Abstract
Pma1-10 is a mutant plasma membrane ATPase defective at the restrictive temperature in stability at the cell surface. At 37°, Pma1-10 is ubiquitinated and internalized from the plasma membrane for degradation in the vacuole. YVH1, encoding a tyrosine phosphatase, is a mutant suppressor of pma1-10; in the absence of Yvh1, Pma1-10 remains stable at the plasma membrane, thereby permitting cells to grow. The RING finger domain of Yvh1, but not its phosphatase domain, is required for removal of mutant Pma1-10 from the plasma membrane. Yvh1 is a novel ribosome assembly factor: in yvh1Δ cells, free 60S and 80S ribosomal subunits are decreased, free 40S subunits are increased, and half-mer polysomes are accumulated. Pma1-10 is also stabilized by deletion of 60S ribosomal proteins Rpl19a and Rpl35a. We propose that changes in ribosome biogenesis caused by loss of Yvh1 or specific ribosomal proteins have effects on the plasma membrane, perhaps by producing specific translational changes.
THE ribosome is a macromolecular assembly of four ribosomal RNAs and >80 different proteins. In eukaryotes, the ribosome is composed of a small 40S subunit and a large 60S subunit that perform the two critical activities of decoding messenger RNA and forming peptide bonds during translation. Ribosomes appear to play an even larger role than simply linking amino acids according to the genetic code. Ribosomes may influence post-translational events because ribosome-associated chaperones affect the folding of nascent polypeptides (Ito 2005; Albanese et al. 2006). Moreover, recent evidence supports a model in which differences in ribosomal protein and RNA composition and modification of ribosomal protein and RNA produce heterogeneous ribosomes, differentially affecting translation of specific mRNAs (Mauro and Edelman 2002; Komili et al. 2007). In eukaryotic cells, the elaborate process of ribosome production begins in the nucleolus, continues in the nucleoplasm and cytoplasm, and is overseen by a growing list of up to 200 factors (Warner 1989; Henras et al. 2008).
We are interested in post-translational sorting mechanisms in the secretory pathway in Saccharomyces cerevisiae. These mechanisms are reflected in the disparate behavior of several different mutants of the polytopic membrane protein, Pma1. Many conformationally defective Pma1 mutants are recognized by an endoplasmic reticulum (ER) quality control mechanism shortly after synthesis and sent for ER-associated degradation (ERAD) (Nakamoto et al. 1998; Han et al. 2007). By contrast, other misfolded Pma1 mutants escape detection by ER quality control and are sent via vesicular transport into the secretory pathway (Pizzirusso and Chang 2004). One such mutant, Pma1-10, is delivered properly to the plasma membrane but fails to remain stable at the cell surface like wild-type Pma1. Instead, Pma1-10 is ubiquitinated and undergoes rapid endocytosis from the plasma membrane followed by vacuolar degradation (Gong and Chang 2001). Because Pma1 activity at the cell surface is essential for growth (Serrano et al. 1986), Pma1-10 is a temperature-sensitive mutant.
After detailed analysis of the behavior of Pma1-10 (Gong and Chang 2001), we reasoned that genetic selection for mutant suppressors of pma1-10 cells should reveal factors involved in recognition of misfolded protein at the cell surface. Indeed, loss of function in Yvh1, a tyrosine phosphatase, allows Pma1-10 to escape ubiquitination and remain stable at the plasma membrane (Liu and Chang 2006). Here, we report the unexpected finding that Yvh1 is a novel ribosome assembly factor. We suggest an effect of Yvh1 on ribosome composition and/or function may affect protein translation, leading to profound consequences at the plasma membrane.
MATERIALS AND METHODS
Yeast strains and media:
KKX103-2B and -2D are pma1-10 yvh1 strains derived from the original suppressor of pma1-10 isolated following insertional mutagenesis, using a library generated by random insertions of LEU2 and lacZ; KKX15-1B (MATa yvh1∷LEU2) was generated by backcross of KKX103-2D to L3852 (MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2). KKY55, -56, and -58 are yvh1 (KKX15-1B) cells with pKK78, pKK21, and pKK87, respectively, integrated at the ura3 locus. rpl19a∷LEU2 isolated as a suppressor of pma1-10 was backcrossed with L3852 to generate KKX28. Most deletion strains used in this study are from the systematic deletion project in the BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) or BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) background; these strains include rpl19aΔ, rpl35aΔ, rpl24aΔ, rps25aΔ, slx9Δ, and yvh1Δ. The deletion strains were confirmed by PCR.
Molecular biology:
pKK5 and pKK7 are URA3-marked (pRS316) and LEU2-marked (pRS315) centromeric plasmids (pRS316) bearing MET25-HA-pma1-10, respectively, as described (Liu and Chang 2006). pKK26 is a HIS3-marked centromeric plasmid (pRS313) bearing MET25-HA-pma1-10. pKK42 is a LEU2-marked centromeric plasmid (pRS315) bearing MET3-HA-pma1-10, described in Liu and Chang (2006). pAB7 and pAB100 with HA-tagged YVH1 expressed from its own promoter in a centromeric plasmid and a YEp24 plasmid, respectively (Beeser and Cooper 1999), are gifts from T. Cooper (University of Tennessee). pBS-yvh1C117S is a gift from S. Harashima (Okasa University, Japan). pKK21 is a URA3-marked YIp (pRS306) bearing HA-tagged YVH1; it was constructed by placing the 2.2-kb XbaI–XbaI fragment from pAB7 into pRS316. pKK27 is a URA3-marked 2μ plasmid bearing HA-yvh1-C117S; it was constructed by using SacI–KpnI to move the insert from pBS-yvh1C117S to pRS202. pKK78 is a URA3-marked YIp (pRS306) bearing an HA-tagged yvh1 C-terminal fragment from residues 204–364; it was constructed by using oligos 385 (CATAAGAGCGTAGTCTGGGACG) and 386 (TCTGGAATGTTTAAAGATTCGGAG) to amplify the Yvh1 C-terminal fragment and vector backbone from pAB7, and the XbaI–XbaI insert was then moved to pRS316. pKK87 is a URA3-marked YIp (pRS306) bearing HA-tagged yvh1-C232A; it was constructed by using oligos 357 (CACCGCAGTGAGAGCTAAGAAGTGCAG) and 358 (CTGCACTTCTTAGCTCTCACTGCGGTG) to introduce the C232A mutation into pAB7 and then the 2.2-kb XbaI–XbaI insert was placed into pRS316. pKK103 is a URA3-marked YIp (pRS306) bearing pma1-4D2E/A changes, as described in Liu et al. (2006). pKK109 is a LEU2-marked centromeric plasmid bearing pma1-4D2E/A changes under the control of its native promoter, as described in Liu et al. (2006). URA3 YIp plasmids were integrated at the chromosomal ura3 locus after cutting with NcoI. pRPL25-GFP is a URA3-marked centromeric plasmid, described in Gadal et al. (2001) (gift from Ed Hurt, Biochemie-Zentrum, Heidelberg, Germany).
