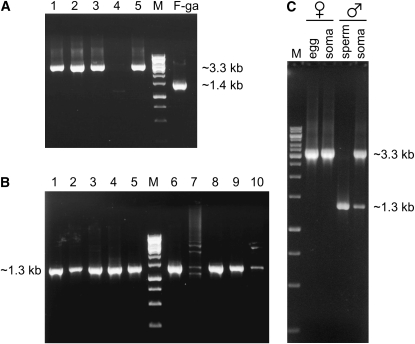
The mere fact that a mitochondrial sequence is extracted from a female or a male individual, or even from a female or a male gonad, cannot be used as evidence that it is a maternally or paternally transmitted genome (Venetis et al. 2006). The reason is that preparations from male gonads are, more often than not, contaminated by DNA from somatic cells that contain maternal mtDNA, and female tissues may contain small amounts of “leaked” paternal mtDNA (Garrido-Ramos et al. 1998). For this reason, we obtained DNA preparations from eggs and somatic tissues of five females and from purified sperm and somatic tissues of 10 males of M. trossulus from Atlantic Canada. The type of CR contained in these tissues was scored as described in materials and methods. No product was obtained from the eggs or the somatic tissue of one female (Figure 1A). The product of eggs from the other four females was ∼3.3 kb, i.e., of the size corresponding to the “long” type CR (including the flanking sequences). The same band size was recovered from the somatic tissues of these four females (Figure 1C). No product was obtained from the sperm of one male (Figure 1B) or from the soma of another male (not shown). The product from the sperm of all 9 males was ∼1.3 kb, i.e., of the size that corresponds to the “short” type CR (including the flanking sequences). The soma of all 8 males produced the 3.3-kb band found in the eggs and the soma of females and, in smaller quantity, the 1.3-kb band found in the purified sperm (Figure 1C). We failed to detect the 3.3-kb zone in any sperm extraction. We have also failed to detect any other mtDNA type in any of these female or male individuals.
Figure 1.—
PCR amplification products from eggs, purified sperm, and somatic tissues of M. trossulus. (A) DNA extracted from eggs. No PCR product was obtained from eggs of one (lane 4) of five females. The product from eggs from a M. galloprovincialis female is shown for comparison (lane F-ga). (B) DNA extracted from sperm after Percoll purification. No clear PCR product was obtained from sperm of one (lane 7) of 10 males. (C) The products from the eggs and the somatic tissues of female no. 1 (lane 1) and from the sperm and the somatic tissues of male no. 8 (lane 8). M: marker DNA. PCR amplification details as in materials and methods.
These findings are fully consistent with the hypothesis that the “short” CR type belongs to the paternal genome and the “long” CR type belongs to the maternal genome of M. trossulus. We, therefore, refer to these two genomes as F-tr and M-tr, respectively. In analogy, we will refer to the maternal (F) and to the paternal (M) genome of M. edulis/M. galloprovincialis as F-ed and M-ed and to those of M. californianus (see below) as F-ca and M-ca. This finding agrees with the results of Rawson (2005) that preceded the work of Breton et al. (2006). The fact that F-tr was found in the somatic tissues of all males and females, but never in the pure sperm, weakens the possibility that somehow it has a dual role, acting both as maternal and as paternal genome.
The CR sequences of the M-tr and F-tr genomes:
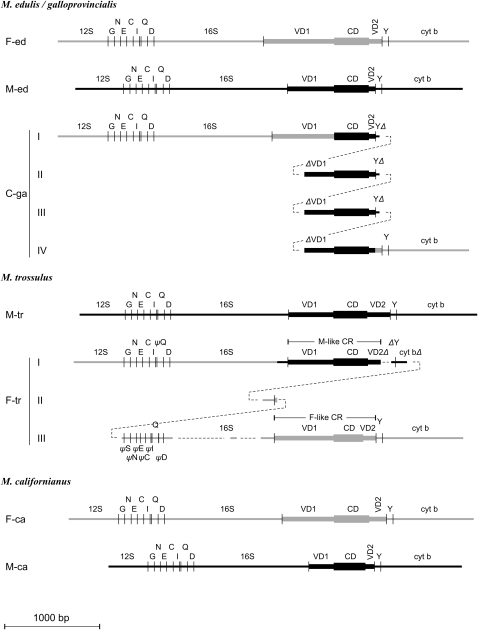
The CR of M-tr has the tripartite structure proposed by Cao et al. (2004) for the mytilid mtDNA (Figure 2). The VD1 is shorter by 18 bp than the VD1 of M-ed and the CD is shorter by 9 bp (Table 3). The most striking difference between the M-tr and M-ed CRs is in the third domain, VD2, which is almost four times as long as the M-ed and consists of two tandem repeats (Figure 3A). This repeat shows relatively high similarity (∼77%) with the VD2 of M-ed illustrating the common descent of the paternal genomes in these two species. We obtained the CR of M-tr from three males and one female individual (Table 1). There is no much variation among the four sequences (Table 3). For the rest of the article we will use the longest sequence from animal w202 (no. 9 in Table 1) for comparisons of the M-tr CR with the CR of other genomes (Table 3).
