Abstract
Purpose
Most gastrointestinal stromal tumors (GISTs) harbor mutant KIT or platelet-derived growth factor receptor α (PDGFRA) kinases, which are imatinib targets. Sunitinib, which targets KIT, PDGFRs, and several other kinases, has demonstrated efficacy in patients with GIST after they experience imatinib failure. We evaluated the impact of primary and secondary kinase genotype on sunitinib activity.
Patients and Methods
Tumor responses were assessed radiologically in a phase I/II trial of sunitinib in 97 patients with metastatic, imatinib-resistant/intolerant GIST. KIT/PDGFRA mutational status was determined for 78 patients by using tumor specimens obtained before and after prior imatinib therapy. Kinase mutants were biochemically profiled for sunitinib and imatinib sensitivity.
Results
Clinical benefit (partial response or stable disease for ≥ 6 months) with sunitinib was observed for the three most common primary GIST genotypes: KIT exon 9 (58%), KIT exon 11 (34%), and wild-type KIT/PDGFRA (56%). Progression-free survival (PFS) was significantly longer for patients with primary KIT exon 9 mutations (P = .0005) or with a wild-type genotype (P = .0356) than for those with KIT exon 11 mutations. The same pattern was observed for overall survival (OS). PFS and OS were longer for patients with secondary KIT exon 13 or 14 mutations (which involve the KIT-adenosine triphosphate binding pocket) than for those with exon 17 or 18 mutations (which involve the KIT activation loop). Biochemical profiling studies confirmed the clinical results.
Conclusion
The clinical activity of sunitinib after imatinib failure is significantly influenced by both primary and secondary mutations in the predominant pathogenic kinases, which has implications for optimization of the treatment of patients with GIST.
INTRODUCTION
The pathogenesis of most gastrointestinal stromal tumors (GISTs) results from activating mutations of KIT or of platelet-derived growth factor receptor α (PDGFRA). More than 80% of GISTs express mutated, constitutively active KIT, and another 5% to 7% express mutated PDGFRA; 10% to 15% of tumors have no associated mutations in these kinases.1-3
Imatinib mesylate, a selective inhibitor of KIT and PDGFRA (and of platelet-derived growth factor receptor β [PDGFRB] and BCR-ABL kinase), has revolutionized the treatment of GIST; however, up to 14% of GISTs exhibit primary resistance to imatinib (defined as progression within 3 to 6 months of initiating therapy),4-6 and another 40% to 50% develop resistance within 2 years of beginning therapy (ie, secondary resistance).5,6 Sunitinib malate (SUTENT; Pfizer, New York, NY), another small-molecule tyrosine kinase inhibitor (TKI) with selectivity for KIT and PDGFRA (and for PDGFRB, all three isoforms of vascular endothelial growth factor receptor [VEGFR], FMS-like tyrosine kinase 3 [FLT3], colony-stimulating factor 1 receptor [CSF-1R], and glial cell line-derived neurotrophic factor receptor [rearranged during transfection; RET; Pfizer, New York, NY; data on file]),7-11 has demonstrated clinical benefit in phase I to phase III trials of patients with imatinib-resistant or -intolerant GIST.12,13 Sunitinib has been approved multinationally for the treatment of patients with GIST for whom prior imatinib therapy failed because of disease progression or drug intolerance.
GIST responsiveness to imatinib varies by primary KIT genotype; exon 11-mutant GISTs are more sensitive than exon 9-mutant or wild-type GISTs (ie, those that lack KIT or PDGFRA mutations).3,14,15 Exons 11 and 9 are the most common sites of KIT mutation in GIST (approximately 70% and 15% of tumors, respectively).3,14 Secondary kinase mutations are common in GISTs that exhibit secondary resistance but not in those that exhibit primary resistance.16,17 Secondary point mutations associated with imatinib resistance usually are located in the drug/adenosine triphosphate (ATP) binding pocket of the receptor (encoded by exons 13 and 14) or in the activation loop (encoded by exon 17).16-28 Two recent studies that used cell-based assays reported that sunitinib inhibited the kinase activity of KIT receptors that contained mutations in the drug/ATP binding pocket that confer resistance to imatinib.29,30 Because these mutations (ie, T670I and V654A [substitutions of isoleucine for threonine at position 670 and alanine for valine at position 654, respectively]) are commonly found in patients with GIST who have secondary imatinib resistance, the results provide a possible basis for sunitinib antitumor activity in patients with imatinib-refractory GIST.
To further explore the relationship between primary and secondary GIST kinase mutations and the response to sunitinib, we determined primary and secondary KIT or PDGFRA mutations in biopsied tissue from patients with imatinib-refractory GIST who received sunitinib as part of a phase I/II trial,12 and we correlated the presence of these mutations with clinical benefit. In addition, in vitro studies assessed the sensitivity of KIT and PDGFRA mutants to sunitinib and imatinib directly.
PATIENTS AND METHODS
Biopsies for genotype analyses were obtained from patients enrolled on a sunitinib phase I/II trial that was described in an earlier report of efficacy/safety results from the study.12 Patients were adults who had histologically confirmed metastatic/unresectable GIST and documented failure of imatinib caused by resistance or intolerance. Most patients (55 of 97) received sunitinib 50 mg/d in 6-week cycles that comprised 4 weeks on, followed by 2 weeks off, treatment. Additional information about methods is listed in the Appendix (online only).
RESULTS
Primary Tumor Genotype and Efficacy
Tissue for pre-imatinib genotype analysis was available for 78 of 97 patients on the trial. These patients overall had bulky metastatic disease and had received a median of 78 weeks of prior imatinib therapy (Table 1). Primary KIT mutations were identified in 83% of tumors, whereas 5% had PDGFRA mutations, and 12% contained wild-type KIT and PDGFRA (Appendix Table A1, online only). The most KIT mutations (69%) were located in exon 11, then in exon 9 (30% of KIT mutations), and then in exon 13 (2% of KIT mutations). PDGFRA mutations were located in exon 12 in one patient's tumor and in exon 18 in the tumors of three patients.
Table 1.
Baseline Characteristics and Prior Imatinib Treatment of Patients With Pre-Imatinib Genotyping Data
| Characteristic | No. of Patients (N = 78) | % of Patients |
|---|---|---|
| Sex | ||
| Male | 53 | 68 |
| Female | 25 | 32 |
| Age, years | ||
| Median | 55 | |
| Range | 26-76 | |
| ECOG performance status | ||
| 0 | 38 | 49 |
| 1 | 24 | 44 |
| 2 | 6 | 8 |
| Time since initial diagnosis, weeks | ||
| Median | 139 | |
| Range | 23-664 | |
| Most common disease present at screening | ||
| Liver metastases | 72 | 92 |
| Soft tissue | 37 | 47 |
| Peritoneal metastases | 36 | 46 |
| Local recurrence | 28 | 36 |
| Prior therapy other than imatinib | ||
| Surgery | 78 | 100 |
| Radiotherapy | 10 | 13 |
| Systemic therapy | 34 | 44 |
| Prior imatinib therapy | ||
| Maximum dose, mg | ||
| Median | 600 | |
| Range | 400-1,000 | |
| Duration of treatment, weeks | ||
| Median | 78 | |
| Range | 10-151 | |
| Reason for discontinuation | ||
| Tumor progression | 74 | 95 |
| Intolerance | 4 | 5 |
Abbreviation: ECOG, Eastern Cooperative Oncology Group.