Pulse chase, protein induction, and Western blot:
Conditions to depress HA-Pma1-10 or HA-Pma1-4D2E using MET25 or MET3 promoters are identical. In some strains, repression of the MET3 promoter is superior to that of the MET25 promoter; however, in this study, there is no difference between the two promoters. Cells were grown to exponential phase in minimal medium with 600 μm methionine. Cells were derepressed by washing with water and resuspending in fresh methionine-free medium for 2 hr. Cells were chased by adding 2 mm methionine. To examine Pma1-10 stability, cells were grown at 25° before derepression and then shifted to 37°. To examine Pma1-4D2E stability, cells were shifted to 30° after derepression.
For Western blot, the cell lysate was prepared by vortexing cells with glass beads as described previously (Chang and Slayman 1991). Samples were normalized to protein content measured by Bradford assay. For blotting with anti-ubiquitin antibody (Zymed Laboratories), nitrocellulose filters were autoclaved as described (Swerdlow et al. 1986).
Fluorescence microscopy:
Cells expressing GFP-tagged Rpl25 were fixed with ethanol for 30 sec, washed with distilled H2O, and stained with DAPI at 1 μg/ml for 10 min, as described in Honma et al. (2006). Cells were visualized with an Olympus fluorescence microscope and images were collected with a Hamamatsu Orca CCD camera.
Polyribosome fractionation:
Ribosomal subunits, 80S monosomes, and polyribosomes were separated on sucrose gradients as described in Fuentes et al. (2007). The cell lysate was prepared and clarified by centrifugation at 10,200 × g for 10 min, and the A260 was measured. Samples were normalized to A260 of clarified lysate for loading onto gradients (15 OD260). After centrifugation of gradients at 96,800 × g for 4 hr at 4°, fractions (600 μl) were collected from 7–47% sucrose gradients and the A254 was continuously monitored using a UA-5 UV/VIS detector (ISCO). After protein precipitation in 10% trichloroacetic acid, fractions were washed and subjected to SDS–PAGE and Western blot. Anti-HA antibody (Covance) and anti-L3 antibody (gift from Jon Warner, Albert Einstein College of Medicine) were used sequentially to detect HA-tagged Yvh1 and the 60S ribosomal protein L3, respectively.
RESULTS
yvh1 is a suppressor of pma1-10:
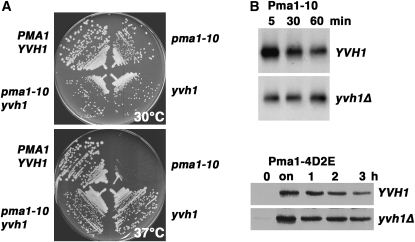
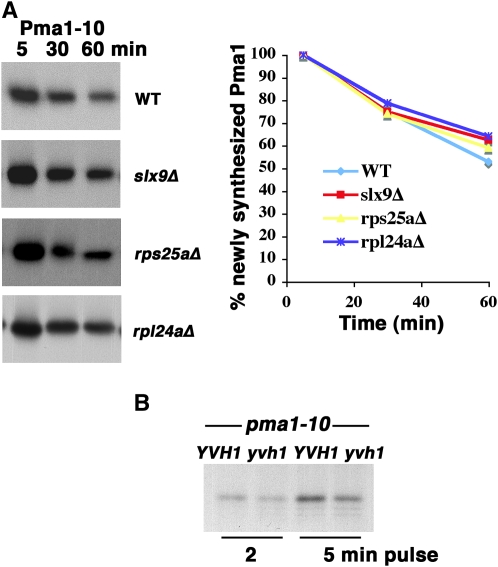
Previously, we characterized Pma1-10, a misfolded membrane protein that is properly targeted to the cell surface, but undergoes ubiquitination and rapid endocytosis for vacuolar delivery and degradation (Gong and Chang 2001; Liu and Chang 2006). A genetic screen for extragenic suppressors of temperature-sensitive pma1-10 cells after insertional mutagenesis led to identification of yvh1. At 37°, pma1-10 yvh1Δ cells grow but pma1-10 cannot (Figure 1A and Liu and Chang 2006). Using HA-tagged Pma1-10 expressed under the control of a methionine-regulated promoter, pulse-chase experiments show that Pma1-10 is stabilized at 37° in yvh1Δ cells compared with YVH1+ cells (Figure 1B, top, and Liu and Chang 2006). These findings suggest that Pma1-10 stabilization in the absence of Yvh1 is the basis for suppression of defective growth at 37°. Previous work has shown that Pma1-10 is stabilized at the plasma membrane in yvh1Δ cells (Liu and Chang 2006). A Pma1 mutant, called 4D2E/A, behaves like Pma1-10: the 4D2E/A mutant is properly targeted to the plasma membrane but undergoes rapid internalization for vacuolar degradation (Liu et al. 2006). [The 4D2E/A mutant has changes in four aspartate (D39–42) and two glutamate (E48–49) residues to alanine.] The 4D2E/A mutant is also stabilized by yvh1Δ cells (Figure 1B, bottom).
