Abstract
Telomeres and subtelomere regions have vital roles in cellular homeostasis and can facilitate niche adaptation. However, information on telomere/subtelomere structure is still limited to a small number of organisms. Prior to initiation of this project, the Neurospora crassa genome assembly contained only 3 of the 14 telomeres. The missing telomeres were identified through bioinformatic mining of raw sequence data from the genome project and from clones in new cosmid and plasmid libraries. Their chromosomal locations were assigned on the basis of paired-end read information and/or by RFLP mapping. One telomere is attached to the ribosomal repeat array. The remaining chromosome ends have atypical structures in that they lack distinct subtelomere domains or other sequence features that are associated with telomeres in other organisms. Many of the chromosome ends terminate in highly AT-rich sequences that appear to be products of repeat-induced point mutation, although most are not currently repeated sequences. Several chromosome termini in the standard Oak Ridge wild-type strain were compared to their counterparts in an exotic wild type, Mauriceville. This revealed that the sequences immediately adjacent to the telomeres are usually genome specific. Finally, despite the absence of many features typically found in the telomere regions of other organisms, the Neurospora chromosome termini still retain the dynamic nature that is characteristic of chromosome ends.
EUKARYOTIC nuclear chromosomes are linear molecules that terminate in specialized sequences known as telomeres. Telomeres are added on to the 3′-end of chromosome ends to prevent loss of DNA from lagging strands during replication. In most eukaryotes, telomeres consist of tandem arrays of simple sequence repeats. Notable exceptions are Drosophila and some other dipterans, which instead possess tandem arrays of retrotransposons at their chromosome ends (Abad et al. 2004). Telomeres made up of simple sequence repeats vary in sequence among organisms, although the strand that reads 5′ to 3′ toward the chromosome end tends to be G-rich. For example, the telomeres of the ascomycete fungus Saccharomyces cerevisiae consist of a TG-rich repeat sequence (Walmsley et al. 1984), plant chromosomes typically end in (TTTAGGG)n (Richards and Ausubel 1988), and chromosomes of mammals (Meyne et al. 1989) and many filamentous fungi (Schechtman 1990; Coleman et al. 1993; Farman and Leong 1995; Bhattacharyya and Blackburn 1997; Keely et al. 2005) end in (TTAGGG)n.
The 3′ strand of the telomere extends as an overhang and is capable of base pairing with itself using non-Watson–Crick interactions (Henderson et al. 1987). It can also invade the TTAGGG duplex region to form a “T-loop” structure (Griffith et al. 1999; Munoz-Jordan et al. 2001; Murti and Prescott 2002). These structures make telomeres refractory to cloning unless the 3′ tails are removed by enzymatic treatment. A consequence of the difficulty in cloning telomeres is that, for most organisms, there is limited information on the organization of chromosome ends. Nevertheless, the characterization of terminal chromosome regions in a few model organisms reveals striking similarities in structure. The sequences that reside adjacent to the telomere repeats generally are also present at other chromosome ends (Pryde et al. 1997). Two domains are often discernible within these subtelomeric sequences. The distal subtelomere domains, located adjacent to the telomere repeats, contain various types of short, tandem repeat motifs and are typically found on several chromosomes (Pryde et al. 1997). The more proximal subtelomere domains tend to be less repeated and commonly contain clusters of related genes.
In microbial eukaryotes, subtelomeric gene clusters often have roles in niche adaptation. For example, the S. cerevisiae subtelomeres contain families of genes involved in sugar utilization, and the types of genes that are amplified in these regions correlate with the niche from which a given strain is isolated (Denayrolles et al. 1997). Other terminally amplified sequences in S. cerevisiae include the FLO and PAU genes, which are involved in flocculation and anaerobic growth, respectively (Rachidi et al. 2000; Verstrepen et al. 2005). These traits also are likely to be adaptive. In microbial pathogens of humans, such as the protists Plasmodium falciparum (malaria) and Trypanosoma brucei (sleeping sickness) and the ascomycete fungus Pneumocystis carinii, the subtelomeres contain families of variant genes coding for surface proteins (Hernandez-Rivas et al. 1997; Barry et al. 2003; Keely et al. 2005). These organisms use various mechanisms to switch expression among different gene copies—a strategy that allows them to evade the immune system (Graham and Barry 1995; Rudenko et al. 1996; Sunkin and Stringer 1996; Wada and Nakamura 1996; Scherf et al. 1998). Interestingly, humans and chickens have large families of olfactory genes, many of which are encoded by subtelomeric gene clusters (Trask et al. 1998; International Chicken Genome Sequencing Consortium 2004).
Such examples suggest that terminal chromosome regions are frequently co-opted for gene amplification and diversification and that some microbes have taken this a step further by developing active mechanisms for switching expression among the subtelomeric genes. Nevertheless, characterization of chromosome ends in additional eukaryotic microbes is needed to determine if this situation is exceptional or the norm.
Telomeres have been cloned from many different microbes but in most cases the number of chromosome ends represented, and the corresponding sequence information, is limited. Consequently, it is usually not possible to detect subtelomeric gene amplifications from such data. Recently, the sequences of all 14 telomeres in the filamentous ascomycete fungus Magnaporthe oryzae were determined (Rehmeyer et al. 2006). Analysis of those sequences revealed the presence of a clearly defined distal subtelomere domain that contains a telomere-linked helicase (TLH) gene. Surprisingly, however, apart from the TLH genes, there was almost no gene duplication near the chromosome termini and, as such, it was not possible to detect a proximal subtelomere domain. At least seven chromosome ends with recognizable distal subtelomeric regions have been identified in another ascomycete fungus, Aspergillus nidulans, but gaps in the genome sequence make it difficult to determine if A. nidulans contains a proximal domain (Clutterbuck and Farman 2007).
Here, we describe the organization and evolution of telomere regions in another ascomycete fungus, Neurospora crassa. The sequenced N. crassa genome (Galagan et al. 2003) contains very little intact, duplicated DNA due to the repeat-induced point mutation (RIP) process, which acts during the sexual cycle and peppers duplicated sequences with polarized transition mutations (G:C to A:T) (Selker 1990). For this reason, it seemed unlikely that N. crassa would possess intact subtelomere domains or terminal gene duplications. Thus, it was of interest to determine the structure of Neurospora chromosome ends.
Prior to the genome sequencing project, only two N. crassa telomere fragments had been cloned and characterized by sequencing. Both contained tandem arrays of the motif TTAGGG (Schechtman 1987, 1990). One segment mapped at the right end of chromosome V (Tel-VR) immediately adjacent to a putative transposable element, Pogo. The standard wild-type strain of N. crassa, Oak Ridge (OR), contains at least nine copies of Pogo but none of the other copies are linked to telomeres. Apart from the terminal TTAGGG repeats, the second telomere sequence exhibited no similarity to Tel-VR and its identity was unknown (Schechtman 1990).
Use of a (TTAGGG)4 probe in a Southern analysis of progeny from a cross between an OR laboratory strain and an exotic wild-type strain, Mauriceville (M), revealed restriction fragment length polymorphisms (RFLPs) at all 14 telomeres and identified map locations for six of them (Schechtman 1989). As the density of the N. crassa map increased, the map positions of a total of 12 telomeres were established (Nelson and Perkins 2000). The genome sequence project promised to provide valuable new insights into the organization and gene content of the OR chromosome ends. However, BLAST searches of the genome assembly revealed only 4 telomeres. Therefore, we used the telomere-mining program TERMINUS (Li et al. 2005) to identify new telomeres among the raw, unassembled sequence reads and to link these new ends to the genome sequence. Cosmid clones containing telomeres obtained from new libraries were then sequenced to close gaps. Through these efforts, we were able to identify which of the genome sequence contigs are close to telomeres, which in turn provided insight into the organization and gene content of these important chromosome regions. Finally, to determine the molecular basis for the high levels of polymorphism at N. crassa chromosome ends, we cloned and characterized several telomeres and subtelomeric regions from the M strain and compared them to their counterparts in OR.
MATERIALS AND METHODS
Neurospora strains:
N. crassa wild-type strains used were Oak Ridge 74-OR23-IVA (FGSC 2489) and Mauriceville 1c-A (FGSC 2225). RFLP mapping strains (FGSC 4450-4487) are progeny isolated from a cross between multicent-2-a (FGSC 4416) and FGSC 2225 (Metzenberg et al. 1984; Metzenberg and Grotelueschen 1995) and were obtained from the Fungal Genetics Stock Center (FGSC), along with the parental strains. The purification of microconidia from macroconidiating strains was performed as described previously (Ebbole and Sachs 1990).
Construction and probing of plasmid/cosmid libraries for telomere-containing clones:
Genomic DNA was isolated using a procedure based on methods previously described (Oakley et al. 1987; Luo et al. 1995). The details are provided in the supplemental information. The pBluescript II KS+ plasmid (Stratagene, La Jolla, CA) was linearized by digestion with SmaI (New England Biolabs, Ipswich, MA), and cosmid vector pMLF4 (see supplemental information) was linearized with EcoRV (New England Biolabs). Both vectors were dephosphorylated by treatment with shrimp alkaline phosphatase (Promega, Madison, WI) for 15 min at 37° and then the enzymes were inactivated by a 20-min incubation at 65°. The linearized vectors were subsequently gel purified using a QIAEX II gel extraction kit (Qiagen, Valencia, CA).
Prior to cloning, the genomic DNA was end repaired with the End-It kit (Epicentre Technologies, Madison, WI). Polished genomic DNA (500 ng) and linearized vector (100 ng) were ligated with T4 DNA ligase (New England Biolabs) overnight at 16°. For the cosmid library construction, half of the ligation mixture (5 μl) was packaged in vitro with λ-phage packaging extract (Epicentre), following the manufacturer's instructions, and used to infect Escherichia coli XL-10 cells (Stratagene). For the plasmid libraries, the ligase was heat inactivated and the DNA was purified by ethanol precipitation. The ligation mixture was then treated with HindIII or BamHI (New England Biolabs), after which the enzymes were heat inactivated and the DNA was precipitated with ethanol. The pellet was dissolved in ligation buffer and then treated with T4 DNA ligase overnight at room temperature. The ligated DNA was ethanol precipitated, resuspended in TE, and transformed into E. coli XL-10 (Stratagene). For both the cosmid and plasmid libraries, the E. coli cells were plated on LB agar with 100 μg/ml carbenicillin at a density of ∼400 transformants/plate.