Figure 2.—
Schematic of the structure of the CR of seven genomes. Parts are aligned according to homology. F-ed, F-tr, and F-ca: the maternal genomes of M. edulis/M. galloprovincialis, M. trossulus, and M. californianus; M-ed, M-tr, and M-ca: the paternal genomes of M. edulis/M. galloprovincialis, M. trossulus, and M. californianus; C-ga: a masculinized (paternally transmitted genome) of M. galloprovincialis. Shaded lines are F-type and heavy lines are M-type sequences, respectively. 12S, 16S, cyt b: 12S-rRNA, 16S-rRNA, and Cyt b; G, N, E, C, I, Q, D, Y: the tRNAGly, tRNAAsn, tRNAGlu, tRNACys, tRNAIle, tRNAGln, tRNAAsp, and tRNATyr. VD1, CD, and VD2: the first variable, the conserved, and the second variable domains of the CR. ψ denotes a pseudogene; Δ denotes a deletion: 5′ (if it precedes) or 3′ (if it follows) the indicated gene or domain. The compound CR of two genomes, C-ga and F-tr, is given in four and three lines, respectively, to demonstrate the homology of genes or domains. Dashed lines serve to join adjacent parts. Lengths of domains are in scale.
TABLE 3.
Mean pairwise divergence (p-distance) of sequences of the same species and of the same transmission mode
| 16S(b)a
|
VD1
|
CD
|
VD2
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Genome | No. of genomes | Length | Divergence | Length | Divergence | Length | Divergence | Length | Divergence |
| M-tr | 4 | 227 | 0.004 (0.003) | 469–472 | 0.011 (0.003) | 358 | 0.007 (0.003) | 243–244 | 0.010 (0.005) |
| F-tr_M-like | 7 | 227b | 0.001 (0.001) | 470–478 | 0.008 (0.002) | 364–365 | 0.002 (0.001) | 134–135 | 0.009 (0.004) |
| F-tr_F-like | 7 | 143–144c | 0.010 (0.004) | 622–623 | 0.013 (0.003) | 313–319 | 0.010 (0.003) | 134–142 | 0.006 (0.004) |
| F-ca | 2 | 229 | 0.004 (0.004) | 550 | 0.005 (0.003) | 373 | 0.008 (0.004) | 184 | 0.005 (0.005) |
| M-ca | 2 | 220–222 | 0.014 (0.008) | 263 | 0.038 (0.012) | 335 | 0.009 (0.005) | 97–162 | 0.093 (0.028) |
Standard error is shown in parentheses.
Refers to a fragment of 16S-rRNA that exists in all compared sequences of Table 1 (see also alignment in supplemental materials).
Refers to the part of 16S-rRNA preceding the M-like CR of F-tr (region I, Figure 2).
Refers to the part of 16S-rRNA between the M-like and F-like CR of F-tr (region III, Figure 2).
Figure 3.—
The second variable domain (VD2) of M-type genomes. (A) The VD2 of M-tr is shown in the middle as two tandem duplications folded one against the other, with stars denoting nucleotide similarity. The VD2 of M-ed is shown in the first row with vertical lines denoting nucleotide similarity with one or both duplications of M-tr. The VD2 of the M-like part of F-tr is shown in the last row with vertical lines denoting nucleotide similarity with one or both duplications of M-tr. (B) The VD2 of two M. californianus males. The first consists of three tandem repeats and the second of six. Stars indicate nucleotide similarity among repeats in the same molecule.
The sequence of the CR of the F-tr genome was given by Rawson (2005) and Breton et al. (2006). We subjected this complex CR to a detailed sequence-similarity analysis (Figure 2). The entire CR consists of one M-like CR and one F-like CR, separated by a sequence of 1013 nucleotides. The M-like and F-like CRs will be discussed below. The intervening sequence consists of the following parts: (a) a piece of tRNATyr truncated at its 5′-end followed by the first 103 bp of Cyt b; (b) a stretch of 125 bp of which the first 97 bp resemble the 3′-end of 16S-rRNA and the remaining 28 bp resemble the 5′-end of CR; and (c) a sequence that corresponds to the last 15 bp of 12S-rRNA, followed by a sequence of 416 bp that contains the seven tRNAs that also occur in the coding part of the genome between 12S-rRNA and 16S-rRNA, and, finally, a sequence of 309 bp that appears to consist of remnants of 16S-rRNA separated by two large gaps (Figure 2; see also supplemental Table 1). Of the above segments, a is M-like in sequence, and b and c are F-like (Figure 2).
The entire CR of F-tr is embedded in sequences that resemble the F-ed more closely than the M-tr, except the 64 bp before the start of the M-like part, which resembles the M-tr genome. This similarity with M-tr continues past the Cyt b sequence of the second part, at which point the sequence turns again to resembling the F-ed genome. These transitions of sequence similarity suggest that the compound CD of the F-tr genome originated from events that involved exchanges between an M-tr genome and an F-like genome. The latter was probably the original M. trossulus maternal genome that was apparently replaced in the population by the present F-tr. A base-by-base alignment of the CR of F-tr with sequences from genes and CR domains from the other genomes is given in the supplemental materials, where we also give a list of lengths for each gene and CR domain in each sequence used in this article. The nucleotide diversity among the six CRs of F-tr and the one reported by Breton et al. (2006) is very minor (Table 3), so we use the longest sequence from animal w2 (no. 3 in Table 1) for comparisons of the M-tr CR with the CR of other genomes (Table 3).