Figure 1.—
yvh1Δ is a suppressor of pma1-10 and pma1-4D2E cells. (A) Suppression of temperature-sensitive growth of pma1-10 by yvh1Δ. Wild-type (L3852), pma1-10 yvh1Δ (KKX103-2D), pma1-10 (XGY32), and yvh1 (KKX15-1D) cells were struck out on plates with synthetic complete medium and incubated at 30° and 37°. (B) Mutant Pma1 is stabilized in yvh1Δ cells. YVH1+ (L3852) and yvh1Δ (KKX15-1B) cells bearing pMET25-HA-pma1-10 (pKK5) or pMET3-HA-pma1-4D2E (pKK103) were grown to midlog in the presence of methionine (0) and then shifted to methionine-free medium to derepress mutant Pma1 synthesis (on). (Top panels) Cells were pulse labeled with Expre35S35S for 10 min and chased for various times. HA-Pma1-10 was immunoprecipitated from lysates normalized to acid-precipitable counts per minute and analyzed by SDS–PAGE and fluorography. (Bottom panels) Cell lysate was prepared at various times after methionine (2 mm) was added to repress synthesis of HA-Pma1-4D2E. Western blot with anti-HA antibody was normalized to protein.
The RING domain plays a critical role in Yvh1 function:
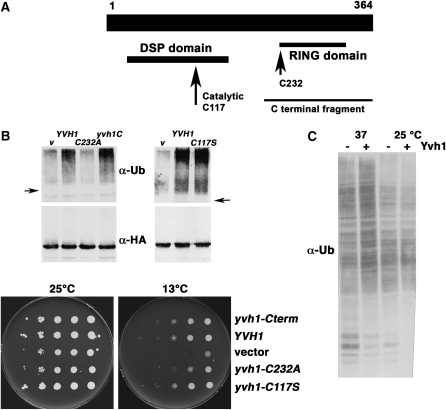
Because Yvh1 is a tyrosine phosphatase with a RING variant domain at its carboxy terminus (Guan et al. 1992; Muda et al. 1999), the role of catalytic phosphatase activity or the RING-like motif was tested. Figure 2A shows the location of the critical catalytic cysteine residue (C117) within the dual specificity phosphatase domain at the N terminus and the RING-like motif at the carboxy terminus of Yvh1. The effect of Yvh1 mutants was tested by assaying ubiquitination of Pma1-10, which disappears in the absence of Yvh1. HA-tagged Pma1-10 was immunoprecipitated from yvh1Δ cells followed by Western blot with anti-ubiquitin. Figure 2B (top) shows a ladder of high-molecular-weight bands extending above the position of nonubiquitinated Pma1-10 in yvh1Δ cells complemented with wild-type YVH1. Like wild-type YVH1, a catalytically inactive C117S Yvh1 mutant (Beeser and Cooper 2000) is able to complement yvh1Δ cells to recover Pma1-10 ubiquitination, suggesting phosphatase activity does not contribute to Pma1-10 ubiquitination. Strikingly, a truncation mutant containing the RING-like domain (yvh1C) complements yvh1Δ cells as well as wild-type YVH1 (Figure 2B); thus, the RING-like domain has sufficient Yvh1 function to result in Pma1-10 ubiquitination. Consistently, mutation of the first cysteine (C232A) of the seven invariant cysteines and one histidine that compose the RING-like domain results in complete loss of function of the otherwise intact Yvh1 protein (Figure 2B).
Figure 2.—
Domain analysis of Yvh1. (A) Yvh1 map showing positions of dual specificity phosphatase (DSP) domain and catalytic cysteine residue 117 (long arrow) and the C-terminal fragment from residue 205–364 with complementing activity. Cysteine 232 (short arrow) is the first of seven invariant cysteines and 1 histidine within a RING-like domain extending to cysteine 326. (B) Ubiquitination of Pma1-10 is impaired in yvh1Δ cells. (Top left panel) yvh1Δ cells with vector (KKY57) or with yvh1-C terminus (KKY 55), wild-type YVH1 (KKY56), or yvh1-C232A (KKY58) and pMET25-HA-pma1-10 (pKK26) were induced to express mutant Pma1 for 2 hr at 37°. (Top right panel) yvh1Δ cells with vector (pRS202), YVH1 (PAB100), or yvh1-C117S (pKK27) and pMET25-HA-pma1-10 (pKK26). HA-Pma1-10 was immunoprecipitated from cell lysate and analyzed by Western blotting with anti-ubiquitin. The nitrocellulose filter was then stripped and reprobed with anti-HA antibody. (Bottom panels) Functional domain of Yvh1 measured by complementation of yvh1Δ cells. Cells were diluted on the basis of optical density, spotted on synthetic complete medium, and incubated at 25° and 13°. (C) Ubiquitination in yvh1Δ cells. Total membranes were isolated by centrifugation of lysate from wild-type and yvh1Δ cells at 100,000 × g for 1 hr. The profile of ubiquitinated proteins was analyzed by Western blot with anti-ubiquitin.
yvh1Δ cells have slightly impaired growth at 30° compared with YVH1+ cells and display cold-sensitive growth as well (Figure 2B, bottom). Domains of Yvh1 responsible for defective growth at 13° were tested. Figure 2B (bottom) shows the Yvh1 RING-like domain is sufficient to complement cold-sensitive growth of yvh1Δ cells. Mutation of the first cysteine residue (C232A) within the RING-like domain of full-length Yvh1 is sufficient to cause cold-sensitive growth (Figure 2B, bottom). These results suggest an essential role for the RING domain and are in agreement with a report that the RING-like domain, but not phosphatase activity, is required for Yvh1 function in sporulation (Beeser and Cooper 2000).
Mutants in ribosomal proteins are suppressors of two pma1 mutants:
A simple explanation for loss of Pma1-10 ubiquitination in the absence of Yvh1 is that Yvh1 functions as a RING domain ubiquitin ligase. To test this hypothesis, a total membrane fraction containing ribosomes was isolated from Yvh1+ and Yvh1− cells by centrifugation of cell lysate at 100,000 × g for 1 hr, and the content of ubiquitinated proteins was analyzed by Western blot with anti-ubiquitin. Figure 2C shows no significant difference in the profile of ubiquitinated proteins in wild-type and yvh1Δ cells. In vitro ubiquitination assays were performed to test this hypothesis; however, no detectable ubiquitination activity was associated with Yvh1 (R. Gardner, University of Washington, unpublished results).