Clinical benefit (partial response [PR] or stable disease [SD] for ≥ 6 months) was observed for the three most common GIST genotypes (Table 2). The clinical benefit rate was 58% for tumors with primary KIT exon 9 mutations, 34% for those with exon 11 mutations, and 56% for those with wild-type KIT and PDGFRA before imatinib therapy. Objective responses (ie, PRs) were significantly more common in patients with KIT exon 9 than exon 11 mutant GISTs (37% v 5%; P = .002). Of the four patients with PDGFRA mutations, none experienced clinical benefit. Among patients classified as imatinib-intolerant (n = 4), tumor genotyping revealed a primary KIT exon 9 mutation in one (who achieved a PR) and a wild-type genotype in the other three patients (who achieved SD, two for > 6 months).
Table 2.
Response to Sunitinib by Primary and Secondary Tumor Genotype
| Response by Tumor Genotype
| |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Primary (n = 77)*
|
Secondary (n = 65)*†
|
||||||||||||
| Mutation Status | No. | Median Duration of Prior IM (months) | RECIST Response
|
Clinical Benefit‡
|
Mutation Status§ | No. | Median Duration of Prior IM (months) | RECIST Response
|
Clinical Benefit‡
|
||||
| No. | % | No. | % | No. | % | No. | % | ||||||
| KIT mutation | 64 | 9 | 14 | 27 | 42 | ||||||||
| KIT exon 9 | 19 | 12.5 | 7 | 37‖ | 11 | 58¶ | KIT 9 → 9 | 13 | 12.2 | 5 | 38 | 8 | 62 |
| KIT 9 → 9 + 13 | 1 | 17.3 | 1 | 100 | 1 | 100 | |||||||
| KIT 9 → 9 + 17 | 2 | 17.7 | 0 | 0 | 0 | 0 | |||||||
| KIT exon 11 | 44 | 22.8 | 2 | 5 | 15 | 34 | KIT 11 → 11 | 10 | 22.1 | 1 | 10 | 1 | 10 |
| KIT 11 → 11 + (13 or 14) | 17 | 20.0 | 1 | 6 | 10 | 59 | |||||||
| KIT 11 → 11 + (17 or 18) | 10 | 23.3 | 0 | 0 | 1 | 10 | |||||||
| KIT exon 13 | 1 | 14.0 | 0 | 0 | 1 | 100 | KIT 13 → 13 + 17 | 1 | 14.0 | 0 | 0 | 1 | 100 |
| PDGFRA mutation | 4 | 0 | 0 | 0 | 0 | ||||||||
| PDGFRA exon 12 | 1 | 18.6 | 0 | 0 | 0 | 0 | PDGFRA 12 → 12 + 18 | 1 | 18.6 | 0 | 0 | 0 | 0 |
| PDGFRA exon 18 | 3 | 7.9 | 0 | 0 | 0 | 0 | PDGFRA 18 → 18 | 2 | 8.5 | 0 | 0 | 0 | 0 |
| No KIT/PDGFRA mutation | 9 | 10.5 | 0 | 0 | 5 | 56 | No mutation → no mutation | 8 | 10.8 | 0 | 0 | 4 | 50 |
Abbreviations: IM, imatinib; RECIST, Response Evaluation Criteria in Solid Tumors; PDGFRA, platelet-derived growth factor receptor α.
One additional patient had baseline pre-imatinib mutations of KIT in both exons 13 and 17 and was excluded from analyses.
One patient included in the primary tumor genotype analysis had a primary exon 11 mutation and secondary exon 13 and 17 mutations in separate lesions and was excluded from secondary tumor genotype analysis.
Clinical benefit is defined as response or stable disease for ≥ 6 months according to Response Evaluation Criteria in Solid Tumors.
Arrows separate primary and secondary genotype results (eg, KIT 11 → 11 is a primary KIT exon 11 mutation with no secondary mutation detected; KIT 11 → 11 + [13 or 14] is a primary KIT exon 11 mutation + secondary KIT exon 13 or 14 mutations).
P = .002 compared with primary KIT exon 11 mutation.
P = .08 compared with primary KIT exon 11 mutation.
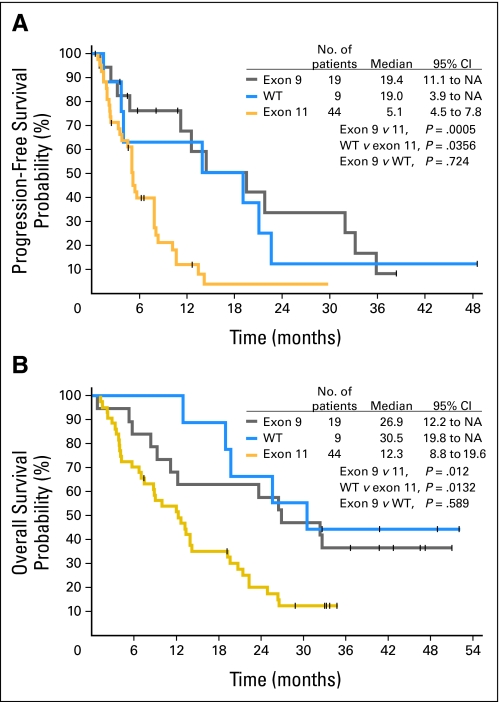
Median progression-free survival (PFS) was significantly longer for patients with primary KIT exon 9 mutations (19.4 months; 95% CI, 11.1 to not yet attained [NA]; P = .0005) or a wild-type genotype (19.0 months; 95% CI, 3.9 to NA; P = .0356) than for those with KIT exon 11 mutations (5.1 months; 95% CI, 4.5 to 7.8; Fig 1A). PFS did not differ significantly between patients with exon 9 mutations and a wild-type genotype. Median overall survival (OS) was also significantly longer for patients with exon 9 mutations (26.9 months; 95% CI, 12.2 to NA; P = .012) or a wild-type genotype (30.5 months; 95% CI, 19.8 to NA; P = .0132) than for those with exon 11 mutations (12.3 months; 95% CI, 8.8 to 19.6; Fig 1B). OS did not differ significantly between patients with exon 9 mutations or a wild-type genotype.
Fig 1.
Impact of primary (pre-imatinib) KIT genotype on efficacy of sunitinib treatment. (A) Progression-free survival. (B) Overall survival. NA, not yet attained; WT, wild type.
Secondary Tumor Genotype and Efficacy
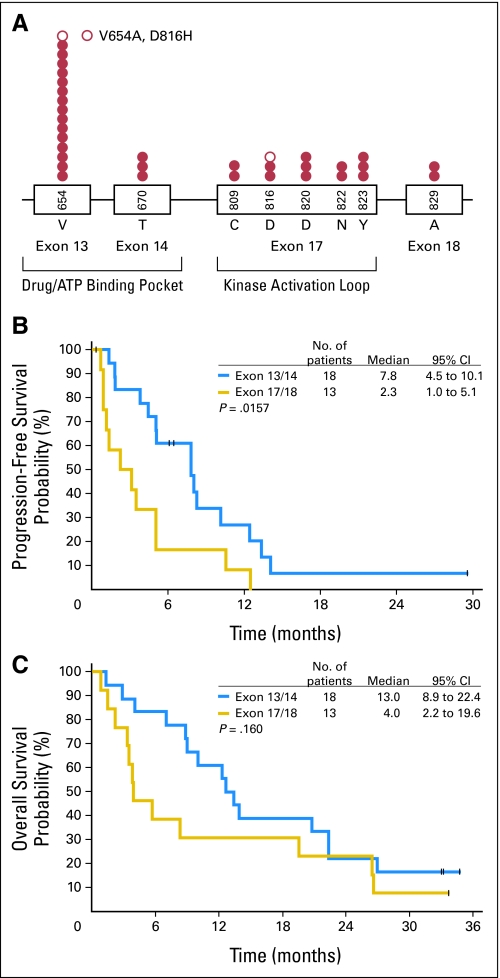
A total of 109 post-imatinib biopsy specimens were available from 67 patients, and secondary KIT mutations were identified in 33 patients (Appendix Table A1). Consistent with prior reports, the mutation distribution was nonrandom, and clusters occurred in exons 13 and 14 that encode the drug/ATP binding pocket of the receptor and exon 17 that encodes the kinase activation loop (Fig 2A). The most commonly identified secondary mutation was V654A in exon 13. Two tumors had secondary KIT exon 18 mutations. One patient had different secondary mutations (exon 13 V654A and exon 17 D816H) in different lesions. Secondary kinase mutations were significantly more common in GISTs with primary KIT exon 11 mutations than in those with exon 9 mutations (73% v 19%; P = .0003). Of the four samples with primary PDGFRA mutations, one had a secondary mutation in exon 18 (primary mutation in exon 12), two lacked secondary mutations (both had primary exon 18 D842V mutations), and the fourth lacked a post-imatinib sample. No secondary mutations were found in the eight post-imatinib samples that lacked primary KIT or PDGFRA mutations.