The CRs of the F-ca and M-ca genomes:
Even though sequence-wise the CRs of M. californianus are quite different from those of the M. edulis species group, a tripartite structure can be also discerned. To delineate the three domains of the CR of M. californianus, we first aligned the F-ca with M-ca sequences to identify the region with the highest similarity, taking advantage of the fact that in the other two species the highest similarity between conspecific genomes with the opposite transmission mode marks the CD of the two genomes. The so-identified region of F-ca was then aligned against the CD of F-ed and that of M-ca against the CD of M-ed. Finally, the whole CR of F-ca and M-ca were aligned against the whole CR of F-ed and M-ed, respectively (see materials and methods). The so-defined CDs of the two M. californianus genomes are 373 bp for F-ca and 335 bp for M-ca. The CR of M-ca is notable for two features. First, its VD1 is much shorter, only 262–263 bp, compared to 490 bp of M-ed and to 469–472 of M-tr (Table 3). Second, its VD2 consists of repeats, like the VD2 of M-tr, but the repeat unit is much smaller, only 28–29 bp long. In the two M-ca CRs that we have sequenced, the number of repeats varied, being three in one and six in the other (Figure 3B). We use the sequences from animals 26 and 15 (nos. 13 and 15, Table 1) for comparisons of the M. californianus CR with the CR of other genomes (Table 3).
A closer look at the CR of the F-tr genome:
A close examination of the CR of the F-tr genome showed that the CD of the F-like part had an internal deletion of ∼30 bp. An alignment of this part of the CD with the corresponding parts of the CDs from all genomes from all three species (Figure 4) showed that the deletion affects the motifs Sp3 and mtTF1, which have sequence analogs in the sea urchin and in the human mtDNA (Cao et al. 2004). These motifs are known to act as regulatory elements for replication and transcription (Fisher et al. 1987; Jacobs et al. 1989; Cantatore et al. 1990). This deletion raises doubts about the functionality of the F-like CD in the F-tr genome.
Figure 4.—
Alignment of the large deletion found in the CD of the F-like part of F-tr against the CD of the other CRs. The deletion affects the Sp3 and the mtTF1 motifs that are assumed to be necessary for the binding of replication and transcription factors.
A similar argument can be made about the M-like VD1 of the F-tr genome. This VD1 shows a high degree of divergence when compared to the VD1 of the M-tr genome. A useful comparison is the divergence between the VD1 of the C genome of M. galloprovincialis (C-ga) and the VD1 of M-ed. The divergence at 16S-rRNA between M-ed and C-ga is 0.140 (SE = 0.010) and the divergence between the VD1 of M-ed and the corresponding M-type VD1 of C-ga is 0.067 (SE = 0.013). The corresponding distances between M-tr and the M-like VD1 of F-tr are 0.131 (SE = 0.009) and 0.216 (SE = 0.019). It can be seen that the divergence at 16S-rRNA is very similar in the two comparisons, yet the divergence at the VD1s of M. trossulus is more than three times larger. In both cases, the M-like sequences were inserted from the paternal genome into the maternal (from M-ed into the F-ga and from M-tr into the F-tr). Judging from the divergence at the 16S rRNA gene, we may assume that these insertions have originated at about the same time. It may not be an accident that the divergence between M-ed and C-ga has remained small. For the C-ga genome there is firm evidence that it acts as a paternal genome; i.e., it is a truly masculinized genome. For the F-tr genome, on the other hand, we have presented evidence that it is maternally transmitted. From this we may suggest that in the F-tr genome the M-like VD1 may not be functional and has entered a state of unconstrained divergence from the corresponding VD1 of the M-tr genome.
The tRNAs of F-tr:
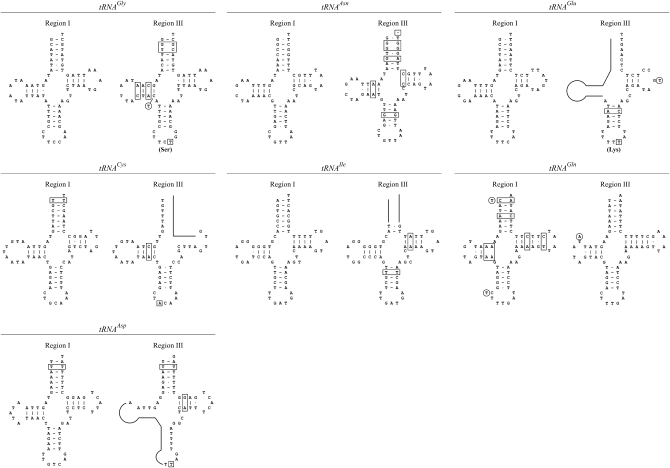
As noted above, the intervening sequence between the M-like and the F-like parts of the CR of F-tr contains parts of 16S-rRNA, Cyt b, and tRNATyr (Figure 2). These parts are obviously nonfunctional. It is of interest, however, to ask whether the same can be said about the seven tRNA sequences. Figure 5 presents the putative secondary structures of these sequences together with the corresponding structures of the tRNAs in the coding part of the F-tr genome. Four tRNAs in the CR (those for the amino acids glutamine, cysteine, isoleucine, and aspartic acid) are clearly nonfunctional and can be characterized as pseudogenes. Proper folding for the tRNAs for the amino acids glycine and asparagine in the CR requires a large number of illegitimate base pairings compared to those in the coding part, so they may also be characterized as nonfunctional. The opposite occurs with the tRNAGln, where proper folding of the sequence in the coding region requires illegitimate pairing at six positions compared to none for the tRNA in the CR. This suggests that after duplication of the seven tRNA cluster, selection has silenced one or the other copy and that for the tRNAGln the silenced copy happened to be the original one.