The E. coli transformants were lifted onto Whatman 541 paper discs and the plates were incubated at 25° overnight to allow the colonies to regrow. The plates were then stored at 4° until positive clones had been identified. The Whatman filters were processed and hybridized as described previously (Gergen et al. 1979). Positive colonies were picked from the original agar plate and restreaked on fresh LB–ampicillin plates; replica filters of these plates were reprobed and the process reiterated until pure clones were obtained. These were then cultured in 100 ml of LB plus 100 μg/ml ampicillin, and cosmid DNA was extracted using the Wizard midi-prep kit (Promega).
Preparation of 32P-labeled telomere probes:
Concatemers of the telomeric repeat sequence (TTAGGG)n were obtained by using the primers TTAGGG and CCCTAA (supplemental Table 1) to perform PCR in the absence of template. Reactions contained 1× buffer (Takara BIO, Madison, WI); 12.5 pmol TTAGGG primer; 12.5 pmol CCCTAA primer; 400 μm dNTPs; and 1.25 units LA Taq polymerase (Takara). The amplification conditions were 94°, 5 min; followed by 35 cycles of 94°, 30 sec; 55°, 30 sec; and 72°, 1 min. Finally, a 5-min extension was performed at 72°. PCR products were resolved by electrophoresis in a 1% agarose gel and the 1.5- to 2-kb DNA products were excised and purified using a QIAEX II gel extraction kit (Qiagen, Valencia, CA). The purified concatemers were then used as templates in a modified primer-directed DNA synthesis reaction (Feinberg and Vogelstein 1984) to generate 32P-labeled telomere probe using an equivalent concentration of oligonucleotide (TAACCC)3 instead of random hexanucleotides.
Sequencing and assembly of telomeric cosmids:
Cosmid DNA was prepared by alkaline lysis and sheared to ∼2-kb fragments using a Hydroshear machine fitted with the standard-sized shearing assembly (Genemachines, Ann Arbor, MI). The fragments were end repaired using the End-It kit (Epicentre), ligated to the pHCamp vector (Lucigen Technologies, Middleton, WI), and electroporated into the E. coli strain EPI300 (Epicentre). Recombinant DNAs were prepared by alkaline lysis and sequenced with Big-Dye V3 chemistry (Applied Biosystems, Foster City, CA), using the SL1 and SR2 primers provided with the vector. Sequencing primers MLF4-T3 and MLF4-T7 (supplemental Table 1) were used for sequencing the ends of the cosmid inserts. Sequences were assembled using the Phred/Phrap software packages (Ewing and Green 1998; Ewing et al. 1998) and manually checked using Consed (Gordon et al. 1998). Small gaps (≤5 kb) between the telomeres and the genome assembly were filled by primer walking, using cosmids as templates.
Southern hybridization analysis:
Genomic DNA isolated from N. crassa and cosmid DNA samples was digested with EcoRV/NotI or HindIII/NotI (New England Biolabs) and fragments were separated by electrophoresis in 0.8% agarose gels. Gels were blotted using standard methods for capillary transfer (Sambrook et al. 1989) or by using an electroblotter (Idea Scientific, Minneapolis). Probes were 32P-labeled by random hexanucleotide priming (Feinberg and Vogelstein 1984) or by specific priming as described below. The hybridization and washing conditions were as described previously (Rountree and Selker 1997). Autoradiography was performed with a Molecular Dynamics or Typhoon PhosphorImager (GE Healthcare, Piscataway, NJ).
Generation of probes for telomere-adjacent sequences:
Telomere-adjacent sequences were PCR amplified from OR genomic DNA using the primers listed in supplemental Table 1. The amplification reaction contained 80 ng genomic DNA, 12.5 pmol of each primer, 1× buffer (Takara), 400 μm dNTPs, and 1.25 units LA Taq polymerase (Takara). Cycling conditions were 94° for 1 min, followed by 35 cycles of 94°, 30 sec; 55°, 30 sec; and 72°, 1 min. The final extension was at 72° for 5 min. Probes were purified by agarose gel electrophoresis and QiaQuick extraction (Qiagen) and then verified by sequencing.
Sliding-window analysis of GC content and RIP indices:
To determine how the nucleotide compositions of the terminal sequences change as a function of the position relative to the telomere, the values of interest were calculated in a “window” of 200 nucleotides. The window was slid in 20-bp increments (in the centromere-to-telomere direction) and values were recalculated for each position. This was reiterated until the right-hand edge of the window met the telomeric end of the sequence. GC content was measured as the percentage of G or C nucleotides in each window. RIP index I was calculated as ApT/TpA, and RIP index II was CpA + TpG/ApC + GpT (Margolin et al. 1998). These operations were performed automatically using a perl script (RIPindex.pl, available from M. Farman upon request).
Terminal gene analysis:
We identified genes that were contained within 50 kb of each telomere by inspecting gene predictions from the N. crassa genome database (Assembly 7, version 3) at the Broad Institute (http://www.broad.mit.edu/annotation/genome/neurospora). In addition, we used blastx to search the nr database at NCBI using the predicted coding sequences as queries. BLAST results were considered significant if the expected value (e-value) was <10−5. The protein sequences were also used to query the online Pfam database (http://pfam.sanger.ac.uk) (Bateman et al. 2004) using default parameters.
RESULTS
N. crassa telomeric restriction fragments:
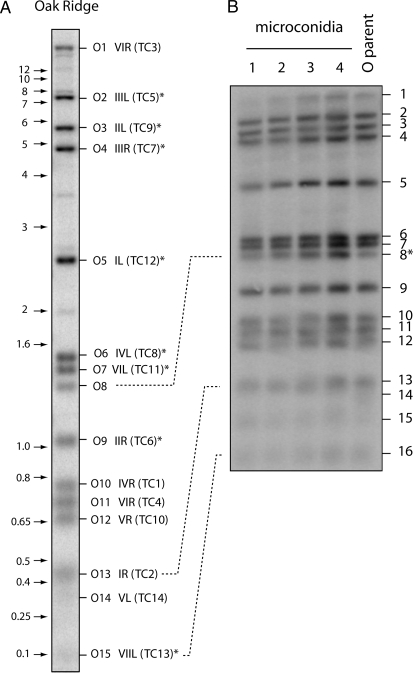
N. crassa has seven chromosomes and thus presumably 14 distinct telomeres. Indeed, Schechtman (1989) reported detecting 10–14 fragments with telomeric sequences in restriction digests of DNA from the standard laboratory wild type (OR 74-OR23-IVA). In this study, we probed HindIII/NotI double-digested OR DNA with a 32P-labeled (TTAGGG)200–300 probe. Unexpectedly, depending on the particular genomic OR DNA sample that was used, we detected 15 or 16 putative telomeric fragments (Figure 1A). Two hybridization signals were fainter than the others (O8 and O14 in Figure 1A), which suggested that our OR stock was a heterokaryon with one or more polymorphic telomeres. To test this idea, we genetically purified the culture by isolating microconidia that contain single haploid nuclei (Maheshwari 1999). Analysis of telomere profiles for the individual microconidial isolates revealed the loss of one telomere-hybridizing fragment that was present in the starting strain (fragment 14, Figure 1B), indicating that the original culture was indeed heterokaryotic for this telomere. However, there still remained 15 hybridization signals in each of the microconidial DNA preparations. Therefore, to address this issue further, we analyzed the segregation of the telomere-hybridizing fragments among progeny of the standard N. crassa mapping cross between multicent-2a and M (Metzenberg et al. 1984) to identify ones that cosegregated with known telomeric markers. The genetic background of multicent-2a is essentially OR, and 13 of the telomere-hybridizing fragments matched between the two strains (supplemental Figure 1A). All of these matching fragments exhibited segregation patterns consistent with telomeric map locations (supplemental Table 2). Surprisingly, even fragment O14 (Figure 1A)—the one that was absent in the microconidial cultures—segregated normally in the cross and mapped to Tel-VL. That O14 corresponds to a bona fide telomere in OR was subsequently confirmed by the cloning of an ∼360-bp telomeric fragment that mapped at Tel-VL (results not shown). The basis for its instability will be discussed later. The telomere-hybridizing fragments O8 and O10 were not present in multicent-2a and, therefore, it was not possible to use the segregation data to determine if either of them corresponded to the 14th telomere. However, the subsequent cloning and characterization of OR telomeres (see below) revealed that O10 corresponds to Tel-IVR and is allelic to the slightly larger Mu10 fragment in multicent-2a (supplemental Figure 1A). Thus, O10 is the elusive telomere 14. The eventual cloning of fragment O8 proved it to be an internal fragment that contains the telomere-like sequence [ctaa(ccctaa)2(ctaaccctaa)7] (data not shown).
Figure 1.—
Telomere restriction profiles of the N. crassa OR strain. Genomic DNA was digested with HindIII plus NotI and fractionated by electrophoresis through agarose gels. The gels were blotted to nylon membranes and probed with the 32P-labeled telomere repeat sequence. The resulting phosphorimages are shown. (A) HindIII/NotI restriction profile of OR. The positions of size markers are shown on the left (in kilobases) and hybridizing bands are numbered, from largest to smallest, on the right. The corresponding TelContig numbers are shown in parentheses. Asterisks mark bands whose TelContig associations were confirmed by characterizing the relevant cosmid clones. Bands 14 and 15 do not reproduce well but were clearly visible upon overexposure of the phosphorimage. (B) Telomere profiles of microconidial isolates of the OR strain. Telomere bands are numbered from largest to smallest. The fragment marked with an asterisk is the only one that did not segregate with the mapping population.
N. crassa telomeres in the genome sequence database:
BLAST searches of the first three Neurospora genome assemblies (V.1–V.3) revealed only five sequences matching the telomere repeat (TTAGGG)n. Subsequent releases (versions 6 and 7) contained four additional telomeres. However, two of the sequences identified in the earlier assemblies were missing from the latest version (V.7), which contains only seven telomeres. All of the telomere sequences in the genome assembly are at the ends of their respective supercontigs, which strongly suggests that there are no major telomeric tracts at internal genomic locations.