Fig 2.
(A) Distribution and frequency of unique secondary (post-imatinib) KIT mutations (per patient) in this study. One patient had different mutations in different biopsy specimens: a V654A mutation in one lesion, a D816H mutation in another (○). Impact of secondary KIT genotype on (B) progression-free survival and (C) overall survival with sunitinib.
Among all patients with KIT mutations, the median PFS with sunitinib was significantly longer for the 18 patients who had secondary KIT exon 13 or 14 mutations (7.8 months; 95% CI, 4.5 to 10.1) than for the 13 patients who had exon 17 or 18 mutations (2.3 months; 95% CI, 1.0 to 5.1; P = .0157; Fig 2B). Likewise, median OS was numerically longer in the former than the latter group (13.0 months [95% CI, 8.9 to 22.4] v 4.0 months [95% CI, 2.2 to 19.6]; P = .160; Fig 2C), and clinical benefit rates were higher (61% v 15%; P = .011; Table 2). Nearly identical results were obtained when only patients with primary exon 11 mutations were considered. For patients with primary exon 11 mutations, there were no significant differences in PFS or OS between those patients with or without secondary mutations.
In Vitro Measures of Activity With Specific Mutants
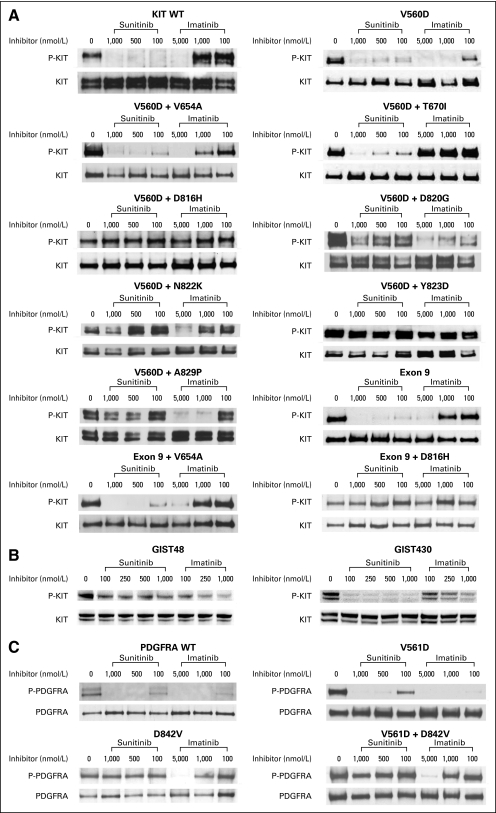
Sunitinib potently inhibited the activity of ligand-activated wild-type KIT, and the KIT exon 11 V560D and exon 9 AY insertion mutants: 50% inhibitory concentration (IC50) values were less than 100 nmol/L for all three kinases (Table 3; Fig 3A). By comparison, the corresponding IC50 values for imatinib were approximately 1,000 nmol/L for wild-type KIT, 100 nmol/L for the V560D mutant, and 1,000 nmol/L for the exon 9 AY mutant. Sunitinib also potently inhibited the phosphorylation of KIT double mutants, in which the second mutation occurred in the drug/ATP binding site of the receptor, such as V560D + V654A (exons 11 + 13) and V560D + T670I (exons 11 + 14). These double mutants were resistant to inhibition by imatinib in vitro. Conversely, KIT double mutants, in which the second mutation occurred in the activation loop (V560D + D816H, V560D + D820G, V560D + N822K, and V560D + Y823D), were resistant to inhibition by sunitinib or imatinib, with sunitinib IC50 values of 1,000 nmol/L or higher. Notably, the V560D + A829P double mutant had an imatinib IC50 that was only two- to three-fold higher than that of V560D alone. In contrast, V560D + A829P was resistant to sunitinib at doses of up to 1,000 nmol/L. The rarity of A829P as a secondary mutation could be caused by its relatively preserved imatinib sensitivity. Similar results to those obtained when exon 11 V560D was used as the primary mutation were obtained when the exon 9 AY insertion was used instead (Table 3; Fig 3A).
Table 3.
In Vitro Effects of Sunitinib and Imatinib on Autophosphorylation of KIT and PDGFRA Mutants Expressed in Chinese Hamster Ovary Cells
| Mutant Construct | Mutation
|
Treatment
|
||||||
|---|---|---|---|---|---|---|---|---|
| 1
|
2
|
Sunitinib
|
Imatinib
|
|||||
| Exon | Function | Exon | Function | Approximate IC50 (nmol/L) | S/R | Approximate IC50 (nmol/L) | S/R | |
| KIT | ||||||||
| Ligand-activated WT | — | — | — | — | < 100 | S | 1,000 | R |
| V560D | 11 | JM | — | — | < 100 | S | 100 | S |
| V560D + V654A | 11 | JM | 13 | ATP BP | < 100 | S | 2,500 | R |
| V560D + T670I | 11 | JM | 14 | ATP BP | < 50 | S | > 5,000 | R |
| V560D + D816H | 11 | JM | 17 | AL | ≥ 1,000 | R | 5,000 | R |
| V560D + D820G | 11 | JM | 17 | AL | ≥ 1,000 | R | 1,000 | R |
| V560D + N822K | 11 | JM | 17 | AL | > 1,000 | R | 2,000 | R |
| V560D + Y823D | 11 | JM | 17 | AL | > 1,000 | R | > 5,000 | R |
| V560D + A829P | 11 | JM | 18 | Extended AL | > 1,000 | R | 200 | I |
| Exon 9 AY | 9 | DM | — | — | < 100 | S | 1,000 | R |
| Exon 9 AY + V654A | 9 | DM | 13 | ATP BP | 100 | S | 3,000 | R |
| Exon 9 + D816H | 9 | DM | 17 | AL | 500 | R | 3,000 | R |
| PDGFRA | ||||||||
| WT | — | — | — | — | < 100 | S | < 100 | S |
| V561D | 12 | JM | — | — | < 100 | S | < 100 | S |
| D842V | 18 | AL | — | — | > 1,000 | R | 2,500 | R |
| V561D + D842V | 12 | JM | 18 | AL | > 1,000 | R | 2,500 | R |
Abbreviations: PDGFRA, platelet-derived growth factor receptor α; S, sensitive; R, resistant; WT, wild type; JM, juxtamembrane region; ATP BP, adenosine triphosphate binding pocket; AL, activation loop; I, intermediate; DM, dimerization.
Fig 3.