Figure 5.—
The most parsimonious two-dimensional structure of the seven tRNAs that are duplicated in the maternal mtDNA genome of M. trossulus. The tRNAs are shown in pairs and in the order in which they occur in the genome, with the tRNA in the coding part first. Missing parts are shown by a solid line and illegitimate pairings with boxes. Substitutions in anticodons are boxed. Circles indicate nucleotides reported in Breton et al. (2006).
DISCUSSION
One phenomenon that has been well demonstrated in the genus Mytilus, but whose presence is still questionable in species outside this genus, is “masculinization.” The phenomenon has been discussed recently in Theologidis et al. (2008). It refers to the presence of mitochondrial genomes whose coding part resembles very closely the sequence of the maternal genome, although they are transmitted paternally. Such genomes were found in M. edulis/M. galloprovincialis and in M. trossulus populations of the Baltic Sea. Originally, it was thought that these genomes were maternal genomes that, for some reason, reversed the transmission route (Saavedra et al. 1997). It is now clear that these genomes result from the exchange of sequences between maternal and paternal genomes in the control region (Venetis et al. 2007). This appears to involve a series of events, such as replication slippage, duplication, deletion, and recombination. It is interesting that, in all reported molecules that have survived the process, the entire core—defined here as the whole molecule except the part between 16S-rRNA and tRNATyr—is of the maternal type and that the control region—defined as the part between these two genes—is a mosaic containing stretches of sequences from the maternal and paternal control regions. An exception has been found in the analysis of the compound CR of the F-tr genome that we report here, where the first 64 bp before the beginning of the CR are from the paternal genome (Figure 2).
The recovery of a genome from males (or even male gonads) with a maternal-type core and a mosaic control region is assumed to be a sure sign of a masculinized genome; i.e., the genome is paternally inherited even though it is maternal-like in sequence. Here we show that this is not enough. The M. trossulus genome sequenced by Breton et al. (2006) has the above characteristics, but has turned out to be the maternal genome of this species. To establish that a genome is masculinized, one needs to demonstrate that the genome is the exclusive mtDNA molecule in the sperm of the male from which it was extracted and also that the male's somatic tissues contain another mtDNA molecule, the presumed maternal genome. As far as we know, these criteria have so far been shown to hold only for the fully sequenced C genome of M. galloprovincialis (Venetis et al. 2006, 2007). Several types of genomes with a mosaic control region and with F-like flanking sequences, which were isolated from male gonads or nonpurified sperm, have been reported in M. trossulus populations of the Baltic Sea (Burzynski et al. 2003, 2006). Many of these might turn out to be paternally transmitted, but this remains to be established.
It is important to have as many truly masculinized genomes as possible, because only then can we start to address the important question of whether there are specific sequences in the mtDNA that are essential for determining whether the genome will follow the maternal or the paternal mode of inheritance. There have been suggestions that these sequences may reside in the major control region. These suggestions were based on the fact that the only marked differences between maternal and paternal genomes are located in the control region (Cao et al. 2004; Mizi et al. 2005) and on the existence in natural populations of genomes of the maternal type with mosaic control regions (Burzynski et al. 2003, 2006; Venetis et al. 2007). It is clear that if some of these genomes are falsely assumed to be masculinized, they will mislead attempts to identify sequences responsible for the reversal of the transmission mode. The same can be said about the search for systematic differences between paternal and maternal genomes in other parts of the molecule. Thus, the alleged amino acid differences between maternal and paternal genomes (Breton et al. 2006) and the assumed interactions between nuclear and mitochondrial genomes (Breton et al. 2007) may need to be reexamined, to the extent they were based on the assumption that the F-tr genome was a masculinized genome.
Given the complex structure of the control region of the maternal genome of M. trossulus, it is interesting to compare its structure with the control regions of other genomes. All male-like domains of this genome resemble the homologous domains of the paternal genome of M. trossulus more closely than those of the paternal genome of M. edulis (Table 4). This suggests that the present-day maternal genome of M. trossulus has resulted from a series of duplication and recombination events that involved the paternal M. trossulus genome and a now-extinct M. trossulus maternal genome. A similar suggestion was made by Rawson (2005). The fact that the same maternal and paternal genomes were found in M. trossulus populations from the northwestern Atlantic and the northeastern Pacific coasts (Table 1) suggests that all North American populations of M. trossulus may contain the same paternal and maternal genomes. Of course, we cannot exclude the possibility that the postulated “old” and allegedly extinct maternal genome might be found in some populations.
TABLE 4.