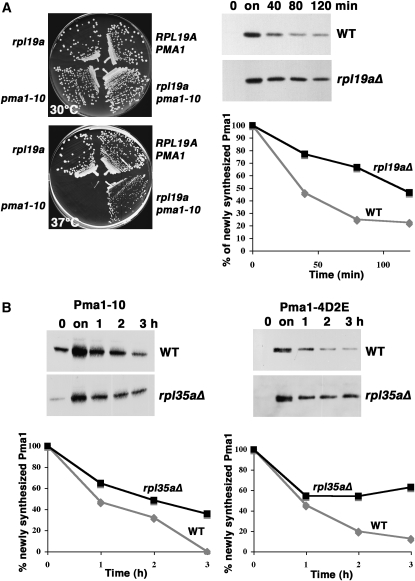
Analysis of other suppressors of pma1-10 led to another proposed function for Yvh1. Genetic screening for suppressors of temperature-sensitive pma1-10 resulted in isolation of mutants in two ribosomal proteins of the large ribosomal subunit, Rpl19a and Rpl35a. Figure 3A (left) shows temperature-sensitive growth of pma1-10 cells is rescued by deletion of RPL19A. A pulse-chase experiment (Figure 3A, right) shows degradation of newly synthesized Pma1-10 is slowed in rpl19aΔ cells. Although an rpl35a mutant generated by insertional mutagenesis was isolated as a suppressor of impaired growth of pma1-10, deletion of RPL35A itself has a growth defect (not shown). However, pulse-chase experiments in Figure 3B show that loss of Rpl35a results in stabilization of both Pma1-10 (Figure 3B, left) and Pma1-4D2E (Figure 3B, right).
Figure 3.—
Stabilization of mutant Pma1 in the absence of ribosomal proteins Rpl19a and Rpl35a. (A) (Left panels) Suppression of temperature-sensitive growth of pma1-10 by rpl19aΔ. Strains were struck out on plates with synthetic complete medium at 30° and 37°. (Right panels) Pma1-10 stabilization. RPL19A+ (BY4741) and rpl19aΔ cells bearing pMET3-HA-pma1-10 (pKK42) were grown to midlog phase in the presence of methionine (0) and then shifted to methionine-free medium to derepress mutant Pma1 synthesis (on). Methionine was then added to repress synthesis, and cells were collected at various times of chase. Cell lysate was analyzed by Western blot with anti-HA antibody. Films were scanned and quantitated using NIH Image. (B) Mutant Pma1 stabilization in rpl35aΔ cells. HA-tagged mutant Pma1-10 (left panel) or Pma1-4D2E (right panel) were derepressed as described in A in wild-type (BY4741) and rpl35aΔ cells bearing pMET25-HA-pma1-10 (pKK7) or pMET3-HA-pma1-4D2E (pKK103). Cells were “chased” by addition of methionine. Lysate was analyzed by Western blot with anti-HA antibody. Films were scanned and quantitated using NIH Image.
The pma1-10 allele has two nucleotide changes resulting in A165G and V197I lesions (Gong and Chang 2001), both of which are necessary to cause temperature sensitivity (Y. Liu, unpublished results). To address the possibility that loss of ribosomal proteins suppresses pma1-10 by affecting translational fidelity, suppression of other pma1 mutants by rpl19aΔ and rpl35aΔ was examined. Figure 3B (right) shows the 4D2E/A mutant (with six residues changed to alanine) is stabilized by rpl35aΔ cells. Because mutants with 3D/A and 2D2E/A changes do not display the phenotype of 4D2E/A (Y. Liu, unpublished results), at least two changes to 4D2E/A would be necessary to generate plasma membrane stability. The 4D2E/A mutant is also stabilized by loss of Yvh1 (Figure 1B, bottom).
60S ribosome assembly is defective in yvh1 cells:
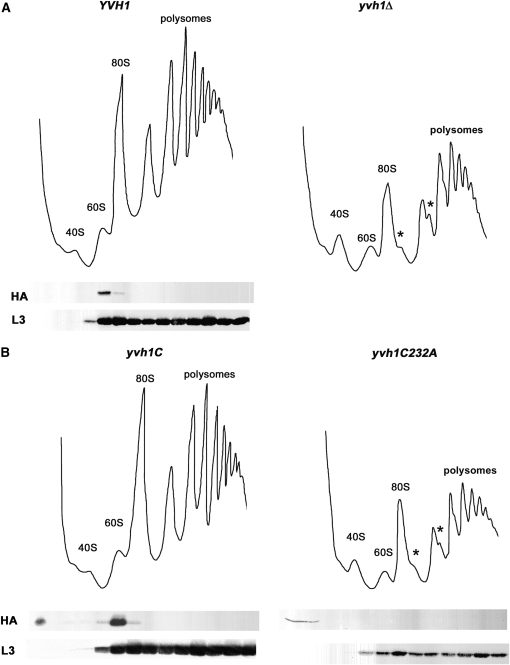
Because loss of Rpl19a and Rpl35a can cause suppression of pma1-10, we considered the possibility that Yvh1 is involved in ribosome assembly. Previously, Yvh1 was found associated with purified 60S preribosomal particles (Nissan et al. 2002) and was also reported to interact by two-hybrid assay with the essential ribosome assembly factor Nop7 (Sakumoto et al. 2001). To test whether Yvh1 plays a role in ribosome assembly, the polysome profile of yvh1Δ cells was analyzed by sucrose density gradients. By contrast with the polysome profile of YVH1+ cells, free 60S and 80S ribosomal subunits are decreased, and free 40S subunits are increased in yvh1Δ cells (Figure 4A). In addition, in yvh1Δ cells, there is accumulation of half-mer polysomes, apparent as shoulders on polysome peaks (Figure 4A, asterisks); these likely reflect initiating 40S subunits stalled on mRNAs due to either a limitation in mature functional 60S subunits or defective subunit joining (Rotenberg et al. 1988). Consistent with its role in 60S ribosomal subunit assembly, HA-tagged Yvh1 cofractionate with 60S subunits, as detected by Western blot across sucrose density gradients; localization is exclusively at 60S subunits as no HA-Yvh1 was detected sedimenting with polysomes (Figure 4A). Ribosomal protein L3 is shown as a marker for 60S and 80S ribosomes and polysomes (Figure 4, A and B, bottom panels).