Effects of sunitinib and imatinib on autophosphorylation of (A) wild-type KIT and KIT mutants transiently expressed in Chinese hamster ovary cells; (B) KIT mutants expressed by gastrointestinal stromal tumor cell lines; or (C) platelet-derived growth factor receptor α mutants transiently expressed in Chinese hamster ovary cells. Wild-type, but not mutant, receptors were ligand-activated. P-KIT, phosphorylated KIT; WT, wild type.
To confirm these findings, we tested the relative potency of imatinib or sunitinib at inhibiting KIT kinase activity in GIST cell lines obtained from imatinib-resistant tumors (Fig 3B). The GIST48 cell line is homozygous for a primary KIT exon 11 V560D mutation and is heterozygous for a secondary exon 17 D820A mutation.17 Concentrations of imatinib greater than 1,000 nmol/L were insufficient to completely inhibit KIT activation in this cell line. (This concentration is 10-fold higher than that necessary to block KIT exon 11-mutant isoforms in GIST cell lines in other studies.24,31,32) Sunitinib was less potent than imatinib at inhibiting KIT autophosphorylation in GIST48 cells. Notably, low doses (100 nmol/L) of either imatinib or sunitinib had a partial inhibitory effect on KIT phosphorylation, presumably because of inhibition of a minority population of V560D homodimers. The GIST430 cell line is heterozygous for a KIT exon 11 deletion mutation and an exon 13 V654A substitution (both on the same allele).17 Sunitinib had significantly greater potency than imatinib for inhibition of KIT autophosphorylation in GIST430 cells (IC50, 1,000 nmol/L for imatinib v < 100 nmol/L for sunitinib).
We also tested the potency of sunitinib at inhibiting the phosphorylation of wild-type PDGFRA or the V561D point mutant: the IC50 values were less than 100 nmol/L for both (Table 3; Fig 3C). V561D, located in the receptor juxtamembrane domain encoded by exon 12, is a relatively common primary PDGFRA mutation in patients with GIST.1 Conversely, D842V, which is the most common PDGFRA mutation in GISTs, which resides in the activation loop encoded by exon 18, and which confers imatinib resistance both as a primary or a secondary mutation,1 conferred resistance to sunitinib in these in vitro experiments (Table 3; Fig 3C). In the clinical study, D842V was detected as a primary mutation in two patients and as a secondary mutation in one patient.
DISCUSSION
These results extend previously reported findings from this study that showed a correlation between sunitinib activity and GIST kinase genotype in patients who have metastatic/unresectable GIST and have experienced imatinib failure.33 Data on the relative responsiveness of different molecular subgroups of imatinib-resistant GIST may help to optimize treatment of patients with GIST and may help to better understand the basis of sunitinib activity in these patients. Such studies may also advance understanding of the mechanisms of resistance and may facilitate development of strategies to circumvent it.
The analyses reported here assessed the effect of tumor kinase genotype on sunitinib activity by using clinical study data complemented by in vitro cellular assays. Although sunitinib demonstrated clinical activity against GISTs of the three most common primary genotypes, both datasets indicated that primary and secondary mutations in the pathogenic kinase strongly influence sunitinib activity. Both the clinical benefit and the objective response rates with sunitinib were higher in patients with primary KIT exon 9 mutations than with exon 11 mutations (clinical benefit rates: 58% v 34%; objective response rates: 37% v 5%; P = .002). Similarly, PFS and OS were significantly longer in patients with primary KIT exon 9 mutations or a wild-type genotype than in those with KIT exon 11 mutations. These results are the converse of those reported for imatinib, in which objective response rates were higher and PFS and OS were longer in patients with GIST who harbored exon 11 mutations than in those who had exon 9 mutations or a wild-type genotype.3,14,15 Notably, the potency of sunitinib against wild-type and exon 9-mutant KIT was superior to that of imatinib in vitro, whereas both drugs exhibited similar potency against KIT exon 11 mutant kinases. A possible explanation is that these mutational sites have different structural effects on KIT, with different consequences for interaction with the two TKIs. Indeed, exon 9 mutations were recently reported to have structural consequences similar to ligand-mediated receptor dimerization.34 This mechanism of kinase activation appears distinct from that caused by mutation of the intracellular juxtamembrane domain encoded by exon 11.35 Others have also observed the impact of mutational site on TKI potency in vitro: by using an isogenic BaF3 model, the imatinib IC50 in cells that expressed exon 9 mutations was found to be approximately eight-fold higher than that obtained in cells that expressed the exon 11 V559D mutation.36 These results suggest that the greater clinical benefit seen for sunitinib-treated patients with exon 9-mutant or wild-type imatinib-resistant GISTs may be related to the greater potency of sunitinib against these kinases. They also suggest that genotypically defined subsets of patients may experience different clinical outcomes when treated with first-line imatinib than with sunitinib. Sunitinib is currently approved only as second-line therapy for GIST, but studies are being planned to evaluate its efficacy and safety as first-line treatment. On the other hand, sunitinib has yet to be tested in imatinib-naïve patients, and the majority of patients in this study with primary KIT exon 11 mutations had acquired secondary KIT mutations that confer imatinib resistance. Studies in imatinib-naïve patients will be required to definitely assess the effect of a primary exon 11 mutation alone on sunitinib activity in vivo.
This study also showed that secondary kinase mutations were significantly more common in GISTs with primary KIT exon 11 than exon 9 mutations and that they did not occur in GISTs with a wild-type genotype, which is consistent with previous reports that secondary kinase mutations are common in GISTs that exhibit secondary imatinib resistance but not in those that exhibit primary resistance.16,17 Moreover, the frequency of secondary mutations is likely to have been underestimated in this analysis, because only one patient in our analysis was found to have different secondary mutations in different lesions, and intra- and interlesion heterogeneity of secondary mutations in GISTs has been documented by others.20,25 Only a limited number of small-needle biopsy specimens were available per patient in our study (mean, 1.4 biopsy specimens per patient; range, 0-3). In particular, it is probable that further sampling would have revealed secondary mutations in those tumors with primary KIT exon 11 mutations that appeared to lack them. Because exon 11 mutants are strongly inhibited by imatinib, secondary resistance is more likely to require the selection and subsequent expansion of clones expressing a second, resistance-conferring mutation than GISTs with exon 9 mutations or a wild-type genotype, which are more likely to be intrinsically resistant to imatinib. Consistent with this, the median duration of prior imatinib treatment for patients who had primary exon 11 mutations was 22.8 months, compared with 12.5 and 10.5 months for patients who had exon 9 mutations or a wild-type genotype, respectively (Table 2). However, it is worth noting that, although the duration of imatinib treatment was a significant prognostic factor for PFS and OS in a univariate analysis, it was not a significant factor in a multivariate analysis (data not shown). Although multivariate analyses performed on such a small sample must be interpreted with caution, they confirmed that primary and secondary KIT genotype were significant prognostic factors for PFS and were marginally significant prognostic factors for OS.
Consistent with previous studies,16,18-28 secondary KIT mutations in patients with imatinib-resistant GIST enrolled on the current study tended to cluster in exons 13 and 14, which encode the drug/ATP binding pocket of the receptor, or in exon 17, which encodes the kinase activation loop. Of note, our in vitro studies showed that sunitinib potently inhibited the kinase activity of KIT receptors that contained secondary mutations in the drug/ATP binding pocket and that are resistant to imatinib, such as V654A (exon 13) and T670I (exon 14). These secondary mutations were coexpressed with a common primary mutation (V560D), which recreated the situation often observed in GISTs that exhibit secondary imatinib resistance. Previous ex vivo studies have also shown that sunitinib inhibits imatinib-resistant KIT receptors that contain mutations in the drug/ATP binding pocket.29,30 However, the in vitro studies performed here also showed that sunitinib was relatively ineffective at inhibiting KIT receptors that contained secondary mutations localized to the activation loop. Consistent with these in vitro findings, PFS and OS were longer and the clinical benefit rate was higher for patients in the clinical trial who had secondary KIT exon 13 or 14 (ie, ATP-binding-pocket) mutations than those with secondary KIT exon 17 or 18 (ie, activation-loop) mutations.