To determine if additional telomeres might have been captured in the genome-sequencing project but had escaped assembly, we used TERMINUS (Li et al. 2005) to identify and assemble telomere-containing sequences present among the raw sequence reads. A total of 301 such reads were identified, with an average CCCTAA repeat length of 120 bp (range: 32–256 bp). These assembled into 14 contigs (TelContigs, or TCs), each of which started with the telomere repeat in the expected orientation: [CCCTAA]n. The sequences of the telomeres previously cloned by Schechtman (1990) were used as queries to search the TelContig sequences using BLAST. This revealed that Tel-VR (GenBank accession no. M37064) is represented by TC10 while the unmapped sequence (M54885) corresponds to TC8. Seven of the TelContigs (including TCs 8 and 10) corresponded to the telomeres that were already present in the Version 7 genome sequence. Using mate-pair sequence information, TERMINUS linked five additional TelContigs to the genome assembly (Table 1). Thus, reliable telomere-to-genome linkages were established for 12 of the 14 TelContigs.
TABLE 1.
Physical and genetic mapping of TelContigs
| TC | No. of reads | Linked to assembly contiga | TERMINUS linkageb | Plasmid clone(s)c | Cosmid clone(s)c | RFLP mapping | Map position inferredd | Gap size (kb)e | M clonef |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 7.247 | 7.247, 7.246 | CW-TT010 | — | IVR | 0 | — | |
| 2 | 3 | — | — | CW-TT012 | — | IR | ? | pCW-TM001 | |
| 3 | 3 | — | — | YSK-A3 | — | VIR | ? | — | |
| 4 | 2 | 7.141 | 7.141 | — | — | VIIR | ? | pCW-TM003 | |
| 5 | 9 | — | 7.74 | JM-TT004 | OR708 | IIIL | ? | MV789 | |
| 6 | 8 | — | 7.77 | CW-TT09 | OR711 | IIR | 0 | MV752 | |
| 7 | 19 | 1.718 | 7.17 | YSK-B8 | OR713 | IIIR | 1.92 | MV762 | |
| 8 | 21 | 7.79 | 7.79 | YSK-A10 | OR712 | IVL | 0 | MV763 | |
| 9 | 12 | — | 7.81 | OR078 | OR709 | IIL | 1.48 | MV756 | |
| 10 | 27 | 1.331 | 7.37 | — | OR703 | VR | 2.13 | — | |
| 11 | 25 | 7.34 | 7.34 | YSK-A9 | OR706 | VIL | 0 | MV791 | |
| 12 | 27 | 7.93 | 7.93 | YSK-A2 | OR726 | IL | 0 | pMV-Tel-IL | |
| 13 | 47 | 7.251 | 7.251 | JM-TT001 | OR710 | VIIL | −0.43g | MV750 | |
| 14 | 5 | 7.162 | 7.162 | CW-TT004 | — | VL | ? | — |
Shows TCs that were already incorporated into at least one of the genome assemblies. The number before the period indicates the assembly version. A dash indicates that the TC was not represented in any assembly.
A dash indicates that the TERMINUS was not able to identify a link to the assembly.
Listed are the IDs of individual clones that represent each TC. A dash indicates that a corresponding clone was not identified.
Map position was inferred on the basis of the physical location of the contig inferred from the genome database (http://www.broad.mit.edu/annotation/genome/neurospora/maps/Index.html).
A zero indicates that the TelContig was already incorporated into the assembly. A question mark denotes remaining gaps.
Clones starting with “p” are plasmids; the remainder are cosmids.
Contigs 7.251 and 7.78 actually overlap.
RFLP mapping of telomere-adjacent sequences to linkage groups:
TERMINUS was unable to link TCs 2 and 3 to the genome assembly. In addition, although TERMINUS successfully placed TC4, TC12, and TC14 adjacent to genomic scaffolds 7.141, 7.93, and 7.162, respectively, the physical and genetic positions of these scaffolds were not known. Therefore, we used RFLP mapping to position each of these sequences. The origins of the RFLP mapping probes and mapping enzymes are listed in supplemental Table 3 and the resulting TelContig segregation data are provided in supplemental Table 4.
Both the TC3 and the TC4 probes yielded a single, major hybridizing fragment in each of several restriction digests of OR DNA. No signals were visible in the M DNA lanes (supplemental Figure 2). Analysis of the segregation of these RFLPs revealed that TC3 is tightly linked to Tel-VIR, while TC4 mapped in a position consistent with Tel-VIIR (supplemental Table 4).
The probe from the telomere-adjacent region of TC2 exhibited weak hybridization to multiple loci in the genomes of both the OR and the M strains, resulting in a background smear in each lane (supplemental Figure 2). However, three enzymes (BamHI, EcoRV, and HindIII) yielded distinct bands in OR DNA, which, again, were not present in the M DNA lanes. Segregation analysis of the two fragments produced by BamHI digestion revealed that they come from two unlinked loci, one of which exhibited complete linkage to markers on the right arm of chromosome I (Tel-IR).
The probe for TC12 hybridized strongly to a single fragment in EcoRV, HindIII, and XbaI digests of OR DNA. Three fainter signals also were detected. Only faint signals were detected in the lanes with M DNA (supplemental Figure 2). The major TC12-hybridizing EcoRV fragment mapped to Tel-IL and the faint signals mapped at unlinked locations.
TC14 contains rDNA sequences, suggesting that it caps the end of the rDNA array, which maps at Tel-VL. To confirm this supposition, a probe adjacent to the telomere in TC14 was used for RFLP analysis. In this case, hybridization signals were detected in both the OR and the M DNA lanes. RFLP segregation analysis confirmed that TC14 maps at Tel-VL.
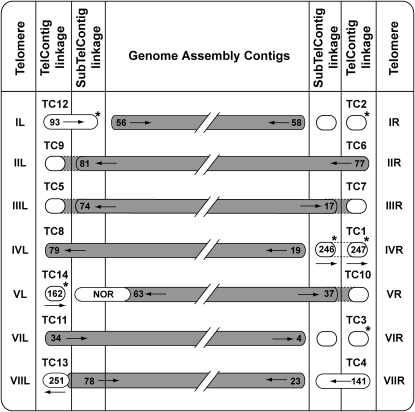
Through the use of TERMINUS and the RFLP mapping efforts described above, we were able to assign chromosomal locations to all 14 TelContigs. This, in turn, resulted in the physical mapping of six genomic contigs whose chromosomal locations previously were unknown. The methods by which each TelContig was linked to the genome assembly are summarized in Figure 2 and Table 1.
Figure 2.—
Relationships between TelContigs and the physical and genetic maps. Capsules containing numbers represent genome assembly contigs and the arrows indicate the sequence orientation. Shaded capsules show contigs whose chromosomal locations are known, while unshaded capsules represent contigs whose genetic or physical map positions were previously unknown. Capsules without numbers depict TelContigs and SubTelContigs that lack overlaps with the genome assembly. Genomic contigs that are shown extending into the TelContig and/or SubTelContig columns exhibited physical overlaps with the respective sequences. A physical overlap between genomic contigs 7.251 and 7.78 is also represented. The NOR spans several sequence contigs, which are not listed due to space limitations. Physical linkages established by TERMINUS are represented by dotted lines, and shading between the dotted lines indicates that we closed the gap by targeted sequencing. Contigs whose chromosomal locations were established or confirmed by RFLP mapping are indicated by asterisks. The outer columns identify the telomeres.
Characterization of plasmid and cosmid clones containing Neurospora chromosome ends:
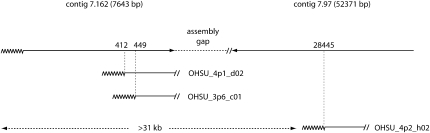
Although TERMINUS identified five telomeres that were not present in any of the genome assemblies and established physical linkages for four of them, sequence gaps still remained between these telomeres and the genome sequence. To close these gaps, we used a 32P-labeled [TTAGGG]n probe to identify telomere-containing clones in “end-enriched” plasmid libraries and in a blunt-ended cosmid library of OR genomic DNA. Telomere-containing plasmid clones were subjected to end sequencing and the resulting reads were assembled. This yielded 17 different telomeric contigs, of which 12 precisely matched TelContigs that had been identified by TERMINUS and 5 corresponded to de novo telomeres that had arisen in the culture. Two of the novel sequences matched TC14 but the location of the telomere repeats in each was different, indicating that these clones contained truncated versions of Tel-VL (Figure 3). The sizes of the HindIII fragments containing these de novo telomeres are predicted to be ∼1.4 kb. Therefore, it seems likely that these ends comigrated with band O8 in the original HindIII/NotI digest (Figure 1A). A third novel sequence also contained rDNA sequences and exhibited a perfect match to contig 7.97, which lies proximal to the telomere-containing contig 7.162 (Figure 3). Consequently, this sequence appears to represent a severely truncated version of Tel-VL. The fourth de novo telomere was attached to a sequence from genomic contig 7.57, which maps in the middle of linkage group II, and the fifth lacked matches to the genome assembly. To determine the origin of the fifth novel telomere, we used the telomere-adjacent sequence as a genetic marker. PCR resulted in differential amplification of the marker in the parental DNAs, with multicent-2a producing a clear PCR product and M yielding a much fainter band. This pattern segregated reliably among the progeny, allowing the marker to be confidently placed at Tel-VIIR (data not shown). Thus, the fifth clone must contain a truncated version of Tel-VIIR.
Figure 3.—
Truncations of Tel-VL. The truncated versions of the rDNA telomere are shown aligned with the intact end. The top line shows the two most terminal genomic contigs with the zig-zag line representing the telomere repeats. The sizes of the contigs are in parenetheses. Truncated telomere clones are drawn below, with dashed lines showing the positions of the truncations relative to the intact chromosome end. Estimated deletion sizes are given. The size of the gap between contigs 7.162 and 7.97 is not known.
Unfortunately, none of the gaps in the assembly were captured in the plasmid telomere library. For this reason, we generated large insert cosmid libraries and screened these for telomere-containing clones. Positive clones were grouped on the basis of their restriction patterns, and representatives were then sequenced using primers flanking each end of the insert. BLAST searches against the genome sequence revealed that four gaps between the TelContigs and the assembly were captured in cosmid clones. Therefore, we performed primer walking using the appropriate cosmid templates to sequence across the missing regions. This resulted in the closure of gaps ranging in size from −400 bp (the analysis showed that contigs 7.251 and 7.78 actually overlap) to 4.7 kb (Table 1).