Figure 4.—
Yvh1 is associated with pre-60S ribosomes. (A and B) Ribosome fractionation. Cell lysate was prepared by vortexing with glass beads from the following strains: wild-type (KKY56) and yvh1Δ (KKY57) cells (A) or cells with yvh1-C terminus (KKY55) and full-length yvh1-C232A (KKY58) (B). Ribosomes were separated on 7–47% sucrose gradients and analyzed as described in materials and methods. Polysomes and 40S, 60S, and 80S ribosomes are indicated; asterisks indicate the positions of half-mer polysomes. Western blots show positions of HA-Yvh1 and the ribosomal protein L3 in gradients (bottom panels).
The C-terminal RING-like domain of Yvh1 (yvh1C) plays a critical role in 60S ribosomal subunit assembly because it is sufficient to complement the defective ribosome fractionation profile of yvh1Δ cells (Figure 4B). By contrast, full-length Yvh1 with a single point mutation C232A in the RING variant domain cannot complement 60S assembly defects of yvh1Δ cells (Figure 4B). Moreover, Yvh1-C232A is no longer associated with 60S ribosomes (Figure 4B, bottom panel).
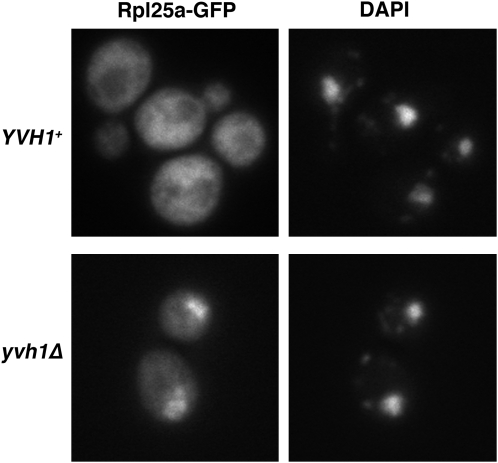
Consistent with its importance in 60S ribosome assembly, yvh1Δ cells have a slight growth defect under normal growth conditions and a more severe growth defect in the presence of the protein synthesis inhibitor cycloheximide (not shown). Defective ribosomal assembly in yvh1Δ cells was also observed using the large ribosomal subunit reporter Rpl25-GFP (Gadal et al. 2001). Figure 5 shows cytoplasmic localization of the reporter in wild-type cells but nuclear accumulation, coincident with DAPI staining, in yvh1Δ cells. Thus, loss of Yvh1 results in impaired nuclear export of 60S subunits.
Figure 5.—
A role for Yvh1 in ribosome assembly. Localization of Rpl25-GFP in yvh1 cells is shown. Wild-type and yvh1Δ cells bearing pRPL25-GFP were visualized by fluorescence microscopy. DAPI staining indicates position of nuclei. Localization of Rpl25 is predominantly nuclear in yvh1Δ cells by contrast with a cytoplasmic localization in YVH1+ cells.
Pma1-10 removal from the plasma membrane is not affected by general defects in ribosome biogenesis:
To determine whether stability of Pma1-10 is affected by defects in biogenesis of the small ribosomal subunit, we examined a mutant in Slx9, a nonessential assembly factor (Bax et al. 2006), and an rps25aΔ mutant. To test whether Pma1-10 stability is affected by general defects in the 60S ribosomal subunit, an rpl24a mutant was assayed because half-mers are accumulated, and there is a decreased rate of translation (Dresios et al. 2000). Figure 6A shows the rate of Pma1-10 degradation at 37° remains unaffected in all mutants tested. From these results, we suggest that the suppressing effect of yvh1 on pma1-10 is not the result of a general defect in ribosome function.
Figure 6.—
Pma1-10 stability and synthesis. (A) Pulse-chase analysis of Pma1-10 in rpl24aΔ, rps25aΔ, and slx9Δ cells. Wild-type (BY4741), rpl24aΔ, rps25aΔ, and slx9Δ cells bearing pMET25-HA-pma1-10 (pKK7) were derepressed to synthesize tagged Pma1-10, as described in materials and methods. Cells were shifted to 37° for 10 min before pulse labeling with Expre35S35S for 5 min and chasing for various times. HA-Pma1-10 was immunoprecipitated and analyzed by SDS–PAGE and fluorography. The results are representative of two to three independent experiments. Autoradiograms were scanned and quantitated using NIH Image. (B) New synthesis of Pma1-10 in YVH1+ and yvh1Δ cells. pma1-10 cells (XGY32 and KKX103-2B) were pulse labeled with Expre35S35S for 2 and 5 min. Pma1-10 was immunoprecipitated with anti-Pma1 antibody and analyzed by SDS–PAGE and fluorography.
Recently, it has been proposed that diversity in ribosome composition, facilitated by specific ribosome assembly factors, may influence translation of specific mRNAs (Komili et al. 2007). This model suggests a plausible mechanism by which loss of Yvh1 leads to increased stability of cell surface Pma1-10 via altered translation of specific mRNAs (see discussion). To test the possibility that translation of Pma1-10 itself is specifically affected by loss of Yvh1, cells were pulse labeled with 35S-cysteine and methionine, and newly synthesized Pma1-10 was immunoprecipitated from lysate. Figure 6B shows that new synthesis of Pma1-10 is only slightly decreased in yvh1Δ cells by comparison with YVH1+ cells. It is possible that decreased translation of misfolded Pma1-10 in the absence of Yvh1 could result in better folding mediated by a relative increase in ER chaperone concentration.