The results of this study provide one explanation for the activity of sunitinib in patients with imatinib-refractory GIST that has been seen in this and other trials.13 However, antiangiogenic effects of sunitinib treatment also may contribute to its effectiveness. In addition to KIT and PDGFRA activity, sunitinib also selectively inhibits PDGFRB and all three isotypes of VEGFR, whereas imatinib inhibits PDGFRB but not VEGFRs. Studies in animal models indicate that dual inhibition of PDGFR and VEGFR produces greater antiangiogenic effects than inhibition of only one or the other,37-39 which suggests that sunitinib may produce greater antiangiogenic effects than imatinib and that these effects may contribute to its activity against imatinib-refractory GISTs.
Of note is our observation that secondary KIT mutants that involve the activation loop are insensitive to both sunitinib and imatinib. Given that different tumor clones in one individual may acquire imatinib resistance because of different secondary mutations, including those involving the KIT activation loop,20,25 not all imatinib-resistant tumors may respond well to sunitinib therapy. Conversely, some GISTs with secondary KIT activation-loop mutations may still be susceptible to sunitinib because of its potent antiangiogenic effects. Additional research of this issue is warranted.
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Xin Huang, Pfizer Inc, (C); Darrel P. Cohen, Pfizer Inc (C); Charles M. Baum, Pfizer Inc (C) Consultant or Advisory Role: Michael C. Heinrich, Novartis (C), Pfizer Inc (U); Christopher L. Corless, Pfizer Inc (C); George D. Demetri, Novartis (C), Pfizer Inc (C), Infinity (C) Stock Ownership: Michael C. Heinrich, Molecular MD; Xin Huang, Pfizer Inc; Darrel P. Cohen, Pfizer Inc; Charles M. Baum, Pfizer Inc Honoraria: Michael C. Heinrich, Novartis; Robert G. Maki, Pfizer Inc; Christopher L. Corless, Pfizer Inc; George D. Demetri, Novartis, Pfizer Inc Research Funding: Michael C. Heinrich, Novartis, Pfizer Inc; Robert G. Maki, Pfizer Inc; George D. Demetri, Novartis, Pfizer Inc, Infinity Expert Testimony: George D. Demetri, Novartis (U), Pfizer Inc (U), Infinity (U) Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Michael C. Heinrich, Wen-Bin Ou, Jonathan A. Fletcher, Darrel P. Cohen, George D. Demetri
Financial support: Michael C. Heinrich, Jonathan A. Fletcher, Charles M. Baum, George D. Demetri
Administrative support: Michael C. Heinrich, Diana Griffith, George D. Demetri
Provision of study materials or patients: Michael C. Heinrich, Robert G. Maki, Jonathan A. Fletcher, George D. Demetri
Collection and assembly of data: Michael C. Heinrich, Robert G. Maki, Cristina R. Antonescu, Amy Harlow, Diana Griffith, Ajia Town, Arin McKinley, Wen-Bin Ou, Jonathan A. Fletcher, Christopher D.M. Fletcher, Xin Huang, Charles M. Baum, George D. Demetri
Data analysis and interpretation: Michael C. Heinrich, Christopher L. Corless, Cristina R. Antonescu, Amy Harlow, Diana Griffith, Ajia Town, Arin McKinley, Wen-Bin Ou, Jonathan A. Fletcher, Xin Huang, Charles M. Baum, George D. Demetri
Manuscript writing: Michael C. Heinrich, Christopher L. Corless, Jonathan A. Fletcher, Darrel P. Cohen, Charles M. Baum, George D. Demetri
Final approval of manuscript: Michael C. Heinrich, Robert G. Maki, Christopher L. Corless, Cristina R. Antonescu, Amy Harlow, Diana Griffith, Ajia Town, Arin McKinley, Wen-Bin Ou, Jonathan A. Fletcher, Christopher D.M. Fletcher, Xin Huang, Darrel P. Cohen, Charles M. Baum, George D. Demetri
Acknowledgments
We thank our colleagues in the multidisciplinary teams that participated in this work: M. Bertagnoli, C. Raut, S. Singer (surgical oncology); J.A. Morgan, S. George, D. D'Adamo, J. Jackson, J. Pokela, A. Dollard, M.T. Quigley, A. Potter, M.L. Keohan, G. Wasilewski (clinical team); C. Bello, A.M. Martino, M. Collier, L. Strawn, Z. Aguilar, and many others (Pfizer Oncology); and T. Harrell, T. Bainbridge, A. Schroeder (Heinrich-Corless Lab). We also thank all of the patients, families, and referring physicians who have been so supportive of this research. Finally, medical writing services were provided by ACUMED (Tytherington, United Kingdom).
Appendix
Methods: mutational analysis.
Archival tumor biopsy specimens collected before initiation of imatinib therapy were obtained with written patient consent, as were additional specimens collected on days 1 and 11 of the current study before sunitinib dosing. Genomic DNA was extracted from formalin-fixed, paraffin-embedded specimens, was amplified by using polymerase chain reaction, and was analyzed for KIT or platelet-derived growth factor receptor α (PDGFRA) mutations, as previously described (Heinrich MC, Corless CL, Duensing A, et al: Science 299:708-710, 2003; Corless CL, McGreevey L, Haley A, et al: Am J Pathol 160:1567-1572, 2002; Rader AE, Avery A, Wait CL, et al: Cancer 93:269-275, 2001; Choy YS, Dabora SL, Hall F, et al: Ann Hum Genet 63:383-391, 1999) by using primer pairs and denaturing high-performance liquid chromatography conditions reported elsewhere (Heinrich MC, Corless CL, Blanke CD, et al: J Clin Oncol 24:4764-4774, 2006).
Methods: tumor responses.
Tumor responses were assessed at baseline and the end of every other cycle on the basis of the Response Evaluation Criteria in Solid Tumors (Therasse P, Arbuck SG, Eisenhauer EA, et al: J Natl Cancer Inst 92:205-216, 2000). Clinical benefit was defined as a partial response or stable disease that lasted 6 months or more. Additional efficacy measures included progression-free survival and overall survival. The current analyses include follow-up through March 2006.
Methods: sensitivity of kinase mutants to tyrosine kinase inhibitors in vitro.
Chinese hamster ovary cells were transiently transfected with mutated KIT or PDGFRA cDNA constructs and treated with various concentrations of sunitinib or imatinib, as previously described (Heinrich MC, Corless CL, Demetri GD, et al: J Clin Oncol 21:4342-4349, 2003; Heinrich MC, Corless CL, Blanke CD, et al: J Clin Oncol 24:4764-4774, 2006). The common exon 11 mutation V560D was selected as prototypic primary KIT mutation, and several representative exon 13, 14, and 17 mutations were selected as secondary mutations, for these constructs. Experiments that involved recombinant DNA were performed under biosafety level 2 conditions in accordance with published guidelines (National Institutes of Health; available at: http://www4.od.nih.gov/oba/rac/guidelines/guidelines.html). The gastrointestinal stromal tumor GIST48 and GIST430 cell lines were established from imatinib-resistant GISTs, as previously described (Heinrich MC, Corless CL, Blanke CD et al: J Clin Oncol 24:4764-4774, 2006). Protein lysates from transfected Chinese hamster ovary cells or GIST cell lines were prepared and were subjected to immunoprecipitation by using anti-KIT or anti-PDGFRA antibodies followed by sequential immunoblotting for phospho-KIT or total KIT, or for phosphotyrosine or total PDGFRA, respectively, as previously reported (Heinrich MC, Corless CL, Duensing A, et al: Science 299:708-710, 2003; Heinrich MC, Corless CL, Demetri GD, et al: J Clin Oncol 21:4342-4349, 2003).