Nucleotide composition at the Neurospora chromosome termini:
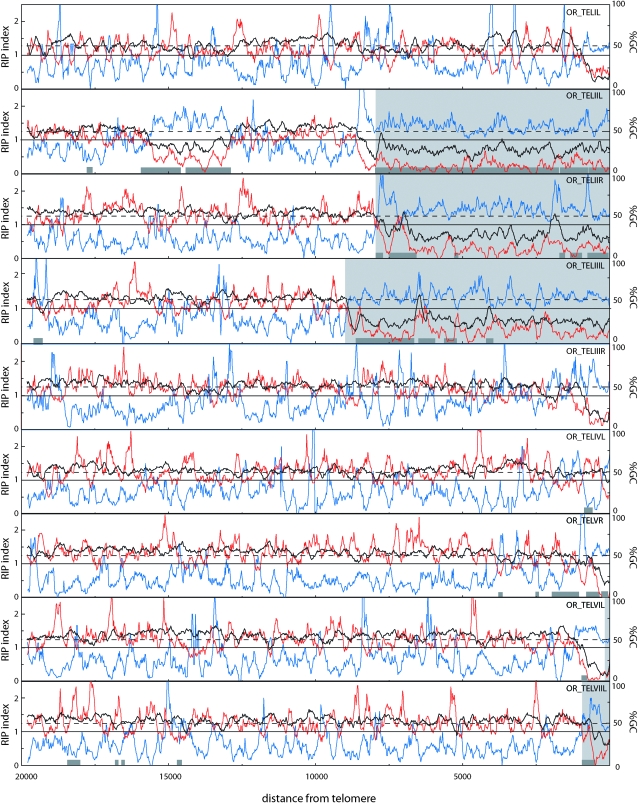
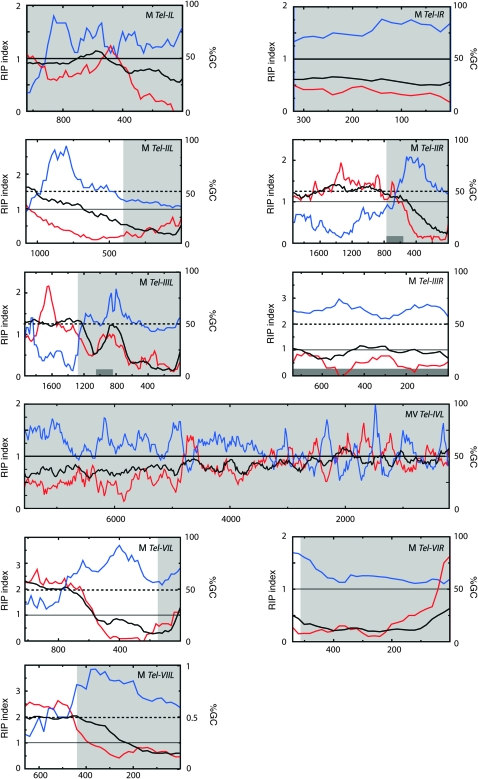
Visual inspection of the TelContig sequences revealed that 12 of them contained extremely AT-rich DNA. In some cases, the AT-rich regions were fully contained within the TelContigs, while in others it extended beyond the contig boundary. Therefore, to determine the extent of the AT-rich sequences and to see if there are additional islands of such DNA in the terminal regions, we used a sliding-window procedure to analyze GC content at the nine chromosome ends for which at least 20 kb of contiguous sequence was available. This revealed that eight of the nine ends had a region with >75% AT within the terminal 1 kb (Figure 4). In most cases, the AT-rich region was restricted to the chromosome tip but we found one telomere with an AT-rich sequence in a proximal region (Tel-IIL, Figure 4). As a test of the significance of the telomeric AT richness, we analyzed the GC content for groups of nine 20-kb sequences sampled randomly from the N. crassa genome. On the basis of 1000 samplings, the average number of 20-kb regions that contained a stretch with GC content <25% was 1.1/group of nine sequences. Furthermore, none of the 1000 groups of nine sequences that were analyzed contained more than five members with <25% GC. Therefore, AT-rich sequences are significantly overrepresented in the telomere-adjacent regions (P ≪ 0.001).
Figure 4.—
Sliding-window analysis of nucleotide composition at OR chromosome ends. The analysis was performed on chromosome ends for which at least 20 kb of sequence was available. GC content (%) and RIP indices I (TpA/ApT) and II (CpA + TpG/ApC + GpT) were calculated in a 200-bp window, which was slid in 20-bp increments across the terminal 20 kb of each chromosome end. The telomere repeats were excluded from the analysis. The black line represents the percentage of GC (%GC), RIP index I is plotted in blue, and RIP index II in red. The x-axis shows the distance from the right edge of the window to the start of the telomere array. The light gray areas highlight sequence regions that are not present at the homologous telomeres of the M strain (see below). Dark gray boxes show the positions of sequences that are closely related to sequences found elsewhere in the genome.
In N. crassa, the presence of AT-rich sequences is often indicative of the action of RIP, a genome defense mechanism that causes G-to-A and C-to-T transition mutations in duplicated sequences (Cambareri et al. 1989). To examine whether the AT-rich sequences at the chromosome termini were caused by RIP, we used a sliding-window analysis to calculate two RIP indices (I and II) across the terminal regions. Regions with a RIP index I >1 and a RIP index II <1 are likely to have been mutated by RIP (Margolin et al. 1998). As shown in Figure 4, both indices suggest that the telomere-associated AT-rich sequences are products of RIP. In addition, there were numerous internal sequences that showed evidence of the action of this mutagenic process. However, nearly all of these regions were short (<400 nucleotides). The only exception was the previously mentioned AT-rich sequence that lies centromere-proximal to Tel-IIL.
Repeated sequences in the terminal regions:
Given that the terminal AT-rich sequences showed hallmarks of RIP, we used BLAST searches to determine if the sequences in question were repeated elsewhere in the Neurospora genome. This resulted in the identification of a number of different repeats (>100 bp) whose copy numbers ranged from 2 to 498 (show as gray boxes in Figure 4). Only two of the repeats showed BLASTX similarity to known transposable elements: one that corresponded to the Pogo element previously identified at Tel-VR (Schechtman 1990) and another that exhibited similarity to the retrotransposons MAGGY (Farman et al. 1996), MGLR-3 (Kang 2001), and Pyret (Nakayashiki et al. 2001) in M. oryzae and Skippy in Fusarium oxysporum (Anaya and Roncero 1995).
RIP normally operates on repeated sequences (Cambareri et al. 1989). Therefore, it was surprising to find a poor correspondence between the locations of AT-rich/RIP-positive DNA and the above-mentioned repeats (Figure 4). In most cases, the AT-rich/RIP-positive DNA either was not associated with a repeat at all or extended well beyond the repeat's boundary. Two examples of this are near Tel-IIR and Tel-IIIL, where there are large tracts of AT-rich/RIP-positive sequences that are single copy. Only one AT-rich/RIP-positive sequence (located at Tel-IIL) coincided perfectly with a repeat.
None of the repeated sequences identified in the above analysis was present in more than one terminal region. This finding suggests that the OR strain lacks a specific subtelomere sequence. In addition to being distributed among several chromosome ends, the subtelomere regions of other organisms are also characterized by the presence of short tandem repeats (Pryde et al. 1997), which could possess important structural and/or functional roles. To determine if the terminal regions of the Neurospora chromosomes retain subtelomere-specific tandem repeats in the absence of a conserved subtelomere sequence, we searched for tandem repeats at the ends of seven chromosomes whose sequences extend out to the telomere repeats. Analysis of 20 kb of terminal sequence from each end identified several tandem repeats (supplemental Table 5). However, the sequences with the largest number of tandem copies were all microsatellites, with repeating units ranging in size from three to six nucleotides. The majority of non-microsatellite, tandem repeats contained fewer than three repeat units. More importantly, however, none of the repeats identified were present at more than one chromosome end, thereby ruling out the existence of conserved, subtelomeric tandem repeat arrays.
Terminally located genes:
Terminal chromosome regions frequently harbor genes involved in niche adaptation. To determine if this is true also for Neurospora, we referred to the N. crassa genome database and retrieved predicted genes that reside within ∼50 kb of the chromosome ends. Some chromosome ends were not analyzed due to uncaptured gaps in the assembly that were also not captured in cosmid clones. Results for 10 chromosome ends are listed in supplemental Table 6.
Inspection of the genes that reside near the Neurospora chromosome ends revealed a number of interesting features. First, as in other fungi, the terminal regions of N. crassa chromosomes were found to harbor genes related to secondary metabolism. Adjacent to Tel-IVL, we identified eight genes belonging to a secondary metabolism gene cluster (SMGC). Listed in the order of their position from the telomere, these encoded a monooyygenase (CYP450), a FAD-binding domain protein, a second CYP450, an O-methyltransferase, a polyketide synthase, a major facilitator superfamily efflux pump, a putative transcription factor, and an oxidoreductase.
Apart from the SMGC, there were no classes of genes that were obviously overrepresented in the terminal chromosome regions, with the possible exception of genes predicted to code for enzymes with plant cell-wall degrading activity. Eight such genes (NCU09904.3, NCU08755.3, NCU08760.3, NCU07134.3, NCU07130.3, NCU09491.3, NCU09518.3, and NCU04997.3) were distributed among 6 of the 10 chromosome ends analyzed. Considering that there are ∼100 glycosyl hydrolases encoded in the 39-Mb genome (Borkovich et al. 2004), the presence of eight genes within just 500 kb of terminal DNA represents a more than fourfold overrepresentation (P < 0.05, Fisher's exact test).