DISCUSSION
A major finding in this work is that Yvh1 is a novel ribosome assembly factor. By polysome fractionation, we show that loss of Yvh1 results in a decrease in 60S ribosomal subunits and 80S monosomes; half-mer polysomes indicate defective or immature 60S ribosomal subunits and/or impaired assembly of 60S with 40S subunits (Figure 4A). Furthermore, HA-tagged Yvh1 is colocalized with 60S ribosomal subunits in fractionation experiments (Figure 4A). Also consistent with a role for Yvh1 in ribosome assembly are the observations that yvh1Δ cells display impaired vegetative growth and growth inhibition in the presence of cycloheximide (Y. Liu, unpublished results). Like many ribosome mutants, yvh1Δ cells are cold sensitive (Sakumoto et al. 2002). Evidence from several previous studies supports our conclusion that Yvh1 participates in 60S ribosomal subunit biogenesis. Yvh1 is associated with cytoplasmic 60S preribosomal particles (Nissan et al. 2002). By two-hybrid assay, Yvh1 interacts with Nop7, a ribosome assembly factor and constituent of several different preribosomal particles (Sakumoto et al. 2001). Another physical interaction study reports Yvh1 interaction with Tif6, Rei1, and Lsg1, all factors involved in 60S ribosome biogenesis (Tarassov et al. 2008).
Genes involved in ribosome biogenesis are coordinately regulated in response to environmental conditions (Gasch et al. 2000). YVH1 has a ribosome biosynthesis regulon: microarray assays show YVH1 has common clustering behavior with ribosome biogenesis factors and the YVH1 promoter is predicted to contain the PAC consensus motif (Wade et al. 2001). In addition, Yvh1 regulates sporulation (Beeser and Cooper 2000), which is analogous to ribosome assembly occurring during morphologic differentiation in bacteria (Karbstein 2007).
Yvh1 is important but not essential for 60S ribosome assembly as cells are viable in the absence of Yvh1. Consistent with defective 60S ribosome biogenesis in yvh1Δ cells, the large ribosomal subunit reporter Rpl25 displays nuclear accumulation (Figure 5). It is not clear how loss of Yvh1 leads to impaired nuclear export of 60S subunits because Yvh1 is localized to the cytosol (Huh et al. 2003). One possible explanation is that Yvh1 assists 60S assembly by shuttling between nucleus and cytoplasm. Human Yvh1 has been shown to localize predominantly to the nucleus but is also detected in the cytosol (Muda et al. 1999); Yvh1 of the malaria parasite Plasmodium falciparum has been shown to undergo nucleocytoplasmic shuttling dependent on developmental stage (Kumar et al. 2004). Another explanation for nuclear accumulation of 60S subunits is that defective function of Yvh1 in the cytoplasm impairs recycling of a biogenesis factor.
Unexpectedly, discovery of Yvh1 as a ribosomal assembly factor came from a genetic screen for suppressors of pma1-10. [In this regard, it is of interest that a genetic screen to isolate multicopy suppressors of a temperature-sensitive mutant defective in the chaperonin Cct complex resulted in isolation of numerous ribosomal proteins, ribosome biogenesis factors, and Yvh1 (Kabir and Sherman 2008).] In the absence of Yvh1, mutant Pma1-10 turnover from the plasma membrane is prevented, permitting cell growth (Figure 1 and Liu and Chang 2006). Analogously, a requirement for 60S assembly in regulating events at the plasma membrane has previously been reported (Miyoshi et al. 2002; Zhao et al. 2003). Even so, it is a challenge to reconcile involvement of Yvh1 in ribosome assembly and its role in turnover of mutant Pma1-10 from the cell surface. One possible explanation is that decreased translational fidelity in the absence of Yvh1 can cause suppression of pma1-10. However, several observations do not readily fit with this possibility: yvh1 is a suppressor of pma1-4D2E/A, but the point mutant Pma1-7 that is routed from the Golgi to the endosomal/vacuolar system for degradation is unaffected by loss of Yvh1 (Liu and Chang 2006). Although absence of Yvh1 suppresses temperature-sensitive growth and several other pma1-10 phenotypes, slow export of Pma1-10 out of the ER is not reversed (Liu and Chang 2006). Together, these results suggest that a decrease in translational fidelity is unlikely the mechanism of yvh1-mediated suppression of pma1-10. Furthermore, it seems unlikely that a changed rate of translation in yvh1Δ cells promotes Pma1-10 stability as slowed translation upon loss of Rpl24a (Dresios et al. 2000) has no effect (Figure 6A).
Recent findings suggest a plausible model to account for pma1-10 suppression by loss of the ribosome assembly factor, Yvh1. Komili et al. (2007) noted that different ribosomal proteins have different assembly requirements and observed using transcriptional profiling that loss of specific ribosomal proteins affects discrete functional categories of genes. These observations, together with previous reports pointing to heterogeneity in ribosome composition (Selker et al. 1985; Mazumder et al. 2003), suggest possible functional heterogeneity. Most recently, a study of yeast lifespan reported a role for the nutrient-responsive transcription factor Gcn4 in promoting longevity; strikingly, the large 60S ribosomal subunit participates in this process as Gcn4 translation becomes increased to varying degrees upon depletion of 60S ribosomal subunits or deletion of different 60S ribosomal proteins (Steffen et al. 2008). Together, such observations support the hypothesis that ribosome composition, including post-translational modifications of ribosomal proteins, and different forms and modifications of rRNA, may influence translation of different mRNAs. If such a model is correct, it is possible that loss of the assembly factor Yvh1 or loss of specific ribosomal proteins, Rpl19a and Rpl35a, may affect ribosome composition and translation of specific mRNAs to affect Pma1-10 behavior at the plasma membrane. [An rpl35a mutant was also isolated as a suppressor of a sec63 mutant, defective in protein translocation into the ER (Ng and Walter 1996).] Also, we cannot exclude the possibility that the pma1-10 suppressors may act by reducing 60S subunits. In this regard, it is of interest that repression of ribosome synthesis in response to a defect in the secretory pathway is suppressed by a deficiency in 60S ribosomal subunits (although the mechanism by which this occurs is not known) (Zhao et al. 2003).