Pairwise p-distances (below diagonal) and standard errors (above diagonal) among homologous domains of CRs from all types of mtDNA genomes
| Genome (length in bp) | F-ed | M-ed | C-ga_I | C-ga_II | C-ga_III | C-ga_IV | M-tr | F-tr_M-like | F-tr_F-like | F-ca | M-ca |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VD1 domain | |||||||||||
| F-ed (654) | 0.021 | 0.007 | 0.027 | 0.026 | 0.026 | 0.022 | 0.023 | 0.018 | 0.021 | 0.029 | |
| M-ed (490) | 0.480 | 0.021 | 0.013 | 0.014 | 0.013 | 0.022 | 0.023 | 0.022 | 0.024 | 0.033 | |
| C-ga_I (654) | 0.032 | 0.472 | 0.027 | 0.027 | 0.026 | 0.022 | 0.023 | 0.018 | 0.021 | 0.029 | |
| C-ga_II (314) | 0.451 | 0.067 | 0.435 | 0.003 | 0.000 | 0.027 | 0.029 | 0.027 | 0.031 | 0.048 | |
| C-ga_III (313) | 0.453 | 0.071 | 0.436 | 0.003 | 0.003 | 0.027 | 0.029 | 0.028 | 0.031 | 0.048 | |
| C-ga_IV (316) | 0.455 | 0.067 | 0.439 | 0.000 | 0.003 | 0.026 | 0.029 | 0.027 | 0.031 | 0.047 | |
| M-tr (472) | 0.518 | 0.423 | 0.511 | 0.355 | 0.356 | 0.356 | 0.019 | 0.022 | 0.022 | 0.036 | |
| F-tr_M-like (478) | 0.487 | 0.445 | 0.494 | 0.387 | 0.385 | 0.384 | 0.216 | 0.023 | 0.023 | 0.035 | |
| F-tr_F-like (622) | 0.320 | 0.485 | 0.329 | 0.472 | 0.474 | 0.476 | 0.527 | 0.524 | 0.021 | 0.029 | |
| F-ca (550) | 0.427 | 0.582 | 0.438 | 0.608 | 0.606 | 0.610 | 0.581 | 0.571 | 0.484 | 0.026 | |
| M-ca (263) | 0.387 | 0.563 | 0.391 | 0.563 | 0.559 | 0.570 | 0.562 | 0.586 | 0.480 | 0.313 | |
| CD domain | |||||||||||
| F-ed (368) | 0.007 | 0.010 | 0.010 | 0.010 | 0.010 | 0.022 | 0.021 | 0.025 | 0.025 | 0.026 | |
| M-ed (367) | 0.016 | 0.008 | 0.008 | 0.008 | 0.008 | 0.022 | 0.021 | 0.025 | 0.025 | 0.026 | |
| C-ga_I (367) | 0.041 | 0.030 | 0.003 | 0.003 | 0.003 | 0.022 | 0.021 | 0.025 | 0.025 | 0.025 | |
| C-ga_II (367) | 0.038 | 0.027 | 0.003 | 0.000 | 0.000 | 0.022 | 0.021 | 0.025 | 0.025 | 0.025 | |
| C-ga_III (367) | 0.038 | 0.027 | 0.003 | 0.000 | 0.000 | 0.022 | 0.021 | 0.025 | 0.025 | 0.025 | |
| C-ga_IV (367) | 0.038 | 0.027 | 0.003 | 0.000 | 0.000 | 0.022 | 0.021 | 0.025 | 0.025 | 0.025 | |
| M-tr (358) | 0.242 | 0.240 | 0.237 | 0.234 | 0.234 | 0.234 | 0.012 | 0.025 | 0.025 | 0.026 | |
| F-tr_M-like (365) | 0.235 | 0.230 | 0.224 | 0.221 | 0.221 | 0.221 | 0.056 | 0.024 | 0.025 | 0.026 | |
| F-tr_F-like (313) | 0.291 | 0.294 | 0.292 | 0.295 | 0.295 | 0.295 | 0.263 | 0.261 | 0.027 | 0.029 | |
| F-ca (373) | 0.341 | 0.336 | 0.345 | 0.342 | 0.342 | 0.342 | 0.342 | 0.324 | 0.409 | 0.024 | |
| M-ca (335) | 0.317 | 0.317 | 0.320 | 0.317 | 0.317 | 0.317 | 0.350 | 0.352 | 0.404 | 0.257 | |
| VD2 domain | |||||||||||
| F-ed (137) | 0.048 | 0.044 | 0.044 | 0.044 | 0.017 | 0.040 | 0.040 | 0.041 | 0.041 | 0.053 | |
| M-ed (71) | 0.194 | 0.028 | 0.028 | 0.028 | 0.048 | 0.052 | 0.052 | 0.060 | 0.059 | 0.083 | |
| C-ga_I (68) | 0.162 | 0.060 | 0.000 | 0.000 | 0.042 | 0.051 | 0.051 | 0.059 | 0.059 | 0.081 | |
| C-ga_II (68) | 0.162 | 0.060 | 0.000 | 0.000 | 0.042 | 0.051 | 0.051 | 0.059 | 0.059 | 0.081 | |
| C-ga_III (68) | 0.162 | 0.060 | 0.000 | 0.000 | 0.042 | 0.051 | 0.051 | 0.059 | 0.059 | 0.081 | |
| C-ga_IV (133) | 0.046 | 0.194 | 0.134 | 0.134 | 0.134 | 0.040 | 0.039 | 0.042 | 0.041 | 0.054 | |
| M-tr (243) | 0.321 | 0.246 | 0.242 | 0.242 | 0.242 | 0.304 | 0.030 | 0.041 | 0.043 | 0.053 | |
| F-tr_M-like (134) | 0.328 | 0.242 | 0.239 | 0.239 | 0.239 | 0.288 | 0.152 | 0.042 | 0.041 | 0.053 | |
| F-tr_F-like (137) | 0.359 | 0.431 | 0.409 | 0.409 | 0.409 | 0.392 | 0.445 | 0.447 | 0.043 | 0.053 | |
| F-ca (184) | 0.382 | 0.387 | 0.397 | 0.397 | 0.397 | 0.378 | 0.402 | 0.375 | 0.469 | 0.049 | |
| M-ca (97) | 0.465 | 0.344 | 0.313 | 0.313 | 0.313 | 0.457 | 0.512 | 0.446 | 0.465 | 0.385 | |