Telomeric restriction fragment variation:
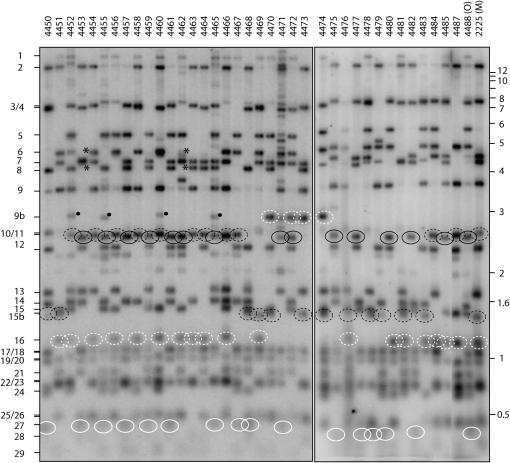
As noted above, the cloning of OR telomeres resulted in the identification of five de novo telomeres: three derived from Tel-VL (the “rDNA end”), one whose telomere-adjacent sequences matched contig 7.57, and another that mapped to Tel-VIIR. Other clear examples of telomere rarrangements were detected while mapping the telomeric RFLPs. Most notably, the M parent appeared to possess two forms of Tel-IL. Examination of progeny isolates 4452–4466 shows that fragment 10 (inherited from M) is the alternate “allele” to fragment 11 (inherited from multicent-2a). However, when looking at progeny 4467–4487, it is clear that fragment 10 is missing and, instead, 15b alternates with 11 (Figure 5). This suggested that the M mapping parent was heterokaryotic and that bands 10 and 15b represent alternate forms of the same telomere. The subsequent cloning and sequencing of these two bands confirmed this notion and revealed that band 10 is a deletion derivative of band 15b (C. Wu, M. S. Sachs and M. L. Farman, unpublished results).
Figure 5.—
Segregation of telomeric RFLPs in the Neurospora mapping cross (Metzenberg et al. 1984). Genomic DNAs were double-digested with HindIII plus NotI and fractionated by electrophoresis through agarose gels. The gels were blotted to nylon membranes and probed with the 32P-labeled telomere repeat sequence. The resulting phosphorimages are shown. Progeny DNA samples are on the left and the parental DNAs are in the two rightmost lanes. Segregating telomere bands are numbered from largest to smallest (left side). Alleles representing Tel-IL are marked by black-outlined ovals: a solid outline for O and a dashed outline for M. An alternate allele for M Tel-IL is labeled as band 15b. Alleles for Tel-VL (the rDNA telomere) are marked by white-outlined ovals: a solid outline for O and a dashed outline for M. An alternate allele for M Tel-VL is labeled as band 9b. Asterisks indicate the loss of both parental markers for Tel-IIIR.
There were additional examples of variation in telomere restriction profiles among progeny from the multicent-2a × M cross [note that multicent-2a has “a nominally ‘Oak Ridge’ genetic background” (Metzenberg et al. 1984) and, therefore, alleles from this parent are referred to as “O”]. Four progeny isolates (4470, 4472, 4473, and 4474) exhibited a novel ∼3-kb telomere fragment that was not observed in either parent (dashed white circles in Figure 5). This fragment appears to represent an alternative M allele for Tel-VL because neither of the parental alleles for this telomere are present in these particular progeny. Weakly hybridizing fragments of a similar size are also visible in progeny 4452, 4455, 4460, and 4465 but Southern hybridization studies with cloned telomere probes revealed that these fragments are partial digestion products of M Tel-IL (data not shown). Progeny isolates 4453 and 4462 inherited neither the O allele nor the M allele for Tel-IIIR (indicated by asterisks in Figure 5). Novel telomere fragments of ∼4.5 kb (comigrating with band 7) and ∼3.9 kb were observed in isolates 4453 and 4462, respectively, and these presumably correspond to differently rearranged versions of Tel-IIIR.
Incomplete digestion of telomere-proximal HindIII sites is also a possible contributor to the variation seen among the mapping progeny. One extreme example of partial cutting was seen at OR Tel-VIR. This telomere has a HindIII site just 133 bp away from the terminal repeat. However, in genomic digests of OR and multicent-2a and in the progeny isolates that inherited the O allele for Tel-VR, this telomere was represented by a fragment that was >20 kb in length (top band in Figure 1A and Figure 5). Thus, the telomere-proximal HindIII site was not cleaved properly. This particular site was not inherently resistant to digestion because amplification of the telomere-adjacent region from genomic DNA yielded a product that was fully cleaved by HindIII (result not shown).
All of the telomere Southern blots that were performed in this study produced numerous weakly hybridizing bands in addition to the strong telomere signals. These weak bands, which sometimes varied between different cultures of the same strain (see supplemental Figure 1) and exhibited varying intensities, are not due to a general problem with restriction digestion as evidenced by a conspicuous absence of “laddering” above the topmost fragments in the blots (see the lane containing DNA from progeny isolate 4471 in Figure 5 for a clear example of a pattern produced by partial digestion). Moreover, reprobing of the same blots with an internal am gene probe produced a single, discrete hybridization signal in all lanes (except for 4471), as did subsequent hybridization experiments in which the same DNA digests were probed with “internal” sequences (results not shown).
Molecular basis for strain-to-strain variation in telomere structure:
Neurospora telomeres are highly polymorphic among strains. For example, the OR and M strains have almost completely different telomeric restriction fragment profiles, as illustrated by the multicent-2a and M parental DNA lanes in Figure 5. Internal loci tend to exhibit much less polymorphism, necessitating use of multiple restriction enzymes to identify RFLPs (Metzenberg et al. 1985). To examine the basis for the high level of sequence variation at the chromosome ends, we cloned telomeres from the M strain in plasmid and cosmid libraries. Four separate telomeres were cloned from the plasmid libraries and another six were cloned in cosmids.
To match each of the M chromosome ends to its homologous counterpart in OR, the clones' end sequences were used to search the TelContigs and the V.7 genome assembly using BLAST. The telomeric ends of one plasmid clone (pJM-TM003, Tel-VIR) and four cosmids (MV750, MV756, MV789, and MV791) exhibited matches to corresponding OR telomeres. Cosmids MV752 (Tel-IIR) and MV763 (Tel-IVL) and plasmid pCW-TM004 (Tel-IL) lacked matches at their telomeric ends. Therefore, primer walking was performed to extend the telomere-proximal sequences to a point at which they could be aligned with their homologous counterparts. This allowed MV752 to be aligned with OR Tel-IIR and pCW-Tm004 with OR Tel-IL. MV763 still lacked a match to the OR genome, even after >10 kb of sequence was obtained.
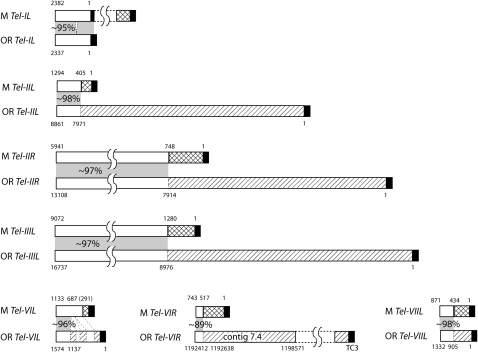
Alignment of homologous telomeres revealed that, in all cases but one, the sequences immediately adjacent to the TTAGGG repeats in one strain are not present at the homologous telomere in the other strain (Figure 6). As an example, an ∼8-kb region next to OR Tel-IIL is replaced by an ∼400-bp sequence at the homologous M telomere (Figure 6, second alignment). The one exception was at Tel-IL, where the M homolog simply has an ∼2.6-kb sequence appended to a short internal telomere repeat array that corresponds to OR Tel-IL (Figure 6, top alignment). One M telomere clone (MV762) had a repetitive element adjacent to the TTAGGG repeats, and this prevented a unique assignment. Nevertheless, the sequence from the opposite end of the insert revealed that this clone contains Tel-IIIR (Table 1). For Tel-IIL and Tel-IIIR, we were able compare ∼20 kb of terminal sequence proximal to the telomere repeats. Although a number of point mutations, small insertion/deletions (indels), and microsatellite expansion/contractions were detected, there was no evidence of any major rearrangements other than at the chromosome tips (results not shown).
Figure 6.—
Alignment of M chromosome ends with their homologous counterparts in OR. Telomere repeats (TTAGGG)n are shown as solid rectangles. Rectangles with diagonal shading represent sequences unique to the OR telomeres, and those with crosshatching are unique to M. The numbers above the M telomeres correspond to the lengths of the novel sequences. The coordinates beneath the OR chromosome ends correspond to the positions within the respective sequence contigs. White rectangles connected by gray shading represent regions of shared terminal sequence and the percentage similarity is noted in the shaded areas. The amount of shared sequence that was compared can be calculated from the coordinates provided.
As was the case with OR, four of the seven M chromosome ends transitioned into highly AT-rich DNA in the telomere-adjacent regions (Figure 7). This led us to question whether the sequence divergence might be the result of recurrent RIP having caused sequences to mutate beyond recognition (by the BLAST algorithm). This possibility was addressed by using ClustalW to search for weak sequence similarities beyond the presumed divergence points. This revealed only one case—involving Tel-VIL—where the homologs could be aligned beyond the initial divergence point detected by BLAST. Interestingly, the new alignments revealed a predominance of the transition mutations that are characteristic of RIP, as well as two large indels (Figure 6). Nevertheless, despite the discovery of extended homology and evidence for RIP, there were still ∼280 bp of sequence adjacent to M-Tel-VIL that were not present in the OR homolog.
Figure 7.—
Sliding-window analysis of nucleotide composition at the chromosome termini of M. Values for %GC (G + C/A + T + G + C), black line; RIP index I (TpA/ApT), blue line; and RIP index II (CpA + TpG / ApC + GpT), red line. These values were calculated across a 200-bp window, which was slid in 20-bp increments across the terminal sequence (excluding the TTAGGG repeats). The x-axis shows the distance from the telomere to the right edge of the window. Horizontal, dashed lines are provided to show the 50% GC mark, while the solid line shows a RIP index value of 1. The light gray area highlights sequence regions that are not present at the homologous telomeres of the OR strain. Dark gray boxes show the positions of repeated sequences.