The Yvh1 RING-like domain is necessary and sufficient for normal levels of 60S and 80S ribosomal subunits and polysome profiles (Figure 4B). Similarly, Beeser and Cooper (2000) reported that the Yvh1 RING-like domain, not its dual-specificity phosphatase activity, is necessary to perform its role in glycogen accumulation, growth, and spore maturation. The molecular activity provided by the RING-like domain of Yvh1 is unclear at present. Ribosomal proteins undergo a variety of post-translational modifications, including phosphorylation (Nusspaumer et al. 2000; Mazumder et al. 2003) and ubiquitination (Spence et al. 2000; Peng et al. 2003). In several instances, ribosomal protein phosphorylation has been shown to have transcript-specific rather than global effects on translation (see discussion in Mazumder et al. 2003). Yvh1 phosphatase activity has been demonstrated in vitro (Guan et al. 1992), and we can speculate that Yvh1 phosphatase activity may have a regulatory function in vivo that affects ribosomal assembly and perhaps synthesis of specific proteins under certain conditions. The ubiquitin–proteasome system also participates in ribosome biogenesis (Kim et al. 2006; Stavrena et al. 2006). Although many ubiquitin ligases have RING finger domains (Fang et al. 2003), there is no evidence at present for a role for Yvh1 in ubiquitination: no significant difference in the profile of ubiquitinated proteins was observed in wild-type and yvh1Δ cells (Figure 2C).
Ubiquitination serves as a signal for endocytosis of a number of plasma membrane proteins and it also mediates removal of mutant Pma1-10 from the plasma membrane (Liu and Chang 2006). Interestingly, Pma1-10 ubiquitination is reversed when endocytosis is blocked (Liu and Chang 2006). Loss of Pma1-10 ubiquitination in yvh1Δ cells may reflect impaired detection by a quality control mechanism and/or enhanced stabilizing interactions at the plasma membrane. Loss of ubiquitination is not a strict prerequisite for suppression as we have isolated a high-copy suppressor, Ykl077w, that promotes growth of pma1-10 cells at 37° and partial stability of Pma1-10 without affecting its ubiquitination (Y. Liu, unpublished results).
Although at present we cannot exclude the possibility that Yvh1 serves another independent function, we propose its role in ribosome biogenesis is important for targeting Pma1-10 for removal from the plasma membrane. An effect on ribosome biogenesis may affect a number of components in a pathway when disruption of a single component of a pathway is not sufficient to achieve suppression. Future work will focus on determining transcriptional changes in the absence of Yvh1 as a first step toward identifying possible preferred translational targets and the cellular processes promoted by the Yvh1 ribosome assembly factor.
Acknowledgments
We thank Terrance Cooper, Satoshi Harashima, Ed Hurt, Jonathan Warner, Janine Maddock, and Katrin Karbstein for advice and reagents and Mengxi Jiang for helping us with polyribosome fractionation. This work was supported by grant GM 58212 from the National Institutes of Health.
References
- Albanese, V., A. Y.-W. Yam, J. Baughman, C. Parnot and J. Frydman, 2006. Systems analysis reveals two chaperone networks with distinct functions in eukaryotic cells. Cell 124 75–88. [DOI] [PubMed] [Google Scholar]
- Bax, R., H. A. Raue and J. C. Vos, 2006. Slx9p facilitates efficient ITS1 processing of pre-rRNA in Saccharomyces cerevisiae. RNA 12 2005–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeser, A. E., and T. G. Cooper, 1999. The dual-specificity protein phosphatase Yvh1p acts upstream of the protein kinase Mck1p in promoting spore development in Saccharomyces cerevisiae. J. Bacteriol. 181 5219–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeser, A. E., and T. G. Cooper, 2000. The dual-specificity protein phosphatase Yvh1p regulates sporulation, growth, and glycogen accumulation independently of catalytic activity in Saccharomyces cerevisiae via the cyclic AMP-dependent protein kinase cascade. J. Bacteriol. 182 3517–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, A., and C. W. Slayman, 1991. Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J. Cell Biol. 115 289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresios, J., I. L. Derkatch, S. W. Liebman and D. Synetos, 2000. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39 7236–7244. [DOI] [PubMed] [Google Scholar]
- Fang, S., K. L. Lorick, J. P. Jensen and A. M. Weissman, 2003. RING finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin. Cancer Biol. 13 5–14. [DOI] [PubMed] [Google Scholar]
- Fuentes, J. L., K. Datta, S. M. Sullivan, A. Walker and J. R. Maddock, 2007. In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1. Mol. Genet. Genomics 278 105–123. [DOI] [PubMed] [Google Scholar]
- Gadal, O., D. Straub, J. Kessl, B. Trumpower, D. Tollervey et al., 2001. Nuclear export of 60S ribosomal subunits depnds on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen et al., 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, X., and A. Chang, 2001. A mutant plasma membrane ATPase, Pma1–10, is defective in stability at the yeast cell surface. Proc. Natl. Acad. Sci. USA 98 9104–9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, K., D. J. Hakes, Y. Wang, H.-D. Park, T. G. Cooper et al., 1992. A yeast protein phosphatase related to the vaccinia virus VH1 phosphatase is induced by nitrogen starvation. Proc. Natl. Acad. Sci. USA 89 12175–12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, S., Y. Liu and A. Chang, 2007. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic-reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, Pma1. J. Biol. Chem. 282 26140–26149. [DOI] [PubMed] [Google Scholar]
- Henras, A. K., J. Soudet, M. Gerus, S. Lebaron, M. Caizergues-Ferrer et al., 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65 2334–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma, Y., A. Kitamura, R. Shioda, H. Maruyama, K. Ozaki et al., 2006. TOR regulates late steps of ribosome maturation in the nucleoplasm via Nog1 in response to nutrients. EMBO J. 25 3832–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.-K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425 686–691. [DOI] [PubMed] [Google Scholar]