The CR of both M. californianus genomes exhibit the standard structure of the mytilid genome: they have simple control regions with a conserved central domain flanked by variable domains. The availability of the maternal and paternal control regions from three species, with a variable degree of phylogenetic relatedness, allows the detection of certain patterns in the evolution of the CR in mytilids. In all three species, the first variable domain is shorter in the paternal genome (Table 4). The difference between paternal and maternal genomes is more dramatic in the second variable domain. This domain has approximately the same length in all maternal genomes (137 bp in M. edulis and M. trossulus, 184 bp in M. californianus). In the paternal genomes of two species, M. trossulus and M. californianus, it consists of tandem repeats. No repeat structure can be discerned in the paternal M. edulis VD2, but there is an obvious sequence similarity with the repeats of M. trossulus. Presently, it is impossible to draw any causal relationship between these differences of maternal and paternal genomes and their mode of inheritance.
There remains the question why the CR in the maternal genome of M. trossulus remains so large. Our analysis points to a complementary hypothesis. We have shown that tRNAGln in the coding part of the genome may not fold properly, whereas its copy in the control region does. If this implies nonfunctionality for tRNAGln in the coding part, it will be the first case of a coding gene embedded within the control region of a mitochondrial genome. We have also presented evidence that the conserved region of the F-like part may not be functional. The evidence for nonfunctionality of the first variable domain of the M-like part is much weaker. This domain has diverged from the corresponding domain of the paternal genome much more than in the analogous cases of the M. galloprovincialis C genome and its paternal genome of M. galloprovincialis, and this may suggest a relaxed selective constraint on the VD1 of the M-like part. Finally, the VD2 of the M-like part is half as long as the corresponding part of the paternal genome. We may therefore hypothesize that, in the composite control region of the maternal genome of M. trossulus, the function of the conserved domain is served by the M-type CD, the function of the first variable domain by the F-type VD1, and the function of the second variable domain by the F-type VD2. At present, the evidence for this supposition is weak and it may serve only as a working hypothesis. It is consistent with the observation that the conserved domain differs very little between maternal and paternal genomes and can therefore be assumed to have the same role in either genome; namely, it facilitates the binding of replication and transcription factors. It also reinforces the suspicion that the most likely site for sequences that may control the mode of inheritance of the mitochondrial genome, if such sequences exist at all, is the first variable domain. This will explain why the maternal genome of M. trossulus cannot be inherited paternally, given that the paternal type VD1 has been altered by mutation.
Acknowledgments
This work was supported by Dalhousie University and Patrick Lett Scholarships and Fisheries and Oceans, Canada (to L.C.); by the Greek General Secretariat for Research and Technology (grant PENED-01ED42 to A.M., E.Z. and G.C.R.); by the Marine Genomics Europe Network of Excellence (to E.Z.); by the National Science Foundation (DEB-0412976); and by the Partnership for the Interdisciplinary Study of Coastal Oceans (to G.P. and B.S.O.) and the University of Athens (grant ELKE 70/4/7805 to G.C.R.).