Calculation of RIP indices for the M chromosome ends revealed a similar pattern to what was observed in OR. Specifically, the telomere-adjacent sequences showed hallmarks of RIP but in most cases the sequences involved were not repetitive (Figure 7). We also examined where the RIP-positive sequences started relative to the points of homolog divergence. Interestingly, these positions tended to be quite close to one another (<1 kb). However, only in the case of M Tel-IIIL was the correspondence perfect. For example, with OR-Tel-IIL, OR-Tel-VIL, M-Tel-IIL, M-Tel-VIL, M-Tel-VIR, and M-Tel-VIIL, the RIP-positive region begins proximal to the point of sequence divergence, while in M-Tel-IIR it starts distally (Figures 4 and 7).
Genome-specific sequences in the telomere-adjacent regions:
As noted above, analysis of the sequences that were immediately adjacent to the M telomeres revealed no matches to the OR assembly. Therefore, we asked whether the reciprocal situation is true. Probes were derived from sequences adjacent to each of the 14 OR telomeres and were used to probe Southern blots of both M and OR DNA. As summarized in Table 2, seven of the telomere-adjacent sequences from OR exhibited no hybridization to M DNA. All of the other probes hybridized to multiple loci in the OR genome and yielded similarly numerous signals in M DNA. In four of these cases, however, the OR lanes yielded a single fragment that gave a much more intense hybridization signal than the others, yet similarly intense signals were absent from the M DNA lanes (supplemental Figure 2).
TABLE 2.
Cross-hybridization between OR telomere-adjacent sequences and M DNA
| TelContig probe | Fragments in OR | Fragments in M | Allele missing in M? |
|---|---|---|---|
| TC1 | 1 | 0 | Yes |
| TC2 | 1–2a | 0a | Yes |
| TC3 | 1 | 0 | Yes |
| TC4 | 1 | 0 | Yes |
| TC5 | 1 | 0 | Yes |
| TC6 | 3–4b | 3c | Yes |
| TC7 | 1 | 0 | Yes |
| TC8 | 2b | 0a | Yes |
| TC9 | >10b | >10d | Yes |
| TC10 | 7b | 6c | Yes |
| TC11 | 6+b | 5+ | Unclear |
| TC12 | 3+b | 1–2c | Yes |
| TC13 | 6 | 6 | Unclear |
| TC14e | 4 | 3 | Unclear |
See supplemental Figure 2 for the Southern blot images.
A smear of background hybridization was observed.
One fragment hybridized much more strongly than the others. This was probably the fragment from which the probe was derived.
Hybridization in M was to weakly hybridizing secondary fragments.
The most strongly hybridizing fragment was clearly not present in M.
rDNA telomere.
DISCUSSION
We present a comprehensive analysis of the organization of chromosome ends in the model filamentous fungus N. crassa. Through a combination of bioinformatic analyses, targeted telomere cloning, and RFLP mapping, we obtained sequences for all 14 of the N. crassa telomeres and assigned them to their respective chromosome ends. For four telomeres, we were able to contribute to the genome-finishing effort by closing gaps between telomeric contigs and the neighboring sequences in the genome assembly. Unfortunately, we were unable to close sequence gaps at another four chromosome ends, despite exhaustive attempts to capture these gaps in plasmid and cosmid clones.
Telomere structure in N. crassa:
On the basis of the analysis of individual sequencing reads, the average length of the (TTAGGG)n telomere tract in N. crassa appears to be ∼120 bp. This is similar to that in M. oryzae (Rehmeyer et al. 2006) but slightly larger than that in A. nidulans (Bhattacharyya and Blackburn 1997). Analysis of the individual sequence reads suggested that the telomere length varies among the different chromosome ends. Most notably, Tel-VL, which caps the ribosomal DNA array, had a significantly shorter tract length (the average is 97 bp). The lengths predicted from the sequence analysis did not match the relative hybridization intensities in Southern blots, but it is difficult to estimate telomere length from hybridization signals, in part because some of the telomere-associated HindIII restriction sites were resistant to digestion and were partially cut. Fragments affected in this manner exhibited weaker-than-expected signals (e.g., band 1 in Figure 5). Another possible explanation for the discrepancies between telomere lengths seen in silico vs. in vivo is that the culture used in the genome project is different from the one used here and, perhaps, telomere lengths can vary between different cultures of the same strain.
Analysis of the telomere-adjacent sequences revealed that the organization of Neurospora chromosome ends is quite different from what has been found in other fungi. For example, S. cerevisiae (Louis and Haber 1992), Ustilago maydis (Sanchez-Alonso and Guzman 1998), Pneumocystis carinii (Keely et al. 2005), Kluyveromyces lactis (Nickles and McEachern 2004), M. oryzae (Gao et al. 2002; Rehmeyer et al. 2006), A. nidulans (Clutterbuck and Farman 2007), Nectria hematococca (M. L. Farman, unpublished results) and Cercospora zeae maydis (L. Dunkle and M. L. Farman, unpublished results) all possess distinct subtelomere regions consisting of sequences that are found at several chromosome ends. By contrast, none of the fully assembled Neurospora chromosome ends have similarity to one another for at least 20 kb from the telomere repeat. Nor were there any matches among the smaller, unassembled, telomere-linked contigs. Consistent with the absence of distinct subtelomeric elements, N. crassa lacks the TLH genes that are present in the subtelomere regions of diverse fungi (Louis and Haber 1992; Sanchez-Alonso and Guzman 1998; Gao et al. 2002; Mandell et al. 2004; Inglis et al. 2005; Rehmeyer et al. 2006). Also missing was any sign of telomere-associated, short tandem repeats. Together, these data indicate that N. crassa lacks a distinct subtelomere domain—or at least one that is defined by specific sequences.
AT-rich sequences in the telomere-adjacent regions and their relationship to RIP:
The N. crassa telomere-adjacent regions are almost universally composed of highly AT-rich DNA, a feature that is common to telomere-adjacent regions of other fungi, including K. lactis (Nickles and McEachern 2004), P. carinii (Keely et al. 2005), and M. oryzae (Rehmeyer et al. 2006). In these organisms, however, the AT-rich sequences are part of a distinct subtelomere domain that is duplicated at multiple chromosome ends. In contrast, the AT-rich sequences adjoining the N. crassa telomeres are quite distinct from one another and appear, instead, to be products of RIP. One possibility is that the terminal sequences are RIP'd relics of once-repeated subtelomere domains that have been rendered unique by the mutagenic process. However, the sequence divergence caused by RIP is limited to ∼20% (Cambareri et al. 1989), which is insufficient for preventing detection of similarity by BLAST. Thus, the distinct products of RIP found near Neurospora telomeres apparently had independent origins.
AT richness does not appear to be essential for Neurospora telomere function and/or for maintenance because Tel-VR has an almost neutral GC content (ranging between 41 and 45%) and yet there was no evidence of meiotic/mitotic instability at this chromosome end (bands 7 and 24 in Figure 5). The same was true for two telomeres (Tel-2 and Tel-12) in M. oryzae (Rehmeyer et al. 2006). Thus, it may be that the nucleotide composition of the telomere-adjacent sequences is important for functions relating to neighboring genes, or it is of no consequence.
Genetic and epigenetic telomere variation:
Although the Neurospora chromosome ends appear to be devoid of most of the structural features that are widely present in the telomere regions of other organisms, they retain the dynamic nature that is characteristic of chromosome ends. The rDNA telomere seems to be highly unstable, as three different truncations of this chromosome end were identified. The dynamic nature of the N. crassa rDNA array is well established. Frequent changes in nucleolar organizer region (NOR) size have been documented in progeny of sexual crosses, with the rearrangements having occurred in the premeiotic phase of the sexual cycle (Butler and Metzenberg 1989, 1990). Subsequently, it was shown that the NOR experiences frequent breakage during vegetative growth of an aneuploid strain containing two rDNA repeat arrays (Butler 1992). Our findings show that the rDNA array is highly prone to alteration even in the normal euploid condition. The rDNA array of M. oryzae also terminates in a telomere and this particular chromosome end also exhibits frequent terminal truncation (C. Rehmeyer, J. Starnes, S. Zhou and M. L. Farman, unpublished results). In both Neurospora and Magnaporthe, most of the rDNA truncations resulted in the loss of very little terminal DNA, which suggests that sequence loss usually arises through gradual attrition of DNA from the chromosome end. Thus, it may be that the protective function of the telomere is compromised by the presence of a neighboring rDNA array. Alternatively, sequences near the rDNA–telomere junction may be unusually prone to endonucleolytic attack.
A rearranged version of OR Tel-IIR consisted of TTAGGG repeats attached to a sequence matching contig 7.57. Although this could have resulted from a truncation of Tel-IIR, the amount of terminal sequence lost would have been >650 kb. Therefore, another type of rearrangement seems more likely. One possibility is that a new telomere formed after a nonreciprocal translocation placed sequences from contig 7.57 near the chromosome end. Alternatively, sequence from contig 7.57 may have been copied to the chromosome terminus following invasion of the internal chromosome region by a degraded end. Corroboratory evidence for such behavior comes from two instances of M. oryzae telomeres capturing (and concomitantly duplicating) internal sequences (C. Rehmeyer, J. Starnes and M. L. Farman, unpublished data).
A new version of OR Tel-VIIR lacked a match to the genome assembly but mapped to the right end of chromosome VII. Therefore, it seems likely that the original telomere was truncated back to sequences that fall in the gap between contigs 7.141 and 7.23 (see Figure 2).
All of the above-described rearrangements were discovered by analyzing telomeres cloned from shotgun libraries. Corresponding changes were not observed in the telomere profiles of the original OR stock, presumably because only a very few nuclei contained each specific telomere variant. As a result, it was not possible to rescue appropriate cultures that would enable us to determine the exact nature of the respective rearrangements.
Changes in telomere regions were also detected in the M strain. The most striking example involved Tel-IL, which segregated as though the M parent possessed two alleles of this telomere. Cloning of the corresponding telomere bands confirmed this supposition and revealed that one of them is a derivative of the other that arose through deletion in a short internal telomere tract (M. L. Farman, C. Wu and M. S. Sachs, unpublished results). Such telomere-mediated chromosome breakage is a well-documented phenomenon in fungi (Surosky and Tye 1985; Kistler and Benny 1992), plants (Yu et al. 2006), and humans (Itzhaki et al. 1992). Another telomere that was rearranged in some progeny isolates, perhaps not surprisingly, was M Tel-VL, the rDNA telomere. On the basis of what is known about the OR strain, it seems likely that these also were truncation events.