- Ito, K., 2005. Ribosome-based protein folding systems are structurally divergent but functionally universal across biological kingdoms. Mol. Microbiol. 57 313–317. [DOI] [PubMed] [Google Scholar]
- Kabir, M. A., and F. Sherman, 2008. Overexpressed ribosomal proteins suppress defective chaperonins in Saccharomyces cerevisiae. FEMS Yeast Res. 8 1236–1244. [DOI] [PubMed] [Google Scholar]
- Karbstein, K., 2007. Role of GTPases in ribosome assembly. Biopolymers 87 1–11. [DOI] [PubMed] [Google Scholar]
- Kim, T.-S., C.-Y. Jang, H. D. Kim, J. Y. Lee, B.-Y. Ahn et al., 2006. Interaction of Hsp90 with ribosomal proteins protects from ubiquitination and proteasome-dependent degradation. Mol. Biol. Cell 17 824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili, S., N. G. Farny, F. P. Roth and P. A. Silver, 2007. Functional specificity among ribosomal proteins affects gene expression. Cell 131 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R., A. Musiyenko, E. Cioffi, A. Oldenburg, B. Adams et al., 2004. A zinc-binding dual-specificity YVH1 phosphatase in the malaria parasite, Plasmodium falciparum, and its interaction with the nuclear protein, pescadillo. Mol. Biochem. Parasitol. 133 297–310. [DOI] [PubMed] [Google Scholar]
- Liu, Y., and A. Chang, 2006. Quality control of a mutant plasma membrane ATPase: ubiquitination prevents cell surface stability. J. Cell Sci. 119 360–369. [DOI] [PubMed] [Google Scholar]
- Liu, Y., S. Sitaraman and A. Chang, 2006. Multiple degradation pathways for misfolded mutants of the yeast plasma membrane ATPase, Pma1. J. Biol. Chem. 281 31457–31466. [DOI] [PubMed] [Google Scholar]
- Mauro, V. P., and G. M. Edelman, 2002. The ribosome filter hypothesis. Proc. Natl. Acad. Sci. USA 99 12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder, B., P. Sampath, V. Seshadri, R. K. Maitra, P. E. DiCorleto et al., 2003. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115 187–198. [DOI] [PubMed] [Google Scholar]
- Miyoshi, K., R. Tsuji, H. Yoshida, Y. Maki, A. Wada et al., 2002. Normal assembly of 60S ribosomal subunits is required for the signaling in response to a secretory defect in Saccharomyces cerevisiae. J. Biol. Chem. 277 18334–18339. [DOI] [PubMed] [Google Scholar]
- Muda, M., E. R. Manning, K. Orth and J. E. Dixon, 1999. Identification of the human Yvh1 protein-tyrosine phosphatase orthologue reveals a novel zinc binding domain essential for in vivo function. J. Biol. Chem. 274 23991–23995. [DOI] [PubMed] [Google Scholar]
- Nakamoto, R. K., S. Verjovski-Almeida, K. E. Allen, A. Ambesi, R. Rao et al., 1998. Substitutions of Aspartate 378 in the phosphorylation domain of the yeast PMA1 H+-ATPase disrupt protein folding and biogenesis. J. Biol. Chem. 273 7338–7344. [DOI] [PubMed] [Google Scholar]
- Ng, D. T. W., and P. Walter, 1996. ER membrane protein complex required for nuclear fusion. J. Cell Biol. 132 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan, T. A., J. Babler, E. Petfalski, D. Tollervey and E. Hurt, 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusspaumer, G., M. Remacha and J. P. Ballesta, 2000. Phosphorylation and N-terminal region of yeast ribosomal protein P1 mediate its degradation, which is prevented by protein P2. EMBO J. 19 6075–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., D. Schwartz, J. E. Elias, C. C. Thoreen, D. Cheng et al., 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21 921–926. [DOI] [PubMed] [Google Scholar]
- Pizzirusso, M., and A. Chang, 2004. Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1–7, to the endosomal/vacuolar system in yeast. Mol. Biol. Cell 15 2401–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg, M. O., M. Moritz and J. L. Woolford, Jr., 1988. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 2 160–172. [DOI] [PubMed] [Google Scholar]
- Sakumoto, N., H. Yamashita, Y. Mukai, Y. Kaneko and S. Harashima, 2001. Dual-specificity protein phosphatase Yvh1p, which is required for vegetative growth and sporulation, interacts with yeast pescadillo homolog in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 289 608–615. [DOI] [PubMed] [Google Scholar]
- Sakumoto, N., I. Matsuoka, Y. Mukai, N. Ogawa, Y. Kaneko et al., 2002. A series of double disruptant for protein phosphatase genes in Saccharomyces cerevisiae and their phenotypic analysis. Yeast 19 587–599. [DOI] [PubMed] [Google Scholar]
- Selker, E. U., J. N. Stevens and R. L. Metzenberg, 1985. Heterogeneity of 5S RNA in fungal ribosomes. Science 227 1340–1343. [DOI] [PubMed] [Google Scholar]
- Serrano, R., M. C. Kielland-Brandt and G. R. Fink, 1986. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+, K+), K+- and Ca2+-ATPases. Nature 319 689–693. [DOI] [PubMed] [Google Scholar]
- Spence, J., R. R. Gali, G. Dittmar, F. Sherman, M. Karin et al., 2000. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102 67–76. [DOI] [PubMed] [Google Scholar]
- Stavrena, D. A., M. Kawasaki, M. Dundr, K. Koberna, W. G. Muller et al., 2006. Potential roles for ubiquitin and the proteasome during ribosome biogenesis. Mol. Cell. Biol. 26 5131–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen, K. K., V. L. MacKay, E. O. Derr, M. Tsuchiya, D. Hu et al., 2008. Yeast life span extension by depletion of 60S ribosomal subunits is mediated by Gcn4. Cell 133 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow, P. S., D. Finley and A. Varshavsky, 1986. Enhancement of immunoblot sensitivity by heating of hydrated filters. Anal. Biochem. 156 147–153. [DOI] [PubMed] [Google Scholar]
- Tarassov, K., V. Messler, C. R. Landry, S. Radinovic, M. M. Molina et al., 2008. An in vivo map of the yeast protein interactome. Science 320 1465–1470. [DOI] [PubMed] [Google Scholar]
- Wade, C. H., K. A. Shea, R. V. Jensen and M. A. McAlear, 2001. EBP2 is a member of the yeast RRB regulon, a transcriptionally coregulated set of genes that are required for ribosome and rRNA biosynthesis. Mol. Cell. Biol. 21 8638–8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, J. R., 1989. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol. Rev. 53 256–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., J.-H. Sohn and J. R. Warner, 2003. Autoregulation in the biosynthesis of ribosomes. Mol. Cell. Biol. 23 699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]