References
- Bierne, N., F. Bonhomme, P. Boudry, M. Szulkin and P. David, 2006. Fitness landscapes support the dominance theory of post-zygotic isolation in the mussels Mytilus edulis and M. galloprovincialis. Proc. Biol. Sci. 273 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore, J. L., M. Medina and L. A. Rosenberg, 2004. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod Graptacme eborea and the bivalve Mytilus edulis. Mol. Biol. Evol. 21 1492–1503. [DOI] [PubMed] [Google Scholar]
- Breton, S., G. Burger, D. T. Stewart and P. U. Blier, 2006. Comparative analysis of gender-associated complete mitochondrial genomes in marine mussels (Mytilus spp.). Genetics 172 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton, S., H. D. Beaupre, D. T. Stewart, W. R. Hoeh and P. U. Blier, 2007. The unusual system of doubly uniparental inheritance of mtDNA: Isn't one enough? Trends Genet. 23 465–474. [DOI] [PubMed] [Google Scholar]
- Burzynski, A., M. Zbawicka, D. O. Skibinski and R. Wenne, 2003. Evidence for recombination of mtDNA in the marine mussel Mytilus trossulus from the Baltic. Mol. Biol. Evol. 20 388–392. [DOI] [PubMed] [Google Scholar]
- Burzynski, A., M. Zbawicka, D. O. Skibinski and R. Wenne, 2006. Doubly uniparental inheritance is associated with high polymorphism for rearranged and recombinant control region haplotypes in Baltic Mytilus trossulus. Genetics 174 1081–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantatore, P., M. Roberti, P. P. Loguercio, A. Mustich and M. N. Gadaleta, 1990. Mapping and characterization of Paracentrotus lividus mitochondrial transcripts: multiple and overlapping transcription units. Curr. Genet. 17 235–245. [DOI] [PubMed] [Google Scholar]
- Cao, L., E. Kenchington, E. Zouros and G. C. Rodakis, 2004. Evidence that the large noncoding sequence is the main control region of maternally and paternally transmitted mitochondrial genomes of the marine mussel (Mytilus spp.). Genetics 167 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curole, J. P., and T. D. Kocher, 2002. Ancient sex-specific extension of the cytochrome c oxidase II gene in bivalves and the fidelity of doubly-uniparental inheritance. Mol. Biol. Evol. 19 1323–1328. [DOI] [PubMed] [Google Scholar]
- Dalziel, A. C., and D. T. Stewart, 2002. Tissue-specific expression of male-transmitted mitochondrial DNA and its implications for rates of molecular evolution in Mytilus mussels (Bivalvia: Mytilidae). Genome 45 348–355. [DOI] [PubMed] [Google Scholar]
- Douris, V., S. Giokas, R. Lecanidou, M. Mylonas and G. C. Rodakis, 1998. Phylogenetic analysis of mitochondrial DNA and morphological characters suggest a need for taxonomic re-evaluation within the Alopiinae (Gastropoda: Clausiliidae). J. Moll. Stud. 64 81–92. [Google Scholar]
- Fisher, R. P., J. N. Topper and D. A. Clayton, 1987. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell 50 247–258. [DOI] [PubMed] [Google Scholar]
- Garrido-Ramos, M. A., D. T. Stewart, B. W. Sutherland and E. Zouros, 1998. The distribution of male-transmitted and female-transmitted mitochondrial DNA types in somatic tissues of blue mussels: implications for the operation of doubly uniparental inheritance of mitochondrial DNA. Genome 41 818–824. [Google Scholar]
- Gossling, E., 1992. Systematics and geographic distribution of Mytilus, pp. 1–20 in The Mussel Mytilus: Ecology, Physiology, Genetics and Aquaculture, edited by E. Gossling. Elsevier, Amsterdam.
- Heath, D. D., P. D. Rawson and T. J. Hilbish, 1995. PCR-based nuclear markers identify alien blue mussel (Mytilus spp.) genotypes on the west coast of Canada. Can. J. Fish. Aquat. Sci. 52 2621–2627. [Google Scholar]
- Hoeh, W. R., D. T. Stewart, B. W. Sutherland and E. Zouros, 1996. Multiple origins of gender associated mitochondrial DNA lineages in bivalves (Mollusca: Bivalvia). Evolution 50 2276–2286. [DOI] [PubMed] [Google Scholar]
- Hoeh, W. R., D. T. Stewart, C. Saavedra, B. W. Sutherland and E. Zouros, 1997. Phylogenetic evidence for role-reversals of gender-associated mitochondrial DNA in Mytilus (Bivalvia: Mytilidae). Mol. Biol. Evol. 14 959–967. [DOI] [PubMed] [Google Scholar]
- Hoeh, W. R., D. T. Stewart and S. I. Guttman, 2002. High fidelity of mitochondrial genome transmission under the doubly uniparental mode of inheritance in freshwater mussels (Bivalvia: Unionoidea). Evolution 56 2252–2261. [DOI] [PubMed] [Google Scholar]
- Hoffmann, R. J., J. L. Boore and W. M. Brown, 1992. A novel mitochondrial genome organization for the blue mussel, Mytilus edulis. Genetics 131 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, H. T., E. R. Herbert and J. Rankine, 1989. Sea urchin egg mitochondrial DNA contains a short displacement loop (D-loop) in the replication origin region. Nucleic Acids Res. 17 8949–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5 150–163. [DOI] [PubMed] [Google Scholar]
- Lecanidou, R., V. Douris and G. C. Rodakis, 1994. Novel features of metazoan mtDNA revealed from sequence analysis of three mitochondrial DNA segments of the land snail Albinaria turrita (Gastropoda: Clausiliidae). J. Mol. Evol. 38 369–382. [DOI] [PubMed] [Google Scholar]