Epigenetic modification of chromosomal DNA also may be responsible for some of the restriction fragment variation seen in the Southern blot data. The progeny blot shown in Figure 5 exhibits numerous weak (and non-stoichiometric) hybridization signals that do not segregate in a Mendelian fashion. Even different cultures of the same strain showed variation of the minor hybridizing bands (result not shown). In some cases, these might reflect major rearrangements that occurred in the mycelium during the growth step prior to DNA extraction, but which are not yet fixed in the culture. However, there were also clear instances where such bands resulted from incomplete digestion of the telomere-proximal HindIII site. This partial digestion was not global, and two affected loci that we examined in detail were not inherently resistant to digestion. Therefore, it seems likely that the genomic DNA is modified in a way that makes certain sites resistant to cleavage. Recently, it has been shown that Neurospora telomere regions are subject to methylation (Smith et al. 2008). This could explain the observed partial cutting of the telomere-linked HindIII sites, as this enzyme cuts more slowly if its recognition sequence is hemi-methylated (http://catalog.takara-bio.co.jp/en/product/basic_info.asp?unitid=U100005229), and not at all when methylated completely (Huang et al. 1982). Methylation-related partial digestion could also explain the differences in hybridization intensities among telomeric HindIII fragments. In both the OR and M telomere profiles, there was a notable drop-off in hybridization intensity of fragments < ∼1.55 kb, and the faint hybridization signals occurred at various positions ranging from ∼2 kb upward. Thus, it may be that HindIII sites close to the telomere tend to be methylated and therefore are partially resistant to digestion. This would result in fewer fully cleaved sites and, hence, a variety of larger partial digestion products. In this regard, it is significant that many of the terminal AT-rich, RIP-positive regions extend ∼1.5 kb in from the telomere and heavily RIP'd sequences are frequently methylated (Galagan et al. 2003; Selker et al. 2003).
Telomere-linked genes in Neurospora:
The extraordinary variability and structure of the chromosome ends may have major implications for the genes found in the subtelomeric regions. As in other fungi, the subtelomeric genes in the OR strain of Neurospora have potential significance in niche adaptation. The relative abundance of genes predicted to encode cell-wall-degrading enzymes (CWDEs) near Neurospora telomeres is interesting. The telomere regions of the plant-pathogenic fungus M. oryzae contains only three CWDE genes, despite the fact that families of such genes are expanded in M. oryzae relative to N. crassa (Dean et al. 2005). This may be related to the different lifestyles of the two fungi. Neurospora is a saprophyte and, in nature, is found primarily on burned vegetation where it presumably derives carbon by degrading the walls of dead and dying plant cells. In contrast, Magnaporthe is a facultative biotroph, and a significant portion of its life cycle involves growing inside plant cells without killing them (Kankanala et al. 2007). Here, the destruction of the host's cell wall clearly would be undesirable.
The identification of a SMGC near Neurospora Tel-IVL is also intriguing. All fungal telomere regions that have been examined thus far, including those of Magnaporthe, Aspergillus, Epichloë, Cercospora, and now Neurospora, possess SMCGs within 50 kb of at least one telomere (and often many more) (M. L. Farman, unpublished data). By definition, secondary metabolism is adaptive and, although the reason(s) for SMGC cluster–telomere associations are unclear, it seems likely that there is an adaptive significance to this arrangement. One possibility is that the dynamic nature of the telomeric “neighborhood” provides abundant opportunities for rapid loss or gain of cluster function, either by physical rearrangement of sequences, as seen at M telomeres Tel-IL and Tel-IIR in this study, or through the epigenetic modification of gene expression. In this regard, it is noteworthy that telomere position effects have been demonstrated in N. crassa (Smith et al. 2008) and in A. nidulans (P. Mirabito, personal communication).
Novel sequences in telomere-adjacent regions:
We investigated the basis for the frequent occurrence of telomeric RFLPs between the OR and M strains (Schechtman 1989) and found that this polymorphism is the result of abrupt sequence divergence in the terminal regions, with the homologous chromosomes of OR and M having completely different sequences between these transition points and the terminal TTAGGG repeats. Furthermore, 11 of the sequences adjacent to the OR telomeres and at least 8 of the sequences neighboring M telomeres were absent from the other strain's genome. In some cases, the amount of novel sequence was quite large (≥8 kb), yet most of the regions contained few, if any, predicted genes (results not shown). The origins of these genome-specific, telomere-adjacent sequences are unclear. Most are AT rich and RIP positive, which raises the possibility that they were once repeated sequences but have experienced RIP, followed by further mutation and/or recombination, which has rendered them unique. However, there are two notable exceptions in M Tel-IL and M Tel-IVL. Both of these telomeres have adjacent sequences whose nucleotide compositions are neutral and show little evidence of RIP (Figure 7). Thus, related sequences are clearly absent from the OR genome and therefore must either have been lost in the OR lineage or gained in M. Interestingly, although the novel sequence near M Tel-IVL lacks nucleotide similarity to the OR genome, it exhibited BLASTX matches to two predicted proteins in OR (results not shown). Thus, it seems likely that the sequence has its origin within Neurospora and was not acquired via horizontal transfer.
Conclusion:
New technologies that allow efficient whole-genome studies rely on complete and accurate genome assemblies, but most genome databases lack repeat-rich telomere and centromere regions. We have made a significant advance in completing the genome sequence of N. crassa, an important model organism for genetic and epigenetic studies. Only 7 of the 14 N. crassa telomere ends were reliably captured by the genome sequencing project, and only 2 of these could be placed on the genetic map. We now have sequence data for all 14 ends and have linked 12 to the genome assembly. Telomere sequencing and mapping also led to interesting discoveries about Neurospora biology. Neurospora lacks telomeric tandem repeats comparable to those found in every other organism studied. We propose that this is due to Neurospora's genome defense system, RIP. Like other fungi, Neurospora has an enrichment of genes with putative roles in niche adaptation near its telomeres, and this may be a general characteristic of telomeric regions. Our findings add to our understanding of genome evolution, provide a critical tool for genomic studies, and will aid in the study of telomere function, known to be important in aging and disease.
Acknowledgments
M.L.F. acknowledges technical support from Sherri Schwartz, Brian King, and David Thornbury and thanks Chris Schardl for helpful discussions. We thank James Galagan for writing the perl script to calculate RIP indices and the Fungal Genetics Stock Center for strains. This research was supported by the National Science Foundation (MCB-0653930 to M.L.F.; MCB-0135462 to M.L.F., C.S., and M.S.S.; and MCB-0131383 to E.U.S.); a subcontract to C.S. from the Kentucky Biomedical Research Infrastructure Network (National Center for Research Resources grant 5P20RR016481-03, awarded to Nigel Cooper of the University of Louisville); and a special grant from the U. S. Department of Agriculture (USDA-CREES 2001-34457-10343). K.M.S. was supported by an American Cancer Society postdoctoral fellowship (PF-04-043-01-GMC). H.M.H. was supported by National Institutes of Health P01 grant GM068087. This is Kentucky Agricultural Experiment Station manuscript no. 09-12-1 and is published with the permission of the director.
References
- Abad, J. P., B. De Pablos, K. Osoegawa, P. J. De Jong, A. Martin-Gallardo et al., 2004. Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Mol. Biol. Evol. 21 1620–1624. [DOI] [PubMed] [Google Scholar]
- Anaya, N., and M. I. Roncero, 1995. Skippy, a retrotransposon from the fungal plant pathogen Fusarium oxysporum. Mol. Gen. Genet. 249 637–647. [DOI] [PubMed] [Google Scholar]
- Barry, J. D., M. L. Ginger, P. Burton and R. McCulloch, 2003. Why are parasite contingency genes often associated with telomeres? Int. J. Parasitol. 33 29–45. [DOI] [PubMed] [Google Scholar]
- Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich et al., 2004. The Pfam protein families database. Nucleic Acids Res. 32 138–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, A., and E. H. Blackburn, 1997. Aspergillus nidulans maintains short telomeres throughout development. Nucleic Acids Res. 25 1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner et al., 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68 1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, D. K., 1992. Ribosomal DNA is a site of chromosome breakage in aneuploid strains of Neurospora. Genetics 131 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, D. K., and R. L. Metzenberg, 1989. Premeiotic change of nucleolus organizer size in Neurospora. Genetics 122 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, D. K., and R. L. Metzenberg, 1990. Expansion and contraction of the nucleolus organizer region of Neurospora: changes originate in both proximal and distal segments. Genetics 126 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri, E. B., M. J. Singer and E. U. Selker, 1989. Recurrence of repeat-induced point mutation (RIP) in Neurospora crassa. Genetics 127 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck, J., and M. L. Farman, 2007. Aspergillusnidulans linkage map and genome sequence: closing gaps and adding telomeres, pp. 57–73 in The Aspergilli: Genomics, Medicine, Biotechnology and Research Methods, edited by G. Goldman and S. Osmani. CRC Press, Boca Raton, FL.