Methods: statistical analyses.
Descriptive statistics were used to summarize clinical responses and mutation status, and the two-sided Fisher's exact test was used to evaluate differences in response by tumor genotype. Kaplan-Meier methods were used to assess progression-free and overall survival by KIT genotype. Multivariate analyses were performed with Cox proportional hazard models.
Table A1.
Patients With Pre-Imatinib Genotyping Data
| Patient | Dose (mg) | Schedule (weeks on/off treatment) | Primary Mutation | Secondary Mutations (mutations/samples) | Best Response | TTP (weeks) | OS (weeks) |
|---|---|---|---|---|---|---|---|
| 1 | 50 | 2/2 | PDGFRA exon 12 V561D | PDGFRA exon 18 D842V (1/1) | PD | 3 | 13 |
| 2 | 50 | 2/2 | KIT exon 11 V560D | KIT exon 13 1-bp insertion (3/3) and V654A (2/3) + exon 17 D816H (1/3) | SD < 6 months | 22 | 62 |
| 3 | 50 | 2/2 | KIT exon 11 deletion KPMYEVQWK550-558 | None (1/1) | SD < 6 months | 14 | 17 |
| 4 | 50 | 2/2 | KIT exon 9 insertion AY502-503 | None (1/1) | PR | 138 | 141 |
| 5 | 50 | 2/2 | Wild type | None (1/1) | SD ≥ 6 months | 210* | 226* |
| 6 | 50 | 2/2 | KIT exon 11 deletion KPMYEVQWK550-558 | KIT exon 14 T670I (1/2) and none (1/2) | SD ≥ 6 months | 61 | 97 |
| 7 | 25 | 2/2 | KIT exon 11 L576P | KIT exon 13 V654A (2/2) | SD < 6 months | 8 | 31 |
| 8 | 25 | 2/2 | KIT exon 11 deletion QWKVVEEINGNNYVYID556-572 | KIT exon 13 V654A (1/1) | PD | 8 | 13 |
| 9 | 25 | 2/2 | KIT exon 9 insertion AY502-503 | None | SD ≥ 6 months | 35* | 103 |
| 10 | 25 | 2/2 | KIT exon 13 K642E | KIT exon 17 D816H (1/2) and none (1/2) | SD ≥ 6 months | 54 | 85 |
| 11 | 25 | 2/2 | KIT exon 11 homozygous deletion WK557-558 | None (1/1) | SD < 6 months | 8 | 10 |
| 12 | 25 | 2/2 | Wild type | None (2/2) | PD | 6 | 86 |
| 13 | 50 | 2/2 | KIT exon 11 deletion YIDPTQL570-576 | KIT exon 17 C809G (2/2) | PD | 6 | 10 |
| 14 | 50 | 2/2 | KIT exon 9 insertion AY502-503 | None (2/2) | PR | 155 | 222* |
| 15 | 50 | 2/2 | KIT exon 9 insertion AY502-503 | None (2/2) | PR | 94 | 186* |
| 16 | 50 | 2/2 | KIT exon 9 insertion AY502-503 | KIT exon 13 V654A (2/2) | PR | 54 | 117 |
| 17 | 50 | 2/2 | KIT exon 11 insertion K558NP | NA | SD ≥ 6 months | 34 | 57 |
| 18 | 50 | 2/2 | Wild type | None (2/2) | SD ≥ 6 months | 98 | 112 |
| 19 | 75 | 2/2 | KIT exon 11 homozygous deletion WKVVEEINGNNYVYIDPT557-574 | KIT exon 18 A829P (1/1) | PD | 4 | 7 |
| 20 | 75 | 2/2 | KIT exon 11 deletion WK557-558Q | NA | SD ≥ 6 months | 46 | 93 |
| 21 | 75 | 2/2 | KIT exon 11 homozygous deletion WKVV557-560C | KIT exon 17 Y823D (1/2) and none (1/2) | PD | 3 | 17 |
| 22 | 75 | 2/2 | KIT exon 9 insertion AY502-503 | KIT exon 17 D820G (1/1) | SD < 6 months | 14 | 36 |
| 23 | 50 | 2/2 | Wild type | None (1/1) | SD ≥ 6 months | 60 | 213* |
| 24 | 50 | 2/2 | KIT exon 11 W557G | KIT exon 17 Y823D (1/2) and none (1/2) | SD < 6 months | 10 | 17 |
| 25 | 50 | 2/2 | KIT exon 9 insertion AY502-503 | None (2/2) | SD ≥ 6 months | 62 | 205* |
| 26 | 50 | 2/2 | Wild type | None (2/2) | SD < 6 months | 15* | 177* |
| 27 | 50 | 2/2 | KIT exon 11 deletion PMYE551-554 | KIT exon 13 V654A (2/2) | SD < 6 months | 19 | 143* |
| 28 | 50 | 2/2 | KIT exon 11 deletion VEEINGNNYVYIDPTQL560-576 | KIT exon 13 V654A (1/2) and none (1/2) | PD | 6 | 6 |
| 29 | 50 | 2/2 | PDGFRA exon 18 D842V | None (2/2) | SD < 6 months | 0.1* | 99 |
| 30 | 50 | 2/2 | KIT exon 11 deletion KPMYEVQWK550-558 | KIT exon 13 V654A (1/1) | SD ≥ 6 months | 36 | 39 |
| 31 | 50 | 2/2 | KIT exon 11 L576P | None (2/2) | SD < 6 months | 23 | 29 |
| 32 | 50 | 2/2 | KIT exon 9 insertion AY502-503 | None (2/2) | SD < 6 months | 20 | 25 |
| 33 | 50 | 2/2 | KIT exon 11 deletion YEVQWK553-558 | KIT exon 13 V654A (1/1) | SD < 6 months | 17 | 18 |
| 34 | 50 | 4/2 | KIT exon 9 insertion AY502-503 | None (1/1) | PR | 166* | 177* |
| 35 | 50 | 4/2 | KIT exon 11 homozygous deletion VQWKVVEE555-562 | KIT exon 14 T670I (1/1) | SD ≥ 6 months | 34 | 58 |
| 36 | 50 | 4/2 | KIT exon 11 deletion VQWKV555-559 | None (2/2) | PD | 10 | 108 |
| 37 | 50 | 4/2 | KIT exon 9 insertion AY502-503 | None (1/1) | SD < 6 months | 18 | 115 |
| 38 | 50 | 4/2 | PDGFRA exon 18 deletion IMHD843-846 | NA | SD < 6 months | 22 | 91 |
| 39 | 50 | 4/2 | KIT exon 11 deletion PM551-552 | NA | SD ≥ 6 months | 34 | 84 |
| 40 | 50 | 4/2 | KIT exon 13 K642E + exon 17 N822H | KIT exon 17 C809G (2/2) | SD < 6 months | 17 | 55 |
| 41 | 50 | 4/2 | Wild type | None (2/2) | SD < 6 months | 16 | 133 |
| 42 | 50 | 4/2 | KIT exon 11 insertion (L)TQLPYDHKWEFPRNR574-588 after R588 | KIT exon 13 V654A (2/2) | SD ≥ 6 months | 34 | 44 |
| 43 | 50 | 4/2 | KIT exon 9 insertion AY502-503 | NA | SD ≥ 6 months | 48 | 202* |