- Liu, H. P., J. B. Mitton and S. K. Wu, 1996. Paternal mitochondrial DNA differentiation far exceeds maternal mitochondrial DNA and allozyme differentiation in the freshwater mussel, Anodonta grandis grandis. Evolution 50 952–957. [DOI] [PubMed] [Google Scholar]
- Merritt, T. J., L. Shi, M. C. Chase, M. A. Rex, R. J. Etter et al., 1998. Universal cytochrome b primers facilitate intraspecific studies in molluscan taxa. Mol. Marine Biol. Biotechnol. 7 7–11. [PubMed] [Google Scholar]
- Miller, S. A., D. D. Dykes and H. F. Polesky, 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizi, A., E. Zouros, N. Moschonas and G. C. Rodakis, 2005. The complete maternal and paternal mitochondrial genomes of the Mediterranean mussel Mytilus galloprovincialis: implications for the doubly uniparental inheritance mode of mtDNA. Mol. Biol. Evol. 22 952–967. [DOI] [PubMed] [Google Scholar]
- Mizi, A., E. Zouros and G. C. Rodakis, 2006. Multiple events are responsible for an insertion in a paternally inherited mitochondrial genome of the mussel Mytilus galloprovincialis. Genetics 172 2695–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata, M., C. Kamiya, K. Kawamura and A. Komaru, 2006. Sperm mitochondrial DNA transmission to both male and female offspring in the blue mussel Mytilus galloprovincialis. Dev. Growth. Differ. 48 253–261. [DOI] [PubMed] [Google Scholar]
- Ort, B. S., and G. H. Pogson, 2007. Molecular population genetics of the male and female mitochondrial DNA molecules of the California sea mussel, Mytilus californianus. Genetics 177 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamonti, M., and V. Scali, 2001. Gender-associated mitochondrial DNA heteroplasmy in the venerid clam Tapes philippinarum (Mollusca Bivalvia). Curr. Genet. 39 117–124. [DOI] [PubMed] [Google Scholar]
- Quesada, H., R. Wenne and D. O. Skibinski, 1995. Differential introgression of mitochondrial DNA across species boundaries in the marine mussel genus Mytilus. Proc. R. Soc. Lond. B 262 57–64. [Google Scholar]
- Quesada, H., R. Wenne and D. O. Skibinski, 1999. Interspecies transfer of female mitochondrial DNA is coupled with role-reversals and departure from neutrality in the mussel Mytilus trossulus. Mol. Biol. Evol. 16 655–665. [DOI] [PubMed] [Google Scholar]
- Rawson, P. D., 2005. Nonhomologous recombination between the large unassigned region of the male and female mitochondrial genomes in the mussel, Mytilus trossulus. J. Mol. Evol. 61 717–732. [DOI] [PubMed] [Google Scholar]
- Rawson, P. D., and T. J. Hilbish, 1995. Evolutionary relationships among the male and female mitochondrial DNA lineages in the Mytilus edulis species complex. Mol. Biol. Evol. 12 893–901. [DOI] [PubMed] [Google Scholar]
- Rawson, P. D., C. L. Secor and T. J. Hilbish, 1996. The effects of natural hybridization on the regulation of doubly uniparental mtDNA inheritance in blue mussels (Mytilus spp.). Genetics 144 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra, C., M. I. Reyero and E. Zouros, 1997. Male-dependent doubly uniparental inheritance of mitochondrial DNA and female-dependent sex-ratio in the mussel Mytilus galloprovincialis. Genetics 145 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serb, J. M., and C. Lydeard, 2003. Complete mtDNA Sequence of the North American freshwater mussel, Lampsilis ornata (Unionidae): an examination of the evolution and phylogenetic utility of mitochondrial genome organization in Bivalvia (Mollusca). Mol. Biol. Evol. 20 1854–1866. [DOI] [PubMed] [Google Scholar]
- Skibinski, D. O., C. Gallagher and C. M. Beynon, 1994. a Mitochondrial DNA inheritance. Nature 368 817–818. [DOI] [PubMed] [Google Scholar]
- Skibinski, D. O., C. Gallagher and C. M. Beynon, 1994. b Sex-limited mitochondrial DNA transmission in the marine mussel Mytilus edulis. Genetics 138 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, D. T., C. Saavedra, R. R. Stanwood, A. O. Ball and E. Zouros, 1995. Male and female mitochondrial DNA lineages in the blue mussel (Mytilus edulis) species group. Mol. Biol. Evol. 12 735–747. [DOI] [PubMed] [Google Scholar]
- Theologidis, I., S. Fodelianakis, M. B. Gaspar and E. Zouros, 2008. Doubly uniparental inheritance (DUI) of mitochondrial DNA in Donax trunculus (Bivalvia: Donacidae) and the problem of its sporadic detection in Bivalvia. Evolution 62 959–970. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venetis, C., I. Theologidis, E. Zouros and G. C. Rodakis, 2006. No evidence for presence of maternal mitochondrial DNA in the sperm of Mytilus galloprovincialis males. Proc. Biol. Sci. 273 2483–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venetis, C., I. Theologidis, E. Zouros and G. C. Rodakis, 2007. A mitochondrial genome with a reversed transmission route in the Mediterranean mussel Mytilus galloprovincialis. Gene 406 79–90. [DOI] [PubMed] [Google Scholar]
- Zouros, E., A. O. Ball, C. Saavedra and K. R. Freeman, 1994. a Mitochondrial DNA inheritance. Nature 368 818. [DOI] [PubMed] [Google Scholar]
- Zouros, E., A. O. Ball, C. Saavedra and K. R. Freeman, 1994. b A new type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proc. Natl. Acad. Sci. USA 91 7463–7467. [DOI] [PMC free article] [PubMed] [Google Scholar]