- Coleman, M. J., M. T. McHale, J. Arnau and A. Watson, R. P. Oliver, 1993. Cloning and characterization of telomere DNA from Cladosporium fulvum. Gene 132 67–73. [DOI] [PubMed] [Google Scholar]
- Dean, R. A., N. J. Talbot, D. J. Ebbole, M. L. Farman, T. K. Mitchell et al., 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434 980–986. [DOI] [PubMed] [Google Scholar]
- Denayrolles, M., E. P. de Villechenon, A. Lonvaud-Funel and M. Aigle, 1997. Incidence of SUC-RTM telomeric repeated genes in brewing and wild wine strains of Saccharomyces. Curr. Genet. 31 457–461. [DOI] [PubMed] [Google Scholar]
- Ebbole, D. J., and M. S. Sachs, 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37 17–18. [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8 186–194. [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. C. Wendl and P. Green, 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8 175–185. [DOI] [PubMed] [Google Scholar]
- Farman, M. L., and S. A. Leong, 1995. Genetic and physical mapping of telomeres in the rice blast fungus, Magnaporthe grisea. Genetics 140 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farman, M. L., Y. Tosa, N. Nitta and S. A. Leong, 1996. MAGGY, a retrotransposon found in the genome of the rice blast fungus, Magnaporthe grisea. Mol. Gen. Genet. 251 665–674. [DOI] [PubMed] [Google Scholar]
- Feinberg, A. P., and B. Vogelstein, 1984. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132 266–267. [DOI] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422 859–868. [DOI] [PubMed] [Google Scholar]
- Gao, W., C. H. Khang, S.-Y. Park, Y.-H. Lee and S. K. Kang, 2002. Evolution and organization of a highly dynamic, subtelomeric helicase gene family in the rice blast fungus Magnaporthe grisea. Genetics 162 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen, J. P., R. H. Stern and P. C. Wensink, 1979. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 7 2115–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., C. Abajian and P. Green, 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8 195–202. [DOI] [PubMed] [Google Scholar]
- Graham, S. V., and J. D. Barry, 1995. Transcriptional regulation of metacyclic variant surface glycoprotein gene expression during the life cycle of Trypanosoma brucei. Mol. Cell. Biol. 15 5945–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi et al., 1999. Mammalian telomeres end in a large duplex loop. Cell 97 503–514. [DOI] [PubMed] [Google Scholar]
- Henderson, E., C. C. Hardin, S. K. Walk, I. J. Tinoco and E. H. Blackburn, 1987. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell 51 899–908. [DOI] [PubMed] [Google Scholar]
- Hernandez-Rivas, R., D. Mattei, Y. Sterkers, D. S. Peterson, T. E. Wellems et al., 1997. Expressed var genes are found in Plasmodium falciparum subtelomeric regions. Mol. Cell. Biol. 17 604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. H., C. M. Famet, C. K. Ehrlich and M. Ehrlich, 1982. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 10 1579–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium, 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432 695–716. [DOI] [PubMed] [Google Scholar]
- Inglis, P. W., D. J. Rigden, L. V. Mello, E. J. Louis and M. C. Valadares-Inglis, 2005. Monomorphic subtelomeric DNA in the filamentous fungus, Metarhizium anisopliae, contains a RecQ helicase-like gene. Mol. Genet. Genomics 274 79–90. [DOI] [PubMed] [Google Scholar]
- Itzhaki, J. E., M. A. Barnett, A. B. MacCarthy, V. J. Buckle, W. R. Brown et al., 1992. Targeted breakage of a human chromosome mediated by cloned human telomeric DNA. Nat. Genet. 2 283–287. [DOI] [PubMed] [Google Scholar]
- Kang, S., 2001. Organization and distribution pattern of MGLR-3, a novel retrotransposon in the rice blast fungus Magnaporthe grisea. Fungal Genet. Biol. 32 11–19. [DOI] [PubMed] [Google Scholar]
- Kankanala, P., K. Czymmek and B. Valent, 2007. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19 706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely, S. P., H. Renauld, A. E. Wakefield, M. T. Cushion, A. G. Smulian et al., 2005. Gene arrays at Pneumocystis carinii telomeres. Genetics 105 1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler, H. C., and U. Benny, 1992. Autonomously replicating plasmids and chromosome rearrangement during transformation of Nectria haematococca. Gene 117 81–89. [DOI] [PubMed] [Google Scholar]
- Li, W., C. J. Rehmeyer, C. Staben and M. L. Farman, 2005. TERMINUS: telomeric end-read mining in unassembled sequences. Bioinformatics 21 1695–1698. [DOI] [PubMed] [Google Scholar]
- Louis, E. J., and J. E. Haber, 1992. The structure and evolution of subtelomeric Y' repeats in Saccharomyces cerevisiae. Genetics 131 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Z., M. Freitag and M. S. Sachs, 1995. Translational regulation in response to changes in amino acid availability in Neurospora crassa. Mol. Cell. Biol. 15 5235–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari, R., 1999. Microconidia of Neurospora crassa. Fungal Genet. Biol. 26 1–18. [DOI] [PubMed] [Google Scholar]
- Mandell, J. G., K. J. Goodrich, J. Bahler and T. R. Cech, 2004. Expression of a RecQ helicase homolog affects progression through crisis in fission yeast lacking telomerase. J. Biol. Chem. 280 5249–5257. [DOI] [PubMed] [Google Scholar]
- Margolin, B. S., P. W. Garrett-Engele, J. N. Stevens, D. Y. Fritz, C. Garrett-Engele et al., 1998. A methylated Neurospora 5s rRNA pseudogene contains a transposable element inactivated by repeat-induced point mutation. Genetics 149 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg, R. L., and J. Grotelueschen, 1995. Restriction polymorphism maps of Neurospora crassa: update. Fungal Genet. Newsl. 42 82–90. [Google Scholar]
- Metzenberg, R. L., J. N. Stevens, E. U. Selker and E. Morzycka-Wrobleska, 1984. A method for finding the genetic map position of cloned DNA fragments. Neurospora Newsl. 31 35–39. [Google Scholar]
- Metzenberg, R. L., J. N. Stevens, E. U. Selker and E. Morzycka-Wrobleska, 1985. Identification and chromosomal distribution of 5S rRNA genes in Neurospora crassa. Proc. Natl. Acad. Sci. USA 82 2067–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyne, J., R. L. Ratliff and R. K. Moyzis, 1989. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl. Acad. Sci. USA 86 7049–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan, J. L., G. A. Cross, T. de Lange and J. D. Griffith, 2001. T-loops at trypanosome telomeres. EMBO J. 20 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murti, K. G., and D. M. Prescott, 2002. Topological organization of DNA molecules in the macronucleus of hypotrichous ciliated protozoa. Chromosome Res. 10 165–173. [DOI] [PubMed] [Google Scholar]
- Nakayashiki, H., H. Matsuo, I. Chuma, K. Ikeda, S. Betsuyaku et al., 2001. Pyret, a Ty3/Gypsy retrotransposon in Magnaporthe grisea contains an extra domain between the nucleocapsid and protease domains. Nucleic Acids Res. 29 4106–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, M. A., and D. D. Perkins, 2000. Restriction polymorphism maps of Neurospora crassa: 2000 update. Fungal Genet. Newsl. 47 25–39. [Google Scholar]
- Nickles, K., and M. J. McEachern, 2004. Characterization of Kluyveromyces lactis subtelomeric sequences including a distal element with strong purine/pyrimidine strand bias. Yeast 21 813–830. [DOI] [PubMed] [Google Scholar]
- Oakley, C. E., C. F. Weil, P. L. Kretz and B. R. Oakley, 1987. Cloning of the riboB locus of Aspergillus nidulans. Gene 53 293–298. [DOI] [PubMed] [Google Scholar]
- Pryde, F. E., H. C. Gorham and E. J. Louis, 1997. Chromosome ends: All the same under their caps? Curr. Opin. Genet. Dev. 7 822–828. [DOI] [PubMed] [Google Scholar]
- Rachidi, N., M. J. Martinez, P. Barre and B. Blondin, 2000. Saccharomyces cerevisiae PAU genes are induced by anaerobiosis. Mol. Microbiol. 35 1421–1430. [DOI] [PubMed] [Google Scholar]
- Rehmeyer, C., W. Li, M. Kusaba, Y.-S. Kim, D. Brown et al., 2006. Organization of chromosome ends in the rice blast fungus Magnaporthe oryzae. Nucleic Acids Res. 34 4685–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, E. J., and F. M. Ausubel, 1988. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53 127–136. [DOI] [PubMed] [Google Scholar]
- Rountree, M. R., and E. U. Selker, 1997. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 11 2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko, G., R. McCulloch, A. Dirks-Mulder and P. Borst, 1996. Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Mol. Biochem. Parasitol. 80 65–75. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanchez-Alonso, P., and P. Guzman, 1998. Organization of chromosome ends in Ustilago maydis: RecQ-like helicase motifs at telomeric regions. Genetics 148 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechtman, M. G., 1987. Isolation of telomere DNA from Neurospora crassa. Mol. Cell. Biol. 7 3168–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechtman, M. G., 1989. Segregation patterns of Neurospora chromosome ends: mapping chromosome tips. Fungal Genet. Newsl. 36 71–73. [Google Scholar]
- Schechtman, M. G., 1990. Characterization of telomere DNA from Neurospora crassa. Gene 88 159–165. [DOI] [PubMed] [Google Scholar]
- Scherf, A., R. Hernandez-Rivas, P. Buffet, E. Bottius, C. Benatar et al., 1998. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 17 5418–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker, E. U., 1990. Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet. 24 579–613. [DOI] [PubMed] [Google Scholar]
- Selker, E. U., N. A. Tountas, S. H. Cross, B. S. Margolin, J. G. Murphy et al., 2003. The methylated component of the Neurospora crassa genome. Nature 422 893–897. [DOI] [PubMed] [Google Scholar]
- Smith, K. M., G. O. Kothe, C. B. Matsen, T. K. Khlafallah, K. K. Adhvaryu et al., 2008. The fungus Neurospora crassa displays telomeric silencing mediated by multiple sirtuins and by methylation of histone H3 lysine 9. Epigenetics Chromatin 1 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkin, S. M., and J. R. Stringer, 1996. Translocation of surface antigen genes to a unique telomeric expression site in Pneumoscystis carinii. Mol. Microbiol. 19 283–295. [DOI] [PubMed] [Google Scholar]
- Surosky, R. T., and B.-K. Tye, 1985. Construction of telocentric chromosomes in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 82 2106–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask, B. J., C. Friedman, A. Martin-Gallardo, L. Rowen, C. Akinbami et al., 1998. Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum. Mol. Genet. 7 13–26. [DOI] [PubMed] [Google Scholar]
- Verstrepen, K. J., A. Jansen, F. Lewitter and G. R. Fink, 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, M., and Y. Nakamura, 1996. Antigenic variation by telomeric recombination of major-surface-glycoprotein genes of Pneumocystis carinii. J. Eukaryot. Microbiol. 43 8S. [DOI] [PubMed] [Google Scholar]
- Walmsley, R. W., C. S. Chan, B. K. Tye and T. D. Petes, 1984. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature 310 157–160. [DOI] [PubMed] [Google Scholar]
- Yu, W., J. C. Lamb, F. Han and J. A. Birchler, 2006. Telomere-mediated chromosomal truncation in maize. Proc. Natl. Acad. Sci. USA 103 17331–17336. [DOI] [PMC free article] [PubMed] [Google Scholar]