| 44 | 50 | 4/2 | KIT exon 9 insertion AY502-503 | None (1/1) | PR | 46* | 142 |
| 45 | 50 | 4/2 | KIT exon 11 deletion V559 or V560 | NA | PD | 10 | 52 |
| 46 | 50 | 2/1 | KIT exon 11 deletion EVQWK554-558 | KIT exon 13 V654A (2/2) | SD ≥ 6 months | 35 | 97 |
| 47 | 50 | 2/1 | KIT exon 11 homozygous deletion KV558-559 | None (2/2) | PD | 5* | 60 |
| 48 | 50 | 2/1 | KIT exon 11 deletion INGNNYVYIDPT563-574(GS) | KIT exon 13 V654A (2/2) | SD ≥ 6 months | 28* | 55 |
| 49 | 50 | 2/1 | KIT exon 9 insertion AY502-503 | None (2/2) | SD < 6 months | 6* | 40 |
| 50 | 50 | 4/2 | KIT exon 11 V560D | KIT exon 17 D820A (2/2) | PD | 5 | 25 |
| 51 | 50 | 4/2 | KIT exon 11 deletion PYD577-579 | KIT exon 13 V654A (1/1) | SD ≥ 6 months | 27* | 39 |
| 52 | 50 | 4/2 | KIT exon 11 deletion KVVEEI558-563NV | KIT exon 17 Y823D (2/2) | SD ≥ 6 months | 46 | 116 |
| 53 | 50 | 4/2 | KIT exon 9 insertion AY502-503 | None (1/1) | SD < 6 months | 25* | 48 |
| 54 | 50 | 4/2 | KIT exon 11 deletion WKVVE557-561 | KIT exon 17 N822K (2/2) | SD < 6 months | 22 | 115 |
| 55 | 50 | 4/2 | KIT exon 11 V559D | KIT exon 17 D820Y (1/1) | SD < 6 months | 22 | 146* |
| 56 | 50 | 2/1 | KIT exon 9 insertion AY502-503 | None (2/2) | SD ≥ 6 months | 143 | 177* |
| 57 | 50 | 4/2 | KIT exon 11 deletion YVYIDPTQL568-576VN | KIT exon 13 V654A (2/2) | SD ≥ 6 months | 58 | 151* |
| 58 | 50 | 4/2 | KIT exon 9 insertion AY502-503 | NA | SD < 6 months | 7* | 53 |
| 59 | 50 | 4/2 | KIT exon 11 V560G | KIT exon 13 V654A (2/2) | PR | 128* | 144* |
| 60 | 50 | 4/2 | KIT exon 11 deletion V559 or V560 | KIT exon 13 V654A (2/2) | SD < 6 months | 22 | 90 |
| 61 | 50 | 4/2 | KIT exon 9 insertion AY502-503 | KIT exon 17 D816A (1/1) | PD | 4 | 4 |
| 62 | 50 | 4/2 | KIT exon 11 L576P | KIT exon 13 V654A (2/2) | SD < 6 months | 22 | 61 |
| 63 | 50 | 4/2 | KIT exon 11 deletion VYIDPTQL569-576 | KIT exon 18 A829P (2/2) | SD < 6 months | 10* | 15 |
| 64 | 50 | 4/2 | KIT exon 11 deletion WK557-558 | KIT exon 14 T670I (2/2) | SD ≥ 6 months | 44 | 53 |
| 65 | 50 | 4/2 | Wild type | NA | SD ≥ 6 months | 91 | 142* |
| 66 | 50 | 4/2 | PDGFRA exon 18 D842V | None (1/1) | PD | 10 | 129* |
| 67 | 50 | 4/2 | KIT exon 11 point mutant L576P | NA | SD < 6 months | 24 | 32* |
| 68 | 50 | 4/2 | KIT exon 9 insertion AY502-503 | None (2/2) | PD | 10 | 23 |
| 69 | 50 | 4/2 | KIT exon 11 deletion WK557-558 | None (1/1) | SD < 6 months | 22 | 38 |
| 70 | 50 | 4/2 | Wild type | None (2/2) | SD ≥ 6 months | 68* | 82 |
| 71 | 50 | 4/2 | KIT exon 11 deletion WKV557-559(C) | NA | SD < 6 months | 20* | 125* |
| 72 | 50 | 4/2 | KIT exon 11 V560D | None (1/1) | PD | 9 | 33 |
| 73 | 50 | 4/2 | KIT exon 11 deletion WK557-558 | None (3/3) | SD < 6 months | 10* | 19 |
| 74 | 50 | 4/2 | KIT exon 11 deletion D579 | None (2/2) | SD < 6 months | 10* | 85 |
| 75 | 50 | 4/2 | KIT exon 11 deletion NGNNYVYIDPTQLPY564-578 | KIT exon 17 N822K (1/1) | PD | 2 | 15 |
| 76 | 50 | 4/2 | Wild type | None (1/1) | SD < 6 months | 17 | 56 |
| 77 | 50 | 2/1 | KIT exon 9 insertion AY502-503 | NA | PR | 84 | 114* |
| 78 | 50 | 2/1 | KIT exon 11 deletion KPMYEVQWKV550-559 | None (1/1) | PR | 54* | 84* |
Abbreviations: TTP, time to progression; OS, overall survival; PDGFRA, platelet-derived growth factor receptor α; bp, base pair; PD, progressive disease; SD, stable disease; PR, partial response, NA, not available.
Censored observation.
published online ahead of print at www.jco.org on October 27, 2008
Supported in part by Pfizer Inc, by National Cancer Institute (NCI) Grant No. CA 47179, by NCI Specialized Program of Research Excellence in Gastrointestinal Cancer Grant No. 1P50CA127003-01, by a Veterans Affairs Merit Review Grant, by the Life Raft Group, and by philanthropic support from the following sources: the Virginia and Daniel Ludwig Trust for Cancer Research, the Rubenstein Foundation, the Katz Foundation, the Quick Family Fund for Cancer Research, the Ronald O. Perelman Fund for Cancer Research at Dana-Farber, the Stutman GIST Cancer Research Fund, Leslie's Links, Abolish Cancer Today, and the Shuman Family Fund for GIST Research.
Presented in part at the 41st Annual Meeting of the American Society of Clinical Oncology, May 13-17, 2005, Orlando, FL; the 13th European Cancer Conference, October 30-November 3, 2005, Paris, France; the 42nd Annual Meeting of the American Society of Clinical Oncology, June 2-6, 2006, Atlanta, GA; and the 1st American Association for Cancer Research Conference on Molecular Diagnostics in Cancer Therapeutic Development, September 12-15, 2006, Chicago, IL.
Authors’ disclosures of potential conflicts of interest and author contributions are found at the end of this article.
REFERENCES
- 1.Corless CL, Schroeder A, Griffith D, et al: PDGFRA mutations in gastrointestinal stromal tumors: Frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol 23:5357-5364, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Heinrich MC, Corless CL, Duensing A, et al: PDGFRA activating mutations in gastrointestinal stromal tumors. Science 299:708-710, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Debiec-Rychter M, Sciot R, Le Cesne A, et al: KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer 42:1093-1103, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Demetri GD, von Mehren M, Blanke CD, et al: Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med 347:472-480, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Verweij J, Casali PG, Zalcberg J, et al: Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: Randomised trial. Lancet 364:1127-1134, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Van Glabbeke M, Verweij J, Casali PG, et al: Initial and late resistance to imatinib in advanced gastrointestinal stromal tumors are predicted by different prognostic factors: A European Organisation for Research and Treatment of Cancer-Italian Sarcoma Group-Australasian Gastrointestinal Trials Group study. J Clin Oncol 23:5795-5804, 2005 [DOI] [PubMed] [Google Scholar]
- 7.O'Farrell AM, Abrams TJ, Yuen HA, et al: SU11248 is a novel FLT3 tyrosine kinase inhibitor with potent activity in vitro and in vivo. Blood 101:3597-3605, 2003 [DOI] [PubMed] [Google Scholar]
- 8.Mendel DB, Laird AD, Xin X, et al: In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: Determination of a pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res 9:327-337, 2003 [PubMed] [Google Scholar]
- 9.Abrams TJ, Lee LB, Murray LJ, et al: SU11248 inhibits KIT and platelet-derived growth factor receptor beta in preclinical models of human small-cell lung cancer. Mol Cancer Ther 2:471-478, 2003 [PubMed] [Google Scholar]
- 10.Murray LJ, Abrams TJ, Long KR, et al: SU11248 inhibits tumor growth and CSF-1R-dependent osteolysis in an experimental breast cancer bone metastasis model. Clin Exp Metastasis 20:757-766, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Kim DW, Jo YS, Jung HS, et al: An orally administered multi-target tyrosine kinase inhibitor, SU11248, is a novel potent inhibitor of thyroid oncogenic RET/papillary thyroid cancer kinases. J Clin Endocrinol Metab 91:4070-4076, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Maki RG, Fletcher JA, Heinrich MC, et al: Results from a continuation trial of SU11248 in patients with imatinib-resistant gastrointestinal stromal tumor (GIST). J Clin Oncol 23:818s, 2005. (suppl; abstr 9011) [Google Scholar]
- 13.Demetri GD, van Oosterom AT, Garrett CR, et al: Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: A randomised controlled trial. Lancet 368:1329-1338, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Heinrich MC, Corless CL, Demetri GD, et al: Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol 21:4342-4349, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Debiec-Rychter M, Dumez H, Judson I, et al: Use of c-KIT/PDGFRA mutational analysis to predict the clinical response to imatinib in patients with advanced gastrointestinal stromal tumours entered on phase I and II studies of the EORTC Soft Tissue and Bone Sarcoma Group. Eur J Cancer 40:689-695, 2004 [DOI] [PubMed] [Google Scholar]
- 16.Antonescu CR, Besmer P, Guo T, et al: Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res 11:4182-4190, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Heinrich MC, Corless CL, Blanke CD, et al: Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol 24:4764-4774, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Bertucci F, Goncalves A, Monges G, et al: Acquired resistance to imatinib and secondary KIT exon 13 mutation in gastrointestinal stromal tumour. Oncol Rep 16:97-101, 2006 [PubMed] [Google Scholar]
- 19.Loughrey MB, Beshay V, Dobrovic A, et al: Pathological response of gastrointestinal stromal tumour to imatinib treatment correlates with tumour KIT mutational status in individual tumour clones. Histopathology 49:99-100, 2006 [DOI] [PubMed] [Google Scholar]
- 20.Wardelmann E, Merkelbach-Bruse S, Pauls K, et al: Polyclonal evolution of multiple secondary KIT mutations in gastrointestinal stromal tumors under treatment with imatinib mesylate. Clin Cancer Res 12:1743-1749, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Grimpen F, Yip D, McArthur G, et al: Resistance to imatinib, low-grade FDG-avidity on PET, and acquired KIT exon 17 mutation in gastrointestinal stromal tumour. Lancet Oncol 6:724-727, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Tamborini E, Gabanti E, Lagonigro MS, et al: KIT/Val654 Ala receptor detected in one imatinib-resistant GIST patient. Cancer Res 65:1115, 2005 [PubMed] [Google Scholar]
- 23.McLean SR, Gana-Weisz M, Hartzoulakis B, et al: Imatinib binding and cKIT inhibition is abrogated by the cKIT kinase domain I missense mutation Val654Ala. Mol Cancer Ther 4:2008-2015, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Debiec-Rychter M, Cools J, Dumez H, et al: Mechanisms of resistance to imatinib mesylate in gastrointestinal stromal tumors and activity of the PKC412 inhibitor against imatinib-resistant mutants. Gastroenterology 128:270-279, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Wardelmann E, Thomas N, Merkelbach-Bruse S, et al: Acquired resistance to imatinib in gastrointestinal stromal tumours caused by multiple KIT mutations. Lancet Oncol 6:249-251, 2005 [DOI] [PubMed] [Google Scholar]
- 26.Chen LL, Trent JC, Wu EF, et al: A missense mutation in KIT kinase domain 1 correlates with imatinib resistance in gastrointestinal stromal tumors. Cancer Res 64:5913-5919, 2004 [DOI] [PubMed] [Google Scholar]
- 27.Tamborini E, Bonadiman L, Greco A, et al: A new mutation in the KIT ATP pocket causes acquired resistance to imatinib in a gastrointestinal stromal tumor patient. Gastroenterology 127:294-299, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Wakai T, Kanda T, Hirota S, et al: Late resistance to imatinib therapy in a metastatic gastrointestinal stromal tumour is associated with a second KIT mutation. Br J Cancer 90:2059-2061, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prenen H, Cools J, Mentens N, et al: Efficacy of the kinase inhibitor SU11248 against gastrointestinal stromal tumor mutants refractory to imatinib mesylate. Clin Cancer Res 12:2622-2627, 2006 [DOI] [PubMed] [Google Scholar]
- 30.Carter TA, Wodicka LM, Shah NP, et al: Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc Natl Acad Sci U S A 102:11011-11016, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuveson DA, Willis NA, Jacks T, et al: STI571 inactivation of the gastrointestinal stromal tumor c-KIT oncoprotein: Biological and clinical implications. Oncogene 20:5054-5058, 2001 [DOI] [PubMed] [Google Scholar]
- 32.Noma K, Naomoto Y, Gunduz M, et al: Effects of imatinib vary with the types of KIT-mutation in gastrointestinal stromal tumor cell lines. Oncol Rep 14:645-650, 2005 [PubMed] [Google Scholar]
- 33.Heinrich MC, Maki RG, Corless CL, et al: Sunitinib response in imatinib-resistant GIST correlates with KIT and PDGFRA mutation status. J Clin Oncol 24:520s, 2006. (suppl; abstr 9502) [Google Scholar]
- 34.Yuzawa S, Opatowsky Y, Zhang Z, et al: Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell 130:323-334, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Dibb NJ, Dilworth SM, Mol CD: Switching on kinases: Oncogenic activation of BRAF and the PDGFR family. Nat Rev Cancer 4:718-727, 2004 [DOI] [PubMed] [Google Scholar]
- 36.Guo T, Agaram NP, Wong GC, et al: Sorafenib inhibits the imatinib-resistant KITT670I gatekeeper mutation in gastrointestinal stromal tumor. Clin Cancer Res 13:4874-4881, 2007 [DOI] [PubMed] [Google Scholar]
- 37.Potapova O, Laird AD, Nannini MA, et al: Contribution of individual targets to the antitumor efficacy of the multitargeted receptor tyrosine kinase inhibitor SU11248. Mol Cancer Ther 5:1280-1289, 2006 [DOI] [PubMed] [Google Scholar]
- 38.Erber R, Thurnher A, Katsen AD, et al: Combined inhibition of VEGF and PDGF signaling enforces tumor vessel regression by interfering with pericyte-mediated endothelial cell survival mechanisms. Faseb J 18:338-340, 2004 [DOI] [PubMed] [Google Scholar]
- 39.Bergers G, Song S, Meyer-Morse N, et al: Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J Clin Invest 111:1287-1295